
CCND2 is a prognostic biomarker and correlates with immune infiltration in lung adenocarcinoma
Highlight box
Key findings
• CCND2 was significantly lower in lung adenocarcinoma (LUAD) than adjacent normal tissues.
• LUAD patients who expressed lower CCND2 had a shorter overall survival and CCND2 was an independent prognostic risk factor for LUAD.
What is known and what is new?
• Previous studies showed CCND2 is the hub target gene of miR-17, and miR-17 is differently expressed between LUAD and normal tissues. CCND2 can act as oncogene in LUAD and has a higher frequency of low CCND2 mRNA expression in LUAD tissue than in adjacent normal tissues.
• In the present study, we found that CCND2 was downregulated in LUAD compared to adjacent normal tissues, the overexpression of CCND2 was associated with longer overall survival in LUAD. Naive B cells, resting dendritic cells, macrophages M1, macrophages M2, activated natural killer cells, and resting CD4+ memory cells, had significantly different proportions based on the expression levels of CCND2 in LUAD.
What is the implication, and what should change now?
• CCND2 can be exploited as a novel prognostic biomarker involved in immune infiltration of LUAD, hence providing new preventative and therapeutic options for LUAD.
Introduction
Lung cancer (LC) is the leading cause of cancer-related death and is projected with approximately 350 deaths per day worldwide. There were expected to be 1,918,030 new cases and 609,360 deaths in the United States in 2022 (1). Lung adenocarcinoma (LUAD) is the most prevalent histological subtype of LC among all pathological subtypes of LC (2). The 5-year overall survival rate of patients with advanced LUAD is less than 5%; however, if these patients receive targeted therapy, their survival rate will increase (3). Consequently, it is crucial to identify a biomarker that can accurately predict the prognosis of LUAD in order to guide the selection of the more appropriate targeted therapy. LUAD is a complex, multi-step process linked to the aberrant expression of several genes. With the development of new therapeutic strategies, oncologists have become increasingly interested in immune-related targeted therapy. Thus, a deeper comprehension of the molecular mechanisms and immune infiltration of LUAD could provide precise prognostic indicators and therapeutic options.
CCND2/Cyclin D2 is a member of the D-type cyclin family and modulates the extracellular signaling environment in cell cycle progression, particularly during the G1/S transition, by regulating cyclin-dependent kinases 4 and 6 4/6 (4). CCND2 has been reported to be up-regulated in the condition of growth arrest, as the ectopic expression of CCND2 (5). The expression of CCND2 expression is required for persistent cancer stem cell growth via the maintenance of an intact cell cycle and proliferation with low levels of DNA damage accumulation (6). In human epithelial ovarian cell cancer decreased CCND2 expression is related to a poor prognosis for survival (7). CCND2 aberrant expression is commonly observed in LC tissues, and CCND2 hypermethylation is substantially more prevalent in invasive LUAD (8). CCND2 can be targeted by miR-646 and miR-4317 in non-small cell lung cancer (NSCLC) to inhibit cancer cell proliferation and metastasis (9,10). In the study of Hung et al., NSCLC patients with low expression CCND2 had poor overall survival (11).
CCND2 is the hub target gene of miR-17, and miR-17 is differently expressed between LUAD and normal tissues, according to our earlier research (12). Thus, we deduced that CCND2 expression might play a significant role in LUAD development and progression. In the present study, based on The Cancer Genome Atlas (TCGA) database we evaluated the association between CCND2 expression and the prognosis of LUAD patients, as well as the relationship between CCND2 expression and tumor immune infiltration in LUAD. The analysis workflow of our study is presented in Figure 1. We present this article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1863/rc).
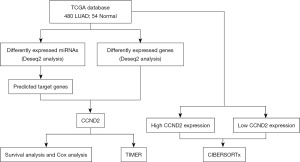
Methods
Clinical samples
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). We downloaded 480 LUAD cases with clinical data, miRNAs expression data, and gene expression data [counts data and transcripts per million (TPM) data] from the TCGA database, as well as 54 adjacent normal tissues. The clinical data comprised gender, age, pathologic stage, pharmaceutical or radiation therapy, overall survival days and survival status. The basic clinical data are displayed in Table 1.
