
Cuproptosis-related molecular classification and gene signature of hepatocellular carcinoma and experimental verification
Highlight box
Key findings
• Two cuproptosis-associated subtypes of hepatocellular carcinoma (HCC) were identified.
• In this study, a novel HCC prognosis prediction signature containing three cuproptosis-related genes (lipoyltransferase 1, dihydrolipoamide s-acetyltransferase, cyclin dependent kinase inhibitor 2A) was constructed and proved to have significant predictive value.
What is known and what is new?
• Cuproptosis is a new type of cell death, which is different from other programmed cell death, and is closely associated with tumor development.
• HCC patients of the two cuproptosis-associated subtypes have different clinical prognosis and immune profiles. The cuproptosis-related gene signature exhibited robust validity in predicting prognosis and the risk score was an independent predictor for overall survival of HCC patients.
What is the implication, and what should change now?
• The promising prognostic accuracy of this signature may facilitate individualized prognosis management and therapeutic intervention.
• More experimental findings are needed in the future to refine the molecular classification and risk signature.
Introduction
Hepatocellular carcinoma (HCC) is a major pathological primary liver malignancy, accounting for more than 90% of all primary liver cancers (1). Its incidence has been increasing annually worldwide, with HCC currently ranking sixth in global tumor incidence and third in cancer-related causes of death (2). HCC can arise from a variety of high-risk factors, among which are chronic hepatitis B or C virus infection, alcoholic liver disease, and non-alcoholic fatty liver disease (3,4). At present, surgical resection is the main treatment option for patients with HCC. Moreover, due to the lack of obvious symptoms and signs of early-stage disease, many patients are usually diagnosed in the middle or late stages, eliminating the opportunity for radical surgical treatment (5). The high heterogeneity of HCC results in poor efficacy of many targeted drugs, or in some patients with HCC, the rapid development of drug resistance (6,7). Therefore, a better understanding of the molecular changes and mechanisms during HCC, as well as the characterization of its occurrence and progression, is of great significance for the identification of novel biomarkers for HCC prognosis and the development of more effective treatment strategies.
Copper is an indispensable mineral nutrient in the human body because it plays important roles in many biological processes, including mitochondrial respiration, kinase signaling, and antioxidation/detoxification (8). Under normal physiological conditions, organisms maintain a low concentration and dynamic balance of copper ions (9). The abnormal accumulation of copper ions can cause copper toxicity and induce cell death (10). Some human gene mutations have been shown to induce various diseases by causing an imbalance in copper homeostasis in the body (11). In recent years, studies have found that the dysregulation of copper homeostasis is crucial in the occurrence and progression of tumors (12,13). Tsvetkov et al. found that copper ions cause an abnormal aggregation of lipoylated proteins by directly binding to them in the tricarboxylic acid cycle pathway, thus interfering with iron-sulfur cluster proteins in respiratory chain complexes and causing a proteotoxic stress response that ultimately leads to cell death. This process is a novel mode of death and is defined as cuproptosis (14). Studies have shown that ten genes including ferredoxin 1 (FDX1), lipoyl syndrome (LIAS), lipoyltransferase 1 (LIPT1), dihydrolipoamide dehydrogenase (DLD), dihydrolipoamide s-acetyltransferase (DLAT), pyruvate dehydrogenase E1 subunit alpha 1 (PDHA1), pyruvate dehydrogenase E1 subunit beta (PDHB), metal regulatory transcription factor 1 (MTF1), glutaminase (GLS), and cyclin-dependent kinase inhibitor 2A (CDKN2A) are key in regulating cuproptosis (14,15). In addition, multiple studies have found that copper ionophores and their chelating agents are promising drug molecules for tumor therapy (16,17). However, the association between the expression of these cuproptosis-related regulatory genes and the prognosis of patients remains largely unknown.
