
HES1 induces ITPR1-mediated autophagy to exert anti-metastatic effects in pituitary adenomas
Highlight box
Key findings
• Through in-depth bioinformatics analysis, we identified ITPR1 as a central hub gene in a pituitary adenoma (PA)-related dataset. ITPR1 expression is reduced in PAs and has significant clinical diagnostic relevance.
What is known and what is new?
• PAs are prevalent intracranial tumors. Molecular intricacies of PA need comprehensive exploration.
• ITPR1’s role as a central hub gene, its clinical diagnostic relevance, and its tumor-suppressive effects in PA.
What is the implication, and what should change now?
• Our findings implied that HES1, ITPR1, and autophagy may be important targets in the treatment of PAs. Relevant actions include the development of new therapeutic approaches focusing on interventions that influence this complex network to improve the outcome of PAs.
Introduction
Pituitary adenomas (PAs), providing for approximately of 15% the majority of intracranial tumors, represent a prevalent and complex pathology (1,2). PA was increasingly recognized in the general population, with an incidence ranging between 3.9 to 7.4 cases annually per 100,000 people. Despite this, their overall prevalence suggests they affect roughly 1 in 1,000 individuals (3). Notably, prolactinomas and nonsecreting PAs make up the majority of these cases. While clinically significant PAs are more common in females, their clinical presentations vary widely. These adenomas can lead to hormone imbalances and visual field defects. In cases with larger tumors, they might also result in hypopituitarism due to the tumor’s mass effect (4). Despite associations with various factors such as genetic alterations and environmental influences, the precise etiology of their pathogenesis remains elusive (5,6). Different hormonal types of PA can manifest a variety of symptoms, often due to excessive hormone secretion (2). For instance, prolactinomas can lead to menstrual irregularities and galactorrhea in females, and impotence in males (7). Adenomas that secrete growth hormone can result in acromegaly, marked by enlarged limbs, altered facial features, and skin changes (8). Adenomas secreting corticotropin result in Cushing’s disease, marked by central obesity, a moon-like facial appearance, hypertension, and purple striae (9). Current treatments, such as surgical intervention, drug therapy, and radiation therapy, vary in effectiveness based on tumor subtype, size, and aggressiveness (10,11). Despite advances in treatment options, PA remains a significant challenge due to the potential for complications and recurrence in long-term prognosis (12). Early detection is crucial for managing PAs effectively. Prolactinomas are typically treated with dopamine agonists initially, while other PAs often require transsphenoidal surgery as the first-line therapy, reserving medical treatment for cases unresponsive to surgical intervention (13). Consequently, researching effective therapeutic strategies is pivotal to unravel the intricate pathogenesis, deepen our understanding of PA, and enhance patient outcomes.
Autophagy, the intracellular process of lysosomal degradation of cellular components, in a number of physiological and pathological situations, plays a critical regulatory function, including tumorigenesis (14). In cancer, autophagy both impedes tumorigenesis by removing damaged proteins and organelles and supports cancer survival under stress by supplying nutrients (15). The crucial involvement of autophagy in the initiation and progression of tumors has become increasingly emphasized., underscoring the potential for therapeutic strategies that target autophagy (16). Recent study by Lyu et al., highlight the function of autophagy autophagy in the pathogenesis of PA. They demonstrate that tetrandrine (Tet) exhibits anti-PA effects via the MAPK/STAT3 signaling pathway, mediating both autophagy and apoptosis (17). In a study by Kun et al., it was shown that inhibiting hypoxia-inducible factor 1α can counteract temozolomide-induced autophagy in rat PA GH3 cells, thereby boosting the treatment effectiveness of temozolomide against PA (18). However, the complex relationship between autophagy and PA pathogenesis remains incompletely understood. Therefore, it is essential to thoroughly investigate the involvement of autophagy in PA.