Table 1
| Items of clinical characteristics | LUAD (N=480) |
|---|---|
| Age, n (%) | |
| ≥65 years | 254 (52.9) |
| <65 years | 207 (43.1) |
| NA | 19 (4.0) |
| Sex, n (%) | |
| Female | 262 (54.6) |
| Male | 218 (45.4) |
| Pathologic stage, n (%) | |
| Stage I | 260 (54.2) |
| Stage II | 116 (24.2) |
| Stage III | 79 (16.5) |
| Stage IV | 25 ( 5.2) |
| Treatment type, n (%) | |
| Radiation therapy | 235 (49.0) |
| Pharmaceutical therapy | 245 (51.0) |
LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas; NA, not available.
Identifying differently expressed miR-17 in LUAD
In our previous study, we demonstrated that miR-17 was differently expressed between LUAD patients and healthy controls (12). In the present study, we further established the different expression of miR-17 in LUAD compared to adjacent normal tissues based on TCGA database using R package “Deseq2”. Adjusted P<0.05 and |log2 fold change (FC)| > 1 were considered as statistical significance. Furthermore, we predicted the target genes of miR-17 using online tool “miRWalk”.
Detecting differently expressed CCND2 in LUAD
We detected differently expressed genes (DEGs) between LUAD and adjacent normal tissues using R package “Deseq2”. Adjusted P<0.05 and |log2 FC| >1 were considered as statistical significance. We selected DEGs, which duplicated the target genes of miR-17, for further analysis. We calculated the mean and standard deviations of CCND2 both in LUAD and adjacent normal tissues. A two-sample t-test was applied to compare the CCND2 expression level between these two groups. The box plot applied to reveal the result by R software.
Survival analysis of CCND2 in LUAD
According to the expression level of CCND2 in LUAD and adjacent normal groups, survival analysis was conducted. After eliminating the survival time of the LUAD patients with less than 90 days, Kaplan-Meier survival curve was performed to estimate the relationship between the overall survival of LUAD patients and CCND2 expression level. Furthermore, we integrated CCND2 expression level and clinical information to identify the independent prognostic factors for LUAD through univariate and multivariate Cox regression analyses.
Correlation between CCND2 and tumor immune infiltration cells
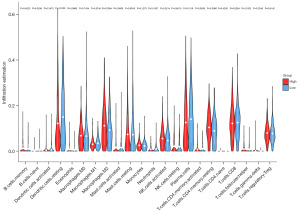
We evaluated the relationship between CCND2 expression and immune cell infiltration (including B cells, CD4 T cells, CD8 T cells, neutrophils, macrophages, and dendritic cells) through gene module using TIMER (Tumor Immune Estimation Resource) online tool (https://cistrome.shinyapps.io/timer/). TIMER is a powerful database which can analyze the immune invasion levels of different tumors in the TCGA (13). We also inferred the relative proportion of infiltrating immune cells from genetic expression profiles (a “signature matrix” of 547 genes) using CIBERSORT (Cell-type Identification by Estimating Relative Subsets of known RNA Transcripts) deconvolution algorithm (https://cibersortx.stanford.edu/). CIBERSORT tool used a gene expression signature matrix (a “signature matrix” of 547 genes) to assess TPM data of 22 tumor infiltrating immune cells (TIICs) in LUAD tissues of the high and low expression level of CCND2 groups (14). We used violin plot to visualize the differences of 22 types of infiltrating immune cells between these two groups.
Statistical analysis
All data analyses and figures were conducted by R software (version 4.0.1). A P value <0.05 was considered to be statistically significant.
Results
MiR-17 was downexpressed in LUAD
We downloaded miRNA expression data of 480 LUAD and 54 adjacent normal tissues from TCGA database, and found miR-17 was downexpressed in LUAD compared to adjacent normal tissues (Adjusted P<0.05 and log2FC =−0.81). Furthermore, we predicted 12,004 target genes of miR-17 using miRWalk online tool.
CCND2 was downregulated in LUAD
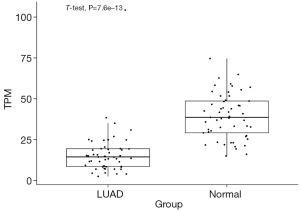
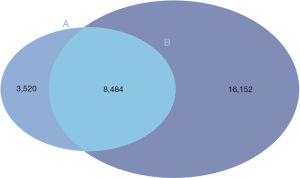
To explore whether CCND2 was differently expressed in LUAD, we analyzed DEGs between LUAD and adjacent normal tissues based on the gene expression data from the TCGA-LUAD. We identified 24,636 DEGs between these two groups and found CCND2 was downregulated in LUAD compared to adjacent normal tissues (Adjusted P<0.05 and log2FC =−1.33, Figure 2). Then, we selected the overlapping genes, containing 8,484 genes which included CCND2, between the target genes of miR-17 and DEGs (Figure 3; the supplementary table is available online: https://cdn.amegroups.cn/static/public/tcr-23-1863-1.xlsx).