In this study, we comprehensively evaluated the gene expression patterns and potential functional enrichment analysis of cuproptosis regulators in HCC using The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) databases. We also subtyped HCC patients using an unsupervised clustering method, explored the differences in immune infiltration and immune checkpoint expression among HCC patients with different subtypes, and predicted their response to immunotherapy and sensitivity to targeted drugs. Finally, we established a prognostic signature to predict the survival status of patients with HCC. Our findings provide a theoretical basis for developing a novel comprehensive treatment strategy for patients with HCC. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1876/rc).
Methods
Data collection and preprocessing
Transcriptome, somatic mutation, and copy number variation (CNV) data of 374 HCC and 50 adjacent normal tissues in The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) dataset, along with the corresponding clinical information, were downloaded from the Genomic Data Commons (GDC) data portal (https://portal.gdc.cancer.gov/). The corresponding mRNA expression data were obtained from the transcriptome expression data by comparing the annotation files of the human genome GRCh38. The mRNA expression data and corresponding clinical information for 232 HCC tissues in the ICGC dataset were downloaded from the International Cancer Genome Consortium database (https://dcc.icgc.org/projects/LIRI-JP). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Access to the unassigned linked dataset was obtained from the TCGA and ICGC databases in accordance with their policies. Immunohistochemistry (IHC) results of normal liver and HCC tissues were obtained from the Human Protein Atlas (HPA; https://www.proteinatlas.org/) database and were used to analyze the protein expression levels of the ten cuproptosis regulators.
Unsupervised clustering for the ten cuproptosis-related regulators
Unsupervised clustering analysis of the TCGA-LIHC dataset was performed using the R package “consensusclusterplus” (18) based on the expression levels of the ten cuproptosis regulators; this was to obtain different regulatory expression profiles of cuproptosis and classify them into corresponding subtypes. The prognosis between the two subtypes was compared using Kaplan-Meier analysis. Chi-square test was used to analyze the correlation between clustering and clinical parameters, and heatmap was used to display the result.
Analysis of the landscape of the tumor immune microenvironment across subtypes
The single-sample gene set enrichment analysis (ssGSEA) algorithm was used with the R package “Gene Set Variation Analysis (GSVA)” (19) to quantify the relative abundance of multiple immune cell infiltrates in the tumor microenvironment (TME) of each HCC sample. The expression of common immune checkpoints in HCC tissues was compared among the different subtypes. Differences in the levels of immune cell infiltration between molecular subtypes were assessed using the Wilcoxon rank-sum test.
Construction and validation of the prognostic signature from cuproptosis-related regulators
First, univariate Cox regression analysis using the R package “survival” (20) was performed to identify the regulatory factors of cuproptosis that were significantly associated with the prognosis of HCC. Subsequently, we used the R package “glmnet” (21) for least absolute shrinkage and selection operator (LASSO) Cox regression to shrink the scope of the gene filtering. Finally, multivariate Cox regression analysis was performed to identify highly correlated genes and to construct a prognostic signature. The regression coefficient (β) from the multivariate Cox regression analysis and the risk score were both calculated using the following formula: (coefficient mRNA1 * expression of mRNA 1) + (coefficient mRNA2 * expression of mRNA 2) + ... + (coefficient mRNA n * expression of mRNA n). We divided all cohort patients into high-risk and low-risk groups according to the TCGA train of patients’ median risk scores. Survival analysis between the two groups was performed using Kaplan-Meier curves and the log-rank test. A receiver operating characteristic (ROC) curve was drawn using the R package “timeROC” (22) to test the predictive ability of the prognostic signature. The prognostic and clinical characteristics of the signature were further validated using TCGA-LIHC and ICGC cohorts.