Within the realm of PAs, it is becoming evident that certain genes may serve as the linchpin in tumor progression and response to therapies. Among these pivotal genes are ITPR1 (inositol 1,4,5-trisphosphate receptor, type 1) and HES1 (hairy and enhancer of split 1). ITPR1, an intracellular calcium release channel, is fundamental for a range of physiological processes, including cellular signaling cascades, apoptosis, and cellular proliferation (19). Dysregulation of this gene may interfere with cell differentiation and growth pathways, which may have important implications for tumor behavior. It is worth mentioning that ITPR1 acts like an application program in cell signaling, controlling various “applications” within the cell, and the interaction between applications affects the overall cell performance. Dysregulation of HES1 interferes with cell differentiation and growth pathways, which may have implications for tumor behavior (20). On the other hand, HES1 is a transcriptional repressor with a renowned function in cellular differentiation and development (21). It is analogous to a regulator that controls gene expression and determines cellular direction. HES1 determines the direction of cell development, which may have far-reaching consequences in terms of tumor behavior. The potential interplay between ITPR1 and HES1, two hub genes, might hold significant implications for the molecular pathogenesis of PAs and the pursuit of effective therapeutic strategies. We found that HES1 regulates the expression of ITPR1, which affects cell signaling cascades, apoptosis and proliferation. This interaction may play a key role in the molecular pathogenesis of PAs. Given this backdrop, our study’s exploration into the interrelation between HES1, ITPR1, and autophagy not only paves the way for a deeper understanding of PAs but also brings into focus the broader role of hub genes in shaping tumor dynamics.
Considering the clinical challenges of PA and the presence of non-responsive cases, the identification of novel molecular targets is important for proper PA management. The purpose of this research is to elucidate the role of HES1 and its interactions with ITPR1 and autophagy in PA cell behavior. In this endeavor, we seek to decipher the intricate regulatory network underlying PA progression, setting the stage for innovative therapeutic approaches for PA patients resistant to current treatments. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1320/rc).
Methods
Retrieval of microarray data and analysis of differential expressed genes (DEGs)
For our investigation into PA, two pertinent microarray datasets were sourced from the gene expression omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) database: GSE36314, with four prolactinoma and three normal pituitary samples, and GSE119063, encompassing five prolactinoma and four normal pituitary samples. Differential gene expression analysis was carried out on both datasets utilizing the “limma” package in R software. We recognized upregulated DEGs with a fold change (FC) >1.5 and downregulated DEGs with an FC <0.67, all meeting importance threshold of P value <0.05. For a holistic view of the gene expression landscape, DEGs were visualized using the “ggplot2” package in R. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Screening and analysis of overlapping DEGs
The overlapping up-regulated and down-regulated DEGs in the GSE36314 and GSE119063 datasets were identified using the “VennDiagram” package in R software. The overlapping DEGs were then analyzed for biological process (BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment using the Database for Annotation, Visualization and Integrated Discovery (DAVID) database (https://david.ncifcrf.gov/tools.jsp). Results with a P value <0.05 were thought to be statistically significant. The overlapping DEGs were subjected to further scrutiny via the construction of a protein-protein interaction (PPI) network using Cytoscape software. Utilizing the Cytohubba plugin, the top 20 genes were identified based on the BottleNeck, Closeness, and EcCentricity algorithms, which were then used to visualize the PPI network corresponding to the selected genes.
Expression and clinical diagnosis analysis of seven overlapping genes in PPI network
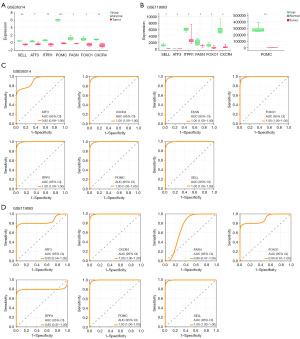
In the PPI network of BottleNeck, Closeness and EcCentricity algorithms, seven overlapping genes were identified, namely SELL, ATF3, ITPR1, POMC, FASN, FOXO1 and CXCR4. Subsequent examination of these seven gene expressions was conducted within the GSE36314 and GSE19063 data samples. Following this, receiver operating characteristic (ROC) curves for seven genes in the GSE36314 and GSE119063 datasets were generated using the “timeROC” package in R. The area under the curve (AUC) was calculated to assess the clinical diagnostic potential of these genes in the context of PA. This analysis ultimately facilitated the selection of the central hub genes for this study.
Cell culture and treatment
The human PA cell lines HP75 and GH3 were obtained from Beijing Zhongyuan Ltd., China. Cells were subsequently subcultured in accordance with the manufacturer’s directions (22). After transfection with the ITPR1 plasmid or control vector, cells were treated with autophagy inhibitors: 3-MA (5 mM) or Baf A1 (100 nM) for 24 hours, based on the aforementioned, in conformity with the manufacturer’s guidelines.