CCND2 expression was associated with longer survival in LUAD patients
Kaplan-Meier analysis indicated LUAD patients with high CCND2 expression had significantly longer overall survival compared to those with low CCND2 expression (P=0.046, Figure 4). Univariate and multivariate Cox regression analyses were performed to identify whether CCND2 was an independent prognostic risk factor for LUAD. The univariate Cox regression revealed that pathologic stage [P<0.001, hazard ratio (HR): 1.61] and CCND2 expression (P=0.039, HR: 0.73) were the prognostic risk factors for LUAD. The multivariate Cox regression showed that pathologic stage (P<0.001, HR: 1.50) and CCND2 expression (P=0.049, HR: 0.77) were also the independent prognostic risk factors for LUAD. The results of the univariate and multivariate Cox regression analysis are shown in Table 2.

Table 2
| Items | Univariate | Multivariate | Deseq2 analysis | |||||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | Adjusted P | log2FC | |||
| Age | 1.19 (0.88–1.62) | 0.254 | 1.28 (0.94–1.75) | 0.303 | ||||
| Sex | 0.94 (0.70–1.27) | 0.704 | 1.02 (0.75–1.37) | 0.768 | ||||
| Stage | 1.61 (1.40–1.84) | <0.001* | 1.50 (1.26–1.80) | <0.001* | ||||
| Pharma | 1.02 (0.76–1.37) | 0.892 | 1.01 (0.75–1.37) | 0.976 | ||||
| Radio | 0.98 (0.73–1.32) | 0.892 | 0.99 (0.73–1.33) | 0.976 | ||||
| CCND2 | 0.73 (0.54–0.99) | 0.039* | 0.77 (0.57–1.03) | 0.049* | <0.05* | −1.33 | ||
*, the P value with statistically significant. HR, hazard ratio; CI, confidence interval; FC, fold change.
CCND2 expression correlated with TIICs in LUAD
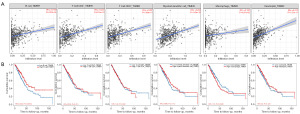
We used TIMER database to explore the possible association between CCND2 expression and TIICs in LUAD. As shown in Figure 5, CCND2 expression was positively associated with the levels of B cells (r=0.159, P=4.00e−04), CD8+ T cells (r=0.287, P=7.88e−11), CD4+ T cells (r=0.301, P=8.14e−12), macrophages (r=0.128, P=4.57e−03), neutrophils (r=0.373, P=1.07e−17), and myeloid dendritic cells (r=0.284, P=1.43e−10), of which the levels of B cells and macrophages had significant relationship with the overall survival of LUAD patients. LUAD patients with longer overall survival had higher level of B cell and lower level of macrophages. Furthermore, we examined whether the tumor immune microenvironment was different between the LUAD patients with high CCND2 expression level and those with low CCND2 expression level via CIBERSORTx analysis. Using the median expression level of CCND2, we separated LUAD patients into high expression and low expression groups. The CIBERSORTx analysis showed that the proportions of naive B cells, resting dendritic cells, and macrophages M1 were higher in the low CCND2 expression level group; whereas the proportions of macrophages M2, activated natural killer (NK) cells, and resting CD4+ memory cells were lower (Figure 6). Therefore, it is suggested that CCND2 may play an important role in regulating immune infiltration in LUAD.