Tumour specimens
Matched cancerous and normal hepatic tissue specimens were obtained from from 30 HCC patients treated at the Department of General Surgery, the Second Affiliated Hospital of Nanchang University from 2021 to 2022. The study was approved by the Ethics Committee of the Second Affiliated Hospital of Nanchang University (No. Review [2021] No. (084)). Informed consent was taken from all the patients.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA from the tissues was extracted using the TRIzol reagent (Invitrogen), and 1 μg of total RNA was used to perform reverse transcription to obtain cDNA using a cDNA synthesis kit (RR037A; Takara). Then qRT-PCR was performed with TB Green Dye (RR430A; Takara) in triplicate and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. Relative quantitation was calculated using the 2−ΔΔCt method. The primers used in this study are listed in Table S1.
Western blotting and IHC staining
Western blotting and immunohistochemical staining were performed as previously described (23). For western blotting, cells were lysed at 4 ℃ in lysis buffer. Total proteins were separated by SDS-PAGE gels and then transferred to 0.22-mm PVDF membranes (Bio-Rad). After blocking, the membranes were incubated with indicated antibodies. The expression was visualized using an ECL detection kit (#RPN2106, GE Healthcare Life Sciences), and semi-quantitative analysis was performed by measuring the density of the bands using ImageQuant LAS 4000 Mini biomolecular imager (GE Healthcare Life Sciences). For IHC staining, cells were fixed with 4% formalin (#163-20145, Wako) for 10 min at room temperature. After blocking, cells were incubated with rabbit anti-human CDKN2A antibody (1:200 dilution), mouse anti-human LIPT1 antibody (1:200 dilution) and mouse anti-human DLAT antibody (1:200 dilution) overnight at 4 ℃. After washing three times with PBS, cells were incubated with anti-rabbit Alexa Fluor 488 (1:10000 dilution) and anti-mouse Alexa Fluor 546 (1:10000 dilution) secondary antibodies at room temperature for 1h in the dark. The immunofluorescence in cells was examined on a laser confocal microscope (FV10i, Olympus).
Statistical analyses
All data were statistically analyzed using R software (version 4.1.3) and GraphPad (version 8.0). Univariate and multivariate Cox regression analyses identified independent predictors of overall survival (OS), their hazard ratio (HR) values, and their 95% confidence intervals (CI). In addition, differences in continuous variables between the two groups were analyzed using the Student’s t-test or Mann-Whitney U-test. Statistical significance was set at P<0.05.
Results
Overview of genetic and expression variation of cuproptosis-related regulators in HCC
A flowchart of the study is shown in Figure 1. In this study, ten regulators of cuproptosis were identified, including seven positive and three negative regulators. We first summarized the prevalence of copy number variations and somatic mutations in the cuproptosis regulators in HCC; mutations in the regulators were present in 16 out of 364 samples in the TCGA-LIHC dataset, with a frequency of 4.4%. Among them, CDKN2A was the most frequently mutated, while no mutations were found in FDX1, LIPT1, or DLAT in the HCC samples (Figure 2A). Investigation of the frequency of CNV alterations revealed a prevalence in the cuproptosis-related regulators; LIAS, GLS, and DLD presented more copy number amplifications, while MTF1, PDHB, FDX1, DLAT, and CDKN2A presented more copy number deletions, particularly of CDKN2A, at greater than 10% (Figure 2B). The locations of CNV alterations in cuproptosis-related regulators on the chromosomes are shown in Figure 2C. To define the expression landscape of these regulators in HCC patients, we compared their mRNA expression levels between normal and HCC samples and found that all but FDX1 were downregulated in the HCC samples (Figure 2D,2E). Notably, the expression of some of the cuproptosis-related regulators lost by CNV was significantly reduced in HCC tissues (e.g., FDX1) and vice versa (e.g., GLS and LIAS), suggesting that CNV alteration could be a key factor contributing to the perturbation of the expression of some of the regulators. The correlation network between these cuproptosis-related regulators is shown in Figure 2F. In addition, we extracted and compared paired expression data of cancer tissues from HCC patients and adjacent noncancerous tissues from the TCGA-LIHC dataset and found that all regulators, except FDX1, were significantly elevated in HCC tissues (Figure 2G, Table 1). IHC staining images from the HPA database further demonstrated the expression of these cuproptosis-related regulators at the protein level, and the results were largely consistent with those at the transcript level (Figure 2H). The above results show that the landscape of genetic and expression alterations of cuproptosis-related regulators is highly heterogeneous between normal liver and HCC tissues, indicating that the imbalanced expression of cuproptosis-related regulators results in different traits in patients with HCC.