Cell transfection
Cell transfection was executed utilizing Lipofectamine 2000 as the transfection agent. Cells were cultured in suitable plates and allowed to reach 70–80% confluency prior to transfection. Subsequently, a transfection mixture, composed of the ITPR1 overexpression plasmid and the transfection agent, was prepared and added to the cells, followed by an incubation period. Simultaneously, to achieve the knockdown of HES1, PA cells were subjected to transfection with specific small interfering RNAs (siRNAs) targeting HES1, referred to as si-HES1-1 and si-HES1-2. These transfections were performed when cells attained a confluency of 50–60%, following which the siRNA transfection mix was added to the cells and permitted to incubate for a designated period.
Western blotting (WB) assay
Cellular proteins were extracted by lysing the cells in RIPA buffer, supplemented with protease and phosphatase inhibitors. ABCA protein assay kit was employed to measure the amount of protein in the lysate. By sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), equal quantities of protein were loaded, separated, and then deposited onto polyvinylidene difluoride (PVDF) membranes. After that, the membranes were blocked with 5% skim milk at room temperature for an hour. They were then using primary antibodies as a probe specific to the target proteins: ITPR1 (CST, Danvers, USA, 1:1,000), Beclin1 (Abcam, Cambridge, USA, 1:1,000), LC3-I (Sigma-Aldrich, St. Louis, USA, 1:1,000), LC3-II (Novus Biologicals, Centennial, USA, 1:1,000), HES1 (CST, 1:1,000), and β-actin (Sigma-Aldrich, 1:2,000) overnight at 4 ℃. After washing, protein bands were visualized using enhanced chemiluminescence (ECL) detection method after incubation with appropriate secondary antibodies (1:5,000) for 1 hour at room temperature. The intensity was measured using ImageJ software.
Cell proliferation assay
Post-transfection, cells were allocated into 96-well plates and incubated under the stipulated conditions. Proliferation rates were examined at the designated time intervals (24, 48, 72, 96 hours) using the cell counting kit-8 (CCK-8) assay. The CCK-8 reagent was introduced into each well, initiating an additional incubation period that facilitates the reaction between the reagent and viable cells. Following this, the optical density (OD) was determined using a microplate reader set to 450 nm.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total cellular RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) as per the manufacturer’s instructions. First-strand cDNA was synthesized from 1 µg of total RNA employing the PrimeScript RT reagent Kit (Takara, Kusatsu, Japan). Subsequently, qRT-PCR was performed using the TB Green Premix Ex Taq (Takara) on a LightCycler 480 Instrument II (Roche, Basel, Switzerland). The primer sequences utilized in this study were as stated below: HES1 forward: (5'-TCAACACGACACCGGATAAA-3'), reverse: (5'-TCAGCTGGCTCAGACTTTCA-3'); ITPR1 forward: (5'-TGCTGTGATTTTAGTGGCGT-3'), reverse: (5'-TCTCCACCCTACCCTTACCT-3'); β-actin forward: (5'-GTCAGTGGTGGACCTGACCT-3'), reverse: (5'-AGGGGAGATTCAGTGTGGTG-3'). Expression levels of HES1 and ITPR1 were standardized to the internal reference gene GAPDH. The relative gene expression was determined using the 2−ΔΔCT method.
Transwell assay
In the invasion assay, the upper chamber of Transwell inserts was covered with Matrigel. Cells (1×105) in serum-free medium were added to the upper chamber, while the lower chamber was filled with media containing 10% fetal bovine serum (FBS) as a chemoattractant. Following a 24-hour incubation, cells that invaded through the Matrigel and reached the underside of the membrane were immobilized using 4% paraformaldehyde and stained cells with DAPI (4',6-diamidino-2-phenylindole) (1:1,000 dilution) for 10 minutes in the dark. Rinse excess DAPI with phosphate-buffered saline (PBS). Observe and quantify the infiltrated cells using a fluorescence microscope. The migration assay was conducted similarly, but without the Matrigel coating.
Dual-luciferase assay
Cells were plated in 24-well dishes and co-transfected with the suitable luciferase reporter. and Renilla control vectors using Lipofectamine 2000. Post 48 h incubation, cell lysis was performed, and luciferase levels was quantified employing the Dual-Luciferase Assay System (Promega, USA). To adjust for transfection efficiency, firefly luciferase activity was normalized to Renilla luciferase activity. Three independent experiments were performed, and the data were presented as fold induction compared to controls.
Statistical analysis
All experimental results were displayed as mean ± standard deviation (SD) from a minimum of three separate experiments. The Student’s t-test was used to compare two groups. Comparisons between multiple groups utilized one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc analysis. A P value <0.05 was considered statistically significant. All statistical analyses were conducted using the SPSS 25.0 software package (IBM, Armonk, NY, USA). Graphs were generated using GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA).