Discussion
With the development of high-throughput sequencing technology over the past decade, molecular prognostic biomarkers of cancers emerged endlessly to predict the treatment response and cancer prognosis. Previously, we discovered the target gene of CCND2 of miR-17 was a hub gene for LUAD via bioinformatics and meta-analysis based on Gene Expression Omnibus (GEO) database. In the present study, we demonstrated that miR-17 and its target gene CCND2 was significantly downregulated in LUAD compared to adjacent normal tissues based on TCGA. LUAD patients with high CCND2 expression exhibited longer overall survival than those with low CCND2 expression. Cox regression analysis verified CCND2 was an independent prognostic risk factor for LUAD. To further explore the probable mechanism of CCND2 in LUAD, we discovered that CCND2 expression was positively correlated with TIICs, and the levels of B cells and macrophages had a significant relationship with the overall survival of LUAD patients. In the low CCND2 expression group, the proportions of naive B cells, resting dendritic cells, and macrophages M1 were higher; whereas macrophages M1, activated NK cells, and resting CD4+ memory cells were lower. Consequently, our study demonstrates CCND2 as a possible molecular prognostic biomarker for LUAD, which may influence the outcome of LUAD progression and tumor immune infiltration in LUAD development.
Gene expression analysis indicated that the changes in expression of CCND2 genes had been evaluated by real-time polymerase chain reaction (RT-PCR) following miR-17 overexpression. The inhibition of miR-17 increases the expression level of CCND2, which is the cell cycle activator, associated with cell proliferation and tumor growth in anaplastic thyroid cancer (15). Western blot was further used to track changes in the expression of CCND2, and the result showed that an increase in expression of CCND2 occurs as a consequence of miR-17-92 cluster overexpression and miR-17 inhibition increases expression of the miR-17-92 cluster, which can regulate cell cycle and enhanced cell proliferation (15,16). A luciferase reporter assay demonstrated that has_cic_105039 promotes cardiomyocyte differentiation by sponging miR-17 to regulate CCND2 expression, which induces cell differentiation, viability and migration (17). Transcriptome and target analyses showed that miR-17 can act on CCND2 which is important for cell proliferation and/or fusion (18). In our present and previous studies, we revealed the similar result that miR-17 affects CCND2 to influence LUAD development and progression. CCND2 is a well-known cyclin which functions in the cell cycle, specifically in G1/S transition. MiR-4317 is a negative regulator of the cell cycle and increased expression of this miRNA results in significant G0/G1 arrest and S phase reduction (10). He et al.’s study indicated that miR-4317 could reduce NSCLC cell growth and metastasis by directly targeting CCND2 and FGR9, especially in lymph node metastasis and advanced clinical stage patients (10). CCND2 is also the direct target of miR-646, the overexpression of miR-646 obviously suppressed the expression of CCND2’s mRNA and protein in NSCLC cell lines. CCND2 overexpression can reverse the suppression of proliferation and invasion induced by miR-646, the suppressive effects of miR646 on NSCLC are mediated by the downregulation of CCND2 (9).
LUAD has inter-patient and intra-tumoral heterogeneity in both tumor microenvironment and tumor cells (19). The tumor microenvironment consists of several TIICs, cancer associated fibroblast, and vascular endothelial. In different tumors, the tumor microenvironment components vary markedly and play important roles in tumor progression and metastasis (20). Some human data from epidemiological studies discovered a relation between increased macrophage level and poor prognosis and response for chemotherapy in LCs (21). Similar to previous studies, we also found LUAD patients with high level macrophage cells showed poor overall survival in present study. Larroquette et al. showed a substantial positive relationship between NSCLC progression and high levels of tumor associated macrophage cell in close proximity to tumor cells (22). High macrophage cell levels were related to poor overall survival in NSCLC. This may be due to overexpression of the angiogenic enzyme thymidine phosphorylase (23). Plasma cells and B cells were shown to be considerably abundant in LUAD tissues with more differentiated states and increased frequencies of somatic hypermutation, and increasing B cell infiltration was associated with a favorable prognosis (24). Perhaps, more studies are required to demonstrate the link between tumor associated immune cell infiltration and prognosis of LUAD patients.