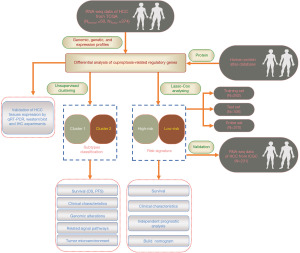
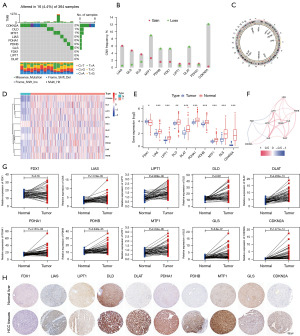
Table 1
| Gene symbol | Gene name | P value |
|---|---|---|
| FDX1 | Ferredoxin 1 | 0.0213 |
| LIAS | Lipoyl syndrome | 1.85E−11 |
| LIPT1 | Lipoyltransferase 1 | 1.39E−14 |
| DLD | Dihydrolipoamide dehydrogenase | 0.00245 |
| DLAT | Dihydrolipoamide s-acetyltransferase | 1.16E−10 |
| PDHA1 | Pyruvate dehydrogenase E1 subunit alpha 1 | 7.54E−11 |
| PDHB | Pyruvate dehydrogenase E1 subunit beta | 4.93E−09 |
| MTF1 | Metal regulatory transcription factor 1 | 2.95E−09 |
| GLS | Glutaminase | 3.87E−12 |
| CDKN2A | Cyclin dependent kinase inhibitor 2A | 1.87E−25 |
Identification of two subtypes of HCC based on the ten cuproptosis-related regulators
Unsupervised consensus clustering analysis of the TCGA-LIHC dataset was performed based on the expression profiles of ten cuproptosis-related regulators. The mean variation coefficient and cluster consensus were determined according to the number of categories (Figure 3A,3B). As presented in Figure 3C, the consensus matrix represented the consensus of k=2, in which the well-defined structure constituted by two blocks was shown. Therefore, we classified all HCC samples into two molecular subtypes based on the differential expression of the ten regulators: cluster 1 (n=284) and cluster 2 (n=86) (Figure 3D). We further analyzed whether there was a difference between the OS of patients with HCC in these two molecular subtypes using the Kaplan-Meier method. The results indicated that cluster 1 had better progression-free survival (PFS) than cluster 2 (Figure 3E). In addition, we investigated the association between disease OS and molecular subtype. Similarly, the results indicated that the cluster 2 subtype had worse OS than the cluster 1 subtype (Figure 3F). These results suggest that patients with cluster 1 HCC have a better prognosis. Next, we analyzed the association between the two molecular subtypes and the clinicopathological characteristics of HCC patients. As shown in Figure 3G, gene expression profiles and clinicopathological parameters including age (≤65 or >65 years), sex, tumor grade (G1–G4), tumor stage (I–IV), and tumor node metastasis (TNM) stage, were illustrated in a heatmap, in which tumor stage differed between the two molecular subtypes (Table 2). To further explore the differences between these two molecular subtypes, we performed mutation signature analysis of the somatic mutation profiles of HCC patients using TCGA-LIHC. Mutation spectrum characteristics showed that missense mutations were the most common type of mutation in both molecular subtypes, in agreement with the results of the mutation analysis in Figure 2. The horizontal histogram on the right shows the top 20 mutated genes in each subtype, with tumor protein P53 (TP53) (39% vs. 24%), LDL receptor related protein 1B (LRP1B) (13% vs. 6%), and FAT atypical cadherin 3 (FAT3) (12% vs. 5%) mutations being more common in cluster 1 (cluster 1 vs. cluster 2). In contrast, catenin beta 1 (CTNNB1) (15% vs. 28%), Xin actin binding repeat containing 2 (XIRP2) (2% vs. 8%), and AT-rich interaction domain 1A (ARID1A) (4% vs. 8%) mutations were more common in the cluster 2 subtype (Figure 3H). Titin (TTN), mucin 16 (MUC16), and albumin (ALB) mutations were the most common gene mutations in both subtypes. To further elucidate the potential signaling pathways or functions of the cuproptosis-related regulators in HCC, we applied GSEA-Kyoto Encyclopedia of Genes and Genomes (KEGG) to clusters 1 and 2, which showed that the metabolic pathways were enriched. Cluster 1 was enriched for linoleic acid and retinol metabolism, while cluster 2 was enriched for amino and nucleotide sugars, purine and sugar, and mannose metabolism, among others (Figure 3I).