Results
Comparative analysis of DEGs in PA
Analysis of the GSE36314 dataset revealed 71 upregulated and 323 downregulated DEGs (Figure 1A). Similarly, the GSE119063 dataset presented 506 upregulated and 3,140 downregulated DEGs (Figure 1B). When comparing both datasets, we identified 18 overlapping upregulated and 140 overlapping downregulated DEGs, yielding a total of 158 overlapping DEGs (Figure 1C).
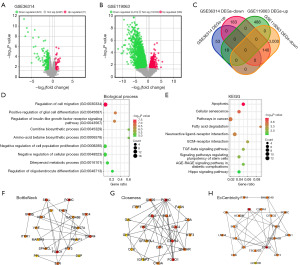
Enrichment analysis of 158 overlapping genes
Enrichment analysis of these DEGs revealed associations with several BP terms. Notably, the top BPs included regulation of cell migration, positive regulation of glial cell differentiation, and carnitine biosynthetic process (Figure 1D). Furthermore, analysis using KEGG pathways showed notable enrichment in diverse pathways, including apoptosis, cellular senescence, fatty acid degradation, extracellular matrix (ECM)-receptor interaction, transforming growth factor (TGF)-beta signaling, Hippo signaling, among others (Figure 1E). To gain further insights into the functional implications of the identified genes, we employed the BottleNeck, Closeness, and EcCentricity algorithms to construct PPI networks for the top 20 genes (Figure 1F-1H) and the analysis revealed intricate connectivity patterns, shedding light on potential key players within the interactome.
Identification of seven genes as potential predictors of PA
We identified seven common genes (SELL, ATF3, ITPR1, POMC, FASN, FOXO1, and CXCR4) within the PPI networks using the BottleNeck, Closeness, and Eccentricity algorithms. Notably, these genes displayed reduced expression in PA samples from both GSE36314 and GSE119063 datasets, pointing to shared molecular mechanisms in PA onset and progression (Figure 2A,2B). To assess the diagnostic value of these genes, ROC curve analyses were conducted for each across the datasets. As shown in Figure 2C,2D, all seven genes showcased high AUC values, underlining their robust predictive capability for PA. Notably, related studies identified ITPR1 as an autophagy-linked gene, positioning it as a central gene for our subsequent in vitro experiments.
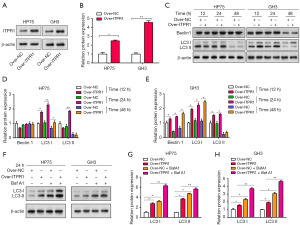
Modulatory effects of ITPR1 overexpression and autophagy inhibition by 3-MA on PA progression
To study the role of ITPR1 in HP75 and GH3 cell lines, we transfected these cells with an ITPR1 overexpression plasmid (Figure 3A,3B). After successful transfection, we assessed the impact of ITPR1 overexpression on autophagy-associated proteins Beclin1, LC3 I, and LC3 II, over different time intervals (12, 24, 48 hours). At 24 hours post-transfection, a marked induction of autophagy was evident, especially in the levels of LC3 I and LC3 II (Figure 3C-3E). Therefore, we selected the 24-hour interval for subsequent experiments. In the next phase, cells with overexpressed ITPR1 were treated with the autophagy inhibitor Baf A1. This treatment resulted in elevated LC3 I and LC3 II protein levels. When combining ITPR1 overexpression with Baf A1 treatment, we noticed even higher autophagy-related protein levels, suggesting a synergistic effect that amplifies autophagy (Figure 3F-3H). Our results pointed to a significant relationship between ITPR1 overexpression and autophagy in PA cells, hinting at a potential pivotal mechanism in PA progression.