CCND2 is a regulator of cell cycle and plays an important role in cell proliferation (25). Few studies has studied CCND2’s role in tumor immune cell infiltration in LUAD, despite its tight association with tumor immune cell infiltration. One study also demonstrated that CCND2 had robust prognostic prediction and potential to evaluate immunotherapy response for LUAD (26). One study indicated that CCND2 acts as an oncogene, which is upregulated by LOC554202 to promote tumor cell proliferation and cell cycle in thyroid cancer (27). With several experiments, CCND2 has been shown to have a unique role in tumor metastasis and LOC554202 increases CCND2 as an oncogene to drive tumor cell proliferation and cell cycle in thyroid cancer and T cell apoptosis in LUAD (27). High-expressed CCND2 can significantly affect cancer cell abilities of migration and invasion and down-regulate, programmed cell death-1 (PD-1)/programmed death-ligand 1 (PD-L1) signaling by causing T cell apoptosis reduction (28). Our results from the transcriptome analysis also implied the possible function of the immune components in tumor microenvironment for the prognosis of LUAD patients. We adopted the CIBERSORTx algorithm to explore the association of CCND2 with the 22 tumor-infiltrating immune cells subsets of immune reaction. We found six types of immune cells, naive B cells, resting dendritic cells, macrophages M1, macrophages M2, activated NK cells, and resting CD4+ memory cells, had significantly different proportions based on the expression levels of CCND2. In addition, TIMER verified a significant positive correlation of the expression of CCND2 with B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and myeloid dendritic cells, only B cells and macrophages had significant relationship with the overall survival of LUAD patients. We found the results from the two databases were partially inconsistent. The different statistical analyses might be one of the reasons. TIMER database used Pearson’s correlation analysis, although the results were significantly statistical, the coefficients of the Pearson’s correlation analysis were weak; the results in our study were based on the online database, which needed large sample prospective research to validation.
CCND2 encodes cyclin D2, a protein involved in cell cycle progression which acts as a regulator of cyclin-dependent kinases 4 and 6 (CDK4/6) in the G1/S transition (29). Analysis of the Infinium Human Methylation 450K BeadChip assay data from TCGA revealed that LUAD showed a higher frequency of low CCND2 mRNA expression than adjacent normal tissues. The association of low CCND2 with LUAD is interesting. CCND2 knockdown in H1299 cells induced cell proliferation and metastasis, suggesting that CCND2 may act as a suppressor gene during some solid tumor formation (11). Similar to previous studies, we demonstrated that the overexpression of CCND2 was associated with longer overall survival in LUAD. However, one study reported that CCND2 acted as an oncogene in LUAD (27). Reduced CCND2 expression promoted cell proliferation, and its overexpression inhibited cancer cell growth (30). Thus the underlying mechanism of CCND2 in LUAD needs further investigation. Mutations or amplifications of CCND2 genes were also found in several cancers, the clinical significance of CCND2 alterations was reported previously. Promoter hypermethylation of CCND2 was expressed in LUAD, especially in invasive peripheral LUAD. Notably, almost no methylation was observed in several CpG sites of CCND2 promoter regions of adjacent normal tissues (8,11).
Conclusions
Our findings indicated that CCND2 was downregulated in LUAD tissues and LUAD patients with downregulated CCND2 had poor overall survival, which was an independent prognostic risk factor for LUAD. The expression level of CCND2 was positively associated with TIICs in the tumor microenvironment, indicating its potential as an immunotherapy target for LUAD.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1863/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1863/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1863/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Oja AE, Piet B, van der Zwan D, et al. Functional Heterogeneity of CD4(+) Tumor-Infiltrating Lymphocytes With a Resident Memory Phenotype in NSCLC. Front Immunol 2018;9:2654. [Crossref] [PubMed]
- Zhang Q, Sakamoto K, Wagner KU. D-type Cyclins are important downstream effectors of cytokine signaling that regulate the proliferation of normal and neoplastic mammary epithelial cells. Mol Cell Endocrinol 2014;382:583-92. [Crossref] [PubMed]