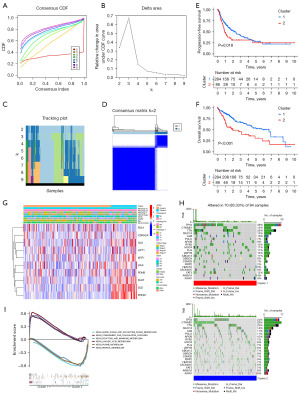
Table 2
| Characteristics | Cluster 1 (n=284) | Cluster 2 (n=86) | P value | Method |
|---|---|---|---|---|
| Albumin (g/dL) | 4.51 (5.49) | 96.42 (694.40) | 0.3263 | Wilcoxon test |
| Child-Pugh classification grade | 0.5271 | Fisher’s exact test | ||
| A | 176 (61.97) | 40 (46.51) | ||
| B | 19 (6.69) | 2 (2.33) | ||
| C | 1 (0.35) | 0 (0) | ||
| NA | 88 (30.99) | 44 (51.16) | ||
| Creatinine (mg/dL) | 2.60 (10.53) | 3.45 (16.13) | 0.704 | Wilcoxon test |
| Fetoprotein (ng/mL) | 16,832 (140,832) | 3,063 (12,057.65) | 0.154 | Wilcoxon test |
| Platelet (/mm3) | 22,525 (70,339.51) | 32,577 (104,691.4) | 0.4828 | Wilcoxon test |
| Prothrombin time (s) | 4.1 (5.06) | 3.8 (4.37) | 0.6465 | Wilcoxon test |
| Body mass index (kg/m2) | 26.54 (9.08) | 24.79 (5.79) | 0.0448 | Wilcoxon test |
| Height (cm) | 167.5 (11.08) | 167 (9.37) | 0.7148 | Wilcoxon test |
| Weight (kg) | 73.84 (19.74) | 69.67 (18.33) | 0.0811 | Wilcoxon test |
| Relative family cancer history | 0.8899 | χ2 test | ||
| Yes | 87 (30.63) | 25 (29.07) | ||
| No | 159 (55.99) | 48 (55.81) | ||
| NA | 38 (13.38) | 13 (15.12) | ||
| Race demographic | 0.7216 | Fisher’s exact test | ||
| American Indian or Alaska Native | 2 (0.70) | 0 (0) | ||
| Asian | 123 (43.31) | 34 (39.53) | ||
| Black or African American | 13 (4.58) | 4 (4.65) | ||
| White | 137 (48.24) | 47 (54.65) | ||
| NA | 9 (3.17) | 1 (1.16) | ||
| Sex | 0.6226 | χ2 test | ||
| Male | 193 (67.96) | 56 (65.12) | ||
| Female | 91 (32.04) | 30 (34.88) | ||
| Age (years) | 0.398 | χ2 test | ||
| ≤65 | 173 (60.92) | 48 (55.81) | ||
| >65 | 111 (39.08) | 38 (44.19) | ||
| Histological grade | 0.728 | Fisher’s exact test | ||
| G1 | 45 (15.85) | 10 (11.63) | ||
| G2 | 137 (48.24) | 40 (46.51) | ||
| G3 | 91 (32.04) | 30 (34.88) | ||
| G4 | 8 (2.82) | 4 (4.65) | ||
| NA | 3 (1.06) | 2 (2.33) | ||
| Clinical stage | 0.006 | Fisher’s exact test | ||
| I | 141 (49.65) | 30 (34.88) | ||
| II | 68 (23.94) | 17 (19.77) | ||
| III | 55 (19.37) | 30 (34.88) | ||
| IV | 5 (1.76) | 0 (0) | ||
| NA | 15 (5.28) | 9 (10.47) | ||
| Pathologic T | 0.078 | Fisher’s exact test | ||
| T1 | 147 (51.76) | 34 (39.53) | ||
| T2 | 72 (25.35) | 21 (24.42) | ||
| T3 | 54 (19.01) | 26 (30.23) | ||
| T4 | 8 (2.82) | 5 (5.81) | ||
| TX/NA | 3 (1.06) | 0 (0) | ||
| Pathologic N | 0.246 | Fisher’s exact test | ||
| N0 | 197 (69.37) | 55 (63.95) | ||
| N1 | 2 (0.70) | 2 (2.33) | ||
| NX/NA | 85 (29.93) | 29 (33.72) | ||
| Pathologic M | 0.542 | Fisher’s exact test | ||
| M0 | 206 (72.54) | 60 (69.77) | ||
| M1 | 4 (1.41) | 0 (0) | ||
| MX | 74 (26.06) | 26 (30.23) | ||
| Survival status | 0.0023 | χ2 test | ||
| Alive | 199 (70.07) | 45 (52.33) | ||
| Dead | 85 (29.93) | 41 (47.67) | ||
Data are presented as mean (SD) or n (%). TCGA-LIHC, The Cancer Genome Atlas Liver Hepatocellular Carcinoma; NA, not applicable.