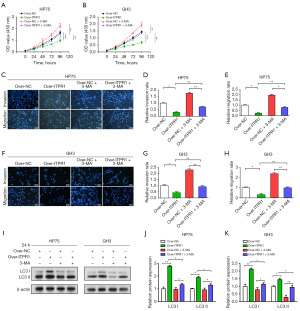
Effect of ITPR1 on autophagy-mediated proliferation, movement, and infiltration of PA cells
In vitro experiments on HP75 and GH3 cells showed that ITPR1 overexpression resulted in significant inhibition of cell proliferation, invasion, and migration, indicating the potential anti-tumor effects of ITPR1. In contrast, the autophagy inhibitor 3-MA was found to promote these cellular behaviors, suggesting that inhibition of autophagy may contribute towards PA progression. Interestingly, a relative attenuation of the competence of these cells was observed when overexpressed ITPR1 was combined with 3-MA, suggesting a potential mitigating effect of ITPR1 on the pro-tumor activity promoted by autophagy inhibition (Figure 4A-4H). Further WB analysis of the autophagy-related proteins LC3 I and LC3 II confirmed these findings. Overexpressed ITPR1 caused an increase in these proteins in PA cells, indicating enhanced autophagy. In contrast, addition of 3-MA resulted in downregulation of these proteins. However, the combination of overexpressed ITPR1 and 3-MA exhibited a regulatory effect that counteracted the inhibitory effect of 3-MA on apoptotic proteins (Figure 4I-4K). Collectively, these findings elucidate the complex interplay between ITPR1, autophagy, and tumorigenesis in PA cells, suggesting that manipulation of this tripartite relationship may provide potential therapeutic strategies for PA.
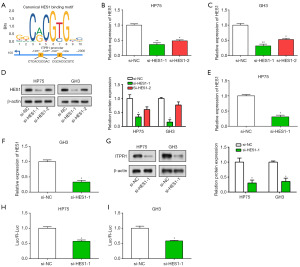
HES1 modulates ITPR1 through transcriptional activation
Utilizing the JASPAR database for bioinformatic analysis, we pinpointed two HES1 responsive elements likely to bind the ITPR1 promoter (Figure 5A). We then used qRT-PCR and WB assays to evaluate HES1 transfection efficiency in PA cells. Notably, si-HES1-2 resulted in a pronounced decrease in HES1 expression, indicating successful HES1 knockdown (Figure 5B-5D). These results position HES1 as an upstream regulator of ITPR1 in PA cells. Additionally, HES1 knockdown corresponded with reduced ITPR1 expression (Figure 5E-5G). The transcriptional activity of the ITPR1 promoter was also diminished, as reflected by the dual luciferase assays (Figure 5H,5I). These findings unveil a sophisticated regulatory interplay between HES1 and ITPR1, underscoring the potential of targeting HES1 for improved PA management.
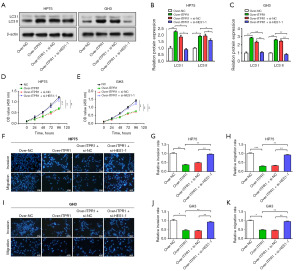
HES1 suppresses metastasis through ITPR1-induced autophagy in PA
After overexpressing ITPR1, we observed upregulation of the autophagy proteins LC3 I and LC3 II using WB, highlighting the function of ITPR1 in autophagy regulation (Figure 6A-6C). Interestingly, HES1 knockdown seemed to negate the ITPR1 overexpression effect on these proteins. Through CCK-8 and Transwell assays, we determined that reduced HES1 expression dampened the enhancing impact of ITPR1 overexpression on PA cell proliferation, invasion, and migration (Figure 6D-6K). Collectively, our findings underscore a nuanced relationship among HES1, ITPR1, and autophagy in PA cell behavior, hinting at the potential of this regulatory triad as a therapeutic target in PA.
Discussion
PA comprise about 15–20% of intracranial tumors, establishing them as a prevalent subtype of intracranial neoplasms (23). The considerable heterogeneity observed in the clinical presentations and biological behaviors of PAs has spurred exhaustive research endeavors to elucidate their pathogenesis and pinpoint novel therapeutic targets (4). Among the many factors involved in the pathogenesis of PA, autophagy, a key cellular mechanism that helps maintain cellular homeostasis, has attracted considerable academic attention (24). Dysregulation of autophagy is linked to a variety of disorders. involving cancer (25). Emerging evidence suggests its potential involvement in the pathogenesis of PA (26). The utilization of bioinformatics has evolved as a formidable instrument for probing the complex molecular underpinnings of disorders such as PA (27). Analyzing high-throughput genetic data facilitates the identification of potential biomarkers and therapeutic targets, encompassing those pertinent to autophagy (28,29). Consequently, the amalgamation of bioinformatics with autophagy research augments the prospects for advancing our comprehension of PA pathobiology and pinpointing innovative therapeutic approaches.