- Wang L, Cui Y, Zhang L, et al. The Silencing of CCND2 by Promoter Aberrant Methylation in Renal Cell Cancer and Analysis of the Correlation between CCND2 Methylation Status and Clinical Features. PLoS One 2016;11:e0161859. [Crossref] [PubMed]
- Park SY, Lee CJ, Choi JH, et al. The JAK2/STAT3/CCND2 Axis promotes colorectal Cancer stem cell persistence and radioresistance. J Exp Clin Cancer Res 2019;38:399. [Crossref] [PubMed]
- Sakuma M, Akahira J, Ito K, et al. Promoter methylation status of the Cyclin D2 gene is associated with poor prognosis in human epithelial ovarian cancer. Cancer Sci 2007;98:380-6. [Crossref] [PubMed]
- Chung JH, Lee HJ, Kim BH, et al. DNA methylation profile during multistage progression of pulmonary adenocarcinomas. Virchows Arch 2011;459:201-11. [Crossref] [PubMed]
- Wang J, Shu H, Guo S. MiR-646 suppresses proliferation and metastasis of non-small cell lung cancer by repressing FGF2 and CCND2. Cancer Med 2020;9:4360-70. [Crossref] [PubMed]
- He X, Chen SY, Yang Z, et al. miR-4317 suppresses non-small cell lung cancer (NSCLC) by targeting fibroblast growth factor 9 (FGF9) and cyclin D2 (CCND2). J Exp Clin Cancer Res 2018;37:230. [Crossref] [PubMed]
- Hung CS, Wang SC, Yen YT, et al. Hypermethylation of CCND2 in Lung and Breast Cancer Is a Potential Biomarker and Drug Target. Int J Mol Sci 2018;19:3096. [Crossref] [PubMed]
- Jia E, Ren N, Zhang R, et al. Circulating miR-17 as a promising diagnostic biomarker for lung adenocarcinoma: evidence from the Gene Expression Omnibus. Transl Cancer Res 2020;9:5544-54. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Attar M, Arefian E, Nabiuni M, et al. MicroRNA 17-92 expressed by a transposone-based vector changes expression level of cell-cycle-related genes. Cell Biol Int 2012;36:1005-12. [Crossref] [PubMed]
- Sweat Y, Ries RJ, Sweat M, et al. miR-17 acts as a tumor suppressor by negatively regulating the miR-17-92 cluster. Mol Ther Nucleic Acids 2021;26:1148-58. [Crossref] [PubMed]
- Yu B, Li M, Han SP, et al. Circular RNA hsa_circ_105039 promotes cardiomyocyte differentiation by sponging miR‑17 to regulate cyclinD2 expression. Mol Med Rep 2021;24:861. [Crossref] [PubMed]
- Kong D, He M, Yang L, et al. MiR-17 and miR-19 cooperatively promote skeletal muscle cell differentiation. Cell Mol Life Sci 2019;76:5041-54. [Crossref] [PubMed]
- Chittezhath M, Dhillon MK, Lim JY, et al. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity 2014;41:815-29. [Crossref] [PubMed]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423-37. [Crossref] [PubMed]
- Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol 2002;196:254-65. [Crossref] [PubMed]
- Larroquette M, Guegan JP, Besse B, et al. Spatial transcriptomics of macrophage infiltration in non-small cell lung cancer reveals determinants of sensitivity and resistance to anti-PD1/PD-L1 antibodies. J Immunother Cancer 2022;10:e003890. [Crossref] [PubMed]
- Koukourakis MI, Giatromanolaki A, Kakolyris S, et al. Different patterns of stromal and cancer cell thymidine phosphorylase reactivity in non-small-cell lung cancer: impact on tumour neoangiogenesis and survival. Br J Cancer 1998;77:1696-703. [Crossref] [PubMed]
- Hao D, Han G, Sinjab A, et al. The Single-Cell Immunogenomic Landscape of B and Plasma Cells in Early-Stage Lung Adenocarcinoma. Cancer Discov 2022;12:2626-45. [Crossref] [PubMed]
- Chiles TC. Regulation and function of cyclin D2 in B lymphocyte subsets. J Immunol 2004;173:2901-7. [Crossref] [PubMed]
- Jin X, Hu Z, Sui Q, et al. A Novel Prognostic Signature Revealed the Interaction of Immune Cells in Tumor Microenvironment Based on Single-Cell RNA Sequencing for Lung Adenocarcinoma. J Immunol Res 2022;2022:6555810. [Crossref] [PubMed]
- Chen C, Qin L, Xiao MF. Long Noncoding RNA LOC554202 Predicts a Poor Prognosis and Correlates with Immune Infiltration in Thyroid Cancer. Comput Math Methods Med 2022;2022:3585626. [Crossref] [PubMed]
- Tian C, Li C, Zeng Y, et al. Identification of CXCL13/CXCR5 axis's crucial and complex effect in human lung adenocarcinoma. Int Immunopharmacol 2021;94:107416. [Crossref] [PubMed]
- Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta 2002;1602:73-87. [PubMed]
- Chen Y, Zhang Q, Wang Q, et al. Genetic association analysis of the RTK/ERK pathway with aggressive prostate cancer highlights the potential role of CCND2 in disease progression. Sci Rep 2017;7:4538. [Crossref] [PubMed]