Immunologic characteristics of the cuproptosis-related subtypes and prediction of immunotherapy responsiveness and targeted drug sensitivity
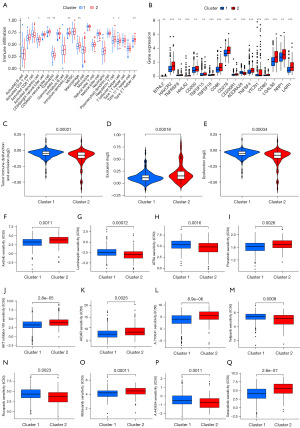
Previous studies have shown that the TME is crucial for immune function and has multiple clinical implications for immunotherapy (24,25). We screened and analyzed immune-related pathways using ssGSEA to understand the differences between the TME immune cell infiltration signatures in the two cuproptosis-related subtypes. The results showed increased infiltration of activated CD4+ T cells, activated dendritic cells (DCs), cd56bright natural killer (NK) cells, cd56dim NK cells, immature DCs, macrophages, regulatory T cells, T follicular helper cells, and type 2 T helper cells in the cluster 2 subtype, whereas eosinophils were more abundant in the cluster 1 subtype (Figure 4A). Heatmaps were used to demonstrate the correlation between the cuproptosis-related regulators in molecular subtypes and the immune cell infiltrates (Figure S1). Immune checkpoint blockade has become an effective treatment for many tumors; therefore, as these two molecular subtypes differed significantly in immune cell infiltration, we analyzed the expression differences of some known immune checkpoints between them. As shown in Figure 4B, most immunomodulatory targets [such as hepatitis A virus cellular receptor 2 (HAVCR2), CD200 molecule (CD200), CD86 molecule (CD86), CD276 molecule (CD276), TNF superfamily member 4 (TNFSF4), galectin 9 (LGALS9), neuropilin 1 (NRP1), and leukocyte associated immunoglobulin like receptor 1 (LAIR1)] were more highly expressed in cluster 2, suggesting that HCC patients in cluster 2 may respond better to immunotherapy than those in cluster 1. Tumor Immune Dysfunction and Exclusion (TIDE) is a computational framework for evaluating tumor immune escape potential based on the gene expression profiles of tumor samples that can provide predictive results for immunotherapy (26). We calculated the TIDE scores of the two molecular subtypes using the above algorithm, and the results showed that the TIDE scores of the cluster 1 subtype were significantly higher than those of cluster 2, indicating that cluster 2 was more likely to respond to immunotherapy, which was consistent with the immune checkpoint analysis (Figure 4C). In addition, we calculated the infiltrated cytotoxic T lymphocyte rejection and dysfunction scores in HCC tumor tissues. The T-cell rejection scores of the cluster 1 subtype were significantly lower than those of cluster 2, and the T-cell dysfunction scores of cluster 1 were significantly higher than those of cluster 2. These results were consistent with the TIDE score results, further suggesting that patients with the cluster 2 subtype might have better immunotherapy responses compared to those with the cluster 1 subtype (Figure 4D,4E). Finally, based on the Genomics of Drug Sensitivity in Cancer (GDSC) database and “pRRophetic” algorithm, we predicted the difference in chemosensitivity to chemotherapeutic drugs between the two molecular subtypes of HCC and found that there was a significant difference in the sensitivity to multiple chemotherapeutic drugs, namely axitinib (P=0.0011), luminespib (P<0.001), all-trans-retinoicacid (ATRA) (P=0.0016), ponatinib (P=0.0026), AKT.inhibitor.VIII (P<0.001), Acadesine (AICAR; P=0.0025), A.770041 (P<0.001), veliparib (P<0.001), rucaparib (P=0.0023), motesanib (P<0.001), pan-Akt inhibitor (A.443654; P=0.0011), and saracatinib (P<0.001) (Figure 4F-4Q). These results suggest that the signature derived from a cuproptosis-related regulators has the potential to predict the response to immunotherapy and sensitivity to multiple targeted drugs in patients with HCC.
Construction and verification of the cuproptosis-related gene prognosis signature
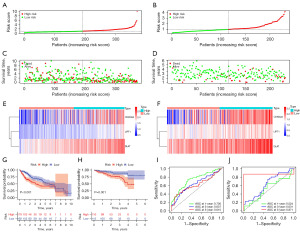
First, using the mRNA expression and clinical data of HCC patients in the TCGA-LIHC dataset, a univariate Cox regression analysis was performed on the previously mentioned ten cuproptosis-related regulators, and five genes were identified as independent factors of OS in HCC patients (Figure 5A). A cuproptosis-related gene prognosis signature was created based on the best value of λ from the LASSO Cox regulation analysis (Figure 5B). The signature comprised LIPT1, DLAT, and CDKN2A, and the risk score was calculated as follows: (0.7808 × LIPT1 EXP) + (0.5432 × DLAT EXP) + (0.2419 × CDKN2A EXP) (Table 3). After the risk scores of each patient in the TCGA-LIHC cohort were calculated, the median value was selected as the cutoff value and the patients in the training and testing cohorts were divided into two groups, the low-risk group and the high-risk group (Figure 5C,5D). The distribution of survival status in both cohorts revealed a higher number of deaths and shorter lives in the high-risk group (Figure 5E,5F). The heatmap indicated a significantly increased expression of the three risk genes among patients in the high-risk group (Figure 5G,5H). Principal components analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE) analyses were also used to test the accuracy of the prognostic signature, which showed two cohorts of patients in the low-risk and high-risk groups with two orientations (Figure S2A,S2B). Kaplan-Meier curves and time-dependent ROC curves were used to assess the overall predictive potential of the prognostic signature for OS of patients in both the training and testing cohorts. The Kaplan-Meier curve showed that the OS of patients with HCC in the high-risk group was significantly lower than that in the low-risk group. Poor prognosis of high-risk patients was also found in the test cohort (Figure 5I,5J). The ROC curves also indicated that the cuproptosis-related gene-independent prognostic signature had a better predictive value for the prognosis of patients with HCC in