From the GSE36314 and GSE119063 datasets, we analyzed a total of 158 overlapping DEGs. Bioinformatics analyses illuminated that these DEGs were notably enriched with BPs and pathways integral to the development and progression of PA and autophagy. Notably, the regulation of cell migration, pivotal for tumor invasion and metastasis, is extensively implicated in PA. Dysregulated cell migration could instigate aggressive tumor behaviors, potentially accounting for the observed clinical variability in PA outcomes. Intriguingly, these DEGs demonstrated significant enrichment in several pivotal pathways such as apoptosis, cellular senescence, fatty acid degradation, ECM-receptor interaction, TGF-β signaling, and Hippo signaling. For instance, both the apoptotic pathway and cellular senescence are recognized as fundamental tumor-suppressive mechanisms (30,31). Conversely, disruptions in these mechanisms could potentially foster tumorigenesis (32,33), which implies their potential involvement in PA. Similarly, the Hippo signaling pathway, a paramount regulator of organ size and tumor suppression, is implicated in the pathophysiology of autophagy (34). Together, these observations emphasize the prospective engagement of the identified DEGs in central BPs and pathways pertinent to PA pathogenesis and autophagy. These observations enhance our comprehension of the molecular underpinnings of PA and establish a strong groundwork for future investigations.
Our exploration spotlighted seven genes (SELL, ATF3, ITPR1, POMC, FASN, FOXO1, and CXCR4) as prospective biomarkers for PA, attributed to their notably reduced expression in PA samples from the GSE36314 and GSE119063 datasets. Importantly, ITPR1, recognized as an autophagy-associated gene, emerged as particularly significant. Gu et al. have evidenced its correlation with survival in breast cancer (35), further underscoring its clinical significance. The predictive accuracy of ITPR1, as well as the other genes, was substantiated by the AUC values derived from ROC curve analysis. Furthermore, the role of ITPR1 in PA was more comprehensively elucidated using HP75 and GH3 cell lines. The overexpression of ITPR1 coincided with heightened levels of autophagy-related proteins, namely Beclin1, LC3 I, and LC3 II, implying a pivotal role for ITPR1 in instigating autophagy within PA. Notably, Beclin 1, a central figure in the initiation of autophagy, demonstrated upregulated expression, a common indicator of augmented autophagic activity. Similarly, LC3 I and LC3 II, crucial constituents in autophagosome formation, displayed elevated levels, further affirming the association between ITPR1 overexpression and autophagy induction. Additionally, we explored the use of the autophagy inhibitor Baf A1, an inhibitor of lysosomal ATPase that hinders the merging of autophagosomes and lysosomes (36). Upregulation of LC3 I and LC3 II protein levels after Baf A1 treatment, particularly in conjunction with ITPR1 overexpression, suggests a synergistic effect that amplifies autophagy. This interaction underscores the critical role of ITPR1 overexpression in autophagy regulation and the progression of PA, deepening our insights into its molecular pathogenesis.
In our study on the intricate mechanisms driving PA development and progression, we investigated the effects of ITPR1 overexpression and 3-MA, an autophagy inhibitor. Our results underscore an anti-tumorigenic role for ITPR1; its overexpression notably hindered PA cell behaviors linked to tumorigenesis. In contrast, 3-MA bolstered these pro-tumorigenic behaviors, pointing to the potential of autophagy modulation in PA therapy. Remarkably, combining ITPR1 overexpression with 3-MA treatment led to a subdued cellular response, implying ITPR1 might counterbalance the pro-tumorigenic outcomes of autophagy inhibition. This hypothesis gained traction in our WB analysis, where the joint application of ITPR1 overexpression and 3-MA altered protein expression, reducing the suppressive effects of 3-MA on autophagy-associated proteins.
Extending these observations, we further elucidated the regulatory environment involving HES1, a potential upstream regulator of ITPR1. Using bioinformatics tools, we discovered two HES1-responsive elements within the ITPR1 promoter region. These findings are consistent with study by Perrone et al., which highlighted the critical role of the Notch system target gene HES1 in the pathogenesis of PAs (37). They observed a positive correlation between HES1 expression and NOTCH1, 2, 4 receptors, indicating activation of the Notch pathway in certain tumors. This suggested that targeting the Notch pathway, including HES1, has potential therapeutic advantages in the treatment of PAs. In our study, HES1 knockdown, verified by qRT-PCR and WB assays, corresponded to a reduction in ITPR1 expression, which provides support for the hypothesis of HES1-mediated transcriptional control of ITPR1. This theory was further supported by the dual luciferase assay results, in which HES1 knockdown resulted in reduced ITPR1 promoter activity. Our in-depth exploration of the functional role of HES1 in regulating PA cell behavior and autophagy revealed that HES1 knockdown counteracted the effects of ITPR1 overexpression on autophagy proteins and also weakened the suppressive impact of ITPR1 on PA cell growth, invasion and migration. These observations are consistent with the findings of Monahan et al. (38), who demonstrated that HES1 is associated with PA cell proliferation and affects the expression of cell cycle inhibitors. Collectively, our results elucidate a complex regulatory interplay involving HES1, ITPR1, and autophagy in PA progression. These novel insights not only confirm existing research, while also laying the groundwork for the creation of innovative therapeutic approaches for PA.
Conclusions
In summary, our research elucidates the complex regulatory interactions between HES1, ITPR1, and autophagy in PA progression. We identified the pivotal role of HES1 in modulating ITPR1 expression, driving autophagy in PA cells, and subsequently curbing tumor invasiveness. These insights emphasize the therapeutic potential of targeting the HES1-ITPR1-autophagy pathway to counteract tumor advancement, presenting a new strategy for addressing treatment-resistant PA.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1320/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1320/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1320/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1320/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liu X, Wang R, Li M, et al. Pituitary adenoma or pituitary neuroendocrine tumor: a narrative review of controversy and perspective. Transl Cancer Res 2021;10:1916-20. [Crossref] [PubMed]
- Banskota S, Adamson DC. Pituitary Adenomas: From Diagnosis to Therapeutics. Biomedicines 2021;9:494. [Crossref] [PubMed]
- Daly AF, Beckers A. The Epidemiology of Pituitary Adenomas. Endocrinol Metab Clin North Am 2020;49:347-55. [Crossref] [PubMed]
- Melmed S, Kaiser UB, Lopes MB, et al. Clinical Biology of the Pituitary Adenoma. Endocr Rev 2022;43:1003-37. [Crossref] [PubMed]
- Giuffrida G, D'Argenio V, Ferraù F, et al. Methylome Analysis in Nonfunctioning and GH-Secreting Pituitary Adenomas. Front Endocrinol (Lausanne) 2022;13:841118. [Crossref] [PubMed]
- Aflorei ED, Korbonits M. Epidemiology and etiopathogenesis of pituitary adenomas. J Neurooncol 2014;117:379-94. [Crossref] [PubMed]
- Mahzari M, Alhamlan KS, Alhussaini NA, et al. Epidemiological and clinical profiles of Saudi patients with hyperprolactinemia in a single tertiary care center. Ann Saudi Med 2022;42:334-42. [Crossref] [PubMed]
- Bello MO, Garla VV. Gigantism and Acromegaly. Treasure Island, FL, USA: StatPearls Publishing, 2023.
- Tritos NA, Miller KK. Diagnosis and Management of Pituitary Adenomas: A Review. JAMA 2023;329:1386-98. [Crossref] [PubMed]
- Raverot G, Ilie MD, Lasolle H, et al. Aggressive pituitary tumours and pituitary carcinomas. Nat Rev Endocrinol 2021;17:671-84. [Crossref] [PubMed]
- Arafah BM, Nasrallah MP. Pituitary tumors: pathophysiology, clinical manifestations and management. Endocr Relat Cancer 2001;8:287-305. [Crossref] [PubMed]
- Lin AL, Donoghue MTA, Wardlaw SL, et al. Approach to the Treatment of a Patient with an Aggressive Pituitary Tumor. J Clin Endocrinol Metab 2020;105:3807-20. [Crossref] [PubMed]
- Molitch ME. Diagnosis and Treatment of Pituitary Adenomas: A Review. JAMA 2017;317:516-24. [Crossref] [PubMed]
- Rakesh R. Role and regulation of autophagy in cancer. Biochim Biophys Acta Mol Basis Dis 2022;1868:166400. [Crossref] [PubMed]
- Li C, Wei C, Zhao G, et al. Cancer cells remodeling and quality control are inextricably linked to autophagy. AIMS Molecular Science 2023;10:109-26.