both the training and testing cohorts (Figure 5K,5L). The clinical characteristics of the TCGA training cohort (n=262) and test cohort (n=108) are shown in Table S2. To validate the accuracy of this prognostic signature, patients in the TCGA-LIHC entire cohort and International Cancer Genome Consortium Liver Cancer (ICGC-LIRI) cohort were divided into high- and low-risk groups using the above method. The high-risk group tended to have more deaths and a higher expression of risk genes (Figure 6A-6F). In both validation cohorts, the reliable clustering ability of the risk score was also validated by PCA and t-SNE analyses (Figure S2C,S2D). Kaplan-Meier analysis showed that patients in the low-risk group had better OS in both the validation cohorts (Figure 6G,6H). Furthermore, the ROC curves indicated that the signature was reliable and had an excellent predictive ability (Figure 6I,6J).

Table 3
| Gene | Multivariate Cox regression analysis | Coefficient | ||
|---|---|---|---|---|
| HR | 95% CI | P value | ||
| LIPT1 | 2.1392 | 1.1929–3.8362 | 0.0107 | 0.7808 |
| DLAT | 1.6064 | 1.1672–2.2109 | 0.0036 | 0.5432 |
| CDKN2A | 1.2760 | 1.0720–1.5189 | 0.0061 | 0.2419 |
HR, hazard ratio; CI, confidence interval.
Independent predictive value of the cuproptosis-related gene risk signature
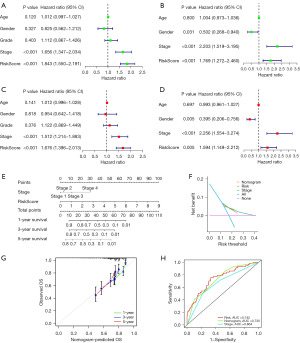
We evaluated whether the risk score of the cuproptosis-related gene signature could serve as an independent prognostic factor for HCC patients using univariate and multivariate Cox regression analyses. Univariate Cox regression analysis indicated that the risk score was an independent predictor of poor survival in patients with HCC in both the TCGA (HR =1.843, 95% CI: 1.550–2.191) and ICGC cohorts (HR =1.769, 95% CI: 1.272–2.460) (Figure 7A,7B, respectively). Multivariate Cox regression analysis showed that the risk score was an independent predictor of OS in HCC patients after inclusion and correction for other confounders (TCGA cohort: HR =1.676, 95% CI: 1.396–2.013, P<0.001; ICGC cohort: HR =1.594, 95% CI: 1.149–2.212, P=0.005) (Figure 7C,7D, respectively). In addition, we generated a heatmap of clinical features for the TCGA cohort based on the signature gene expression profiles and found that the stage and survival status of HCC patients were differentially distributed between low-risk and high-risk subtypes (Figure S3). To improve the clinical usefulness of the signature, we built a statistical nomogram signature in the TCGA cohort according to the risk score and clinical stage of HCC patients, and the decision curve analysis (DCA), calibration and ROC curve indicated that the nomogram signature could effectively predict the probability of survival of HCC patients (Figure 7E-7H).
Experimental validation of the expression levels of cuproptosis related genes in HCC tissues from prognostic signature
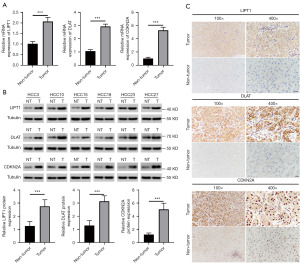
To further validate