- Folkerts H, Hilgendorf S, Vellenga E, et al. The multifaceted role of autophagy in cancer and the microenvironment. Med Res Rev 2019;39:517-60. [Crossref] [PubMed]
- Lyu L, Hu Y, Yin S, et al. Autophagy inhibition enhances anti-pituitary adenoma effect of tetrandrine. Phytother Res 2021;35:4007-21. [Crossref] [PubMed]
- Kun Z, Yuling Y, Dongchun W, et al. HIF-1α Inhibition Sensitized Pituitary Adenoma Cells to Temozolomide by Regulating Presenilin 1 Expression and Autophagy. Technol Cancer Res Treat 2016;15:NP95-NP104. [Crossref] [PubMed]
- Mangla A, Guerra MT, Nathanson MH. Type 3 inositol 1,4,5-trisphosphate receptor: A calcium channel for all seasons. Cell Calcium 2020;85:102132. [Crossref] [PubMed]
- Raimondi L, Ciarapica R, De Salvo M, et al. Inhibition of Notch3 signalling induces rhabdomyosarcoma cell differentiation promoting p38 phosphorylation and p21(Cip1) expression and hampers tumour cell growth in vitro and in vivo. Cell Death Differ 2012;19:871-81. [Crossref] [PubMed]
- Ochi S, Imaizumi Y, Shimojo H, et al. Oscillatory expression of Hes1 regulates cell proliferation and neuronal differentiation in the embryonic brain. Development 2020;147:dev182204. [Crossref] [PubMed]
- Prakash V, R, Patel S, Hariohm K, et al. Importance of squatting and sitting on the floor: perspectives and priorities of rural Indian patients with stroke. Top Stroke Rehabil 2016;23:240-4. [Crossref] [PubMed]
- Pathak K, Pathak M, Patel N, et al. Classification of Brain Tumor Using Convolutional Neural Network. 2019 3rd International conference on Electronics, Communication and Aerospace Technology (ICECA). Coimbatore: IEEE, 2019:128-32.
- Loos B, Klionsky DJ, Du Toit A, et al. On the relevance of precision autophagy flux control in vivo - Points of departure for clinical translation. Autophagy 2020;16:750-62. [Crossref] [PubMed]
- Seranova E, Connolly KJ, Zatyka M, et al. Dysregulation of autophagy as a common mechanism in lysosomal storage diseases. Essays Biochem 2017;61:733-49. [Crossref] [PubMed]
- Schepers J, Behl C. Lipid droplets and autophagy-links and regulations from yeast to humans. J Cell Biochem 2021;122:602-11. [Crossref] [PubMed]
- Papathomas TG, Nosé V. New and Emerging Biomarkers in Endocrine Pathology. Adv Anat Pathol 2019;26:198-209. [Crossref] [PubMed]
- Li N, Zhan X. Mitochondrial Dysfunction Pathway Networks and Mitochondrial Dynamics in the Pathogenesis of Pituitary Adenomas. Front Endocrinol (Lausanne) 2019;10:690. [Crossref] [PubMed]
- Wang FH, Zhang XT, Li YF, et al. The Chinese Society of Clinical Oncology (CSCO): Clinical guidelines for the diagnosis and treatment of gastric cancer, 2021. Cancer Commun (Lond) 2021;41:747-95. [Crossref] [PubMed]
- Di Micco R, Krizhanovsky V, Baker D, et al. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat Rev Mol Cell Biol 2021;22:75-95. [Crossref] [PubMed]
- Delbridge AR, Valente LJ, Strasser A. The role of the apoptotic machinery in tumor suppression. Cold Spring Harb Perspect Biol 2012;4:a008789. [Crossref] [PubMed]
- Puga A, Ma C, Marlowe JL. The aryl hydrocarbon receptor cross-talks with multiple signal transduction pathways. Biochem Pharmacol 2009;77:713-22. [Crossref] [PubMed]
- Li F, Huangyang P, Burrows M, et al. FBP1 loss disrupts liver metabolism and promotes tumorigenesis through a hepatic stellate cell senescence secretome. Nat Cell Biol 2020;22:728-39. [Crossref] [PubMed]
- Tang F, Christofori G. The cross-talk between the Hippo signaling pathway and autophagy:implications on physiology and cancer. Cell Cycle 2020;19:2563-72. [Crossref] [PubMed]
- Gu Y, Li P, Peng F, et al. Autophagy-related prognostic signature for breast cancer. Molecular Carcinogenesis 2016;55:292-9. [Crossref] [PubMed]
- Maharjan Y, Dutta RK, Son J, et al. Intracellular cholesterol transport inhibition Impairs autophagy flux by decreasing autophagosome-lysosome fusion. Cell Commun Signal 2022;20:189. [Crossref] [PubMed]
- Perrone S, Zubeldia-Brenner L, Gazza E, et al. Notch system is differentially expressed and activated in pituitary adenomas of distinct histotype, tumor cell lines and normal pituitaries. Oncotarget 2017;8:57072-88. [Crossref] [PubMed]
- Monahan P, Rybak S, Raetzman LT. The notch target gene HES1 regulates cell cycle inhibitor expression in the developing pituitary. Endocrinology 2009;150:4386-94. [Crossref] [PubMed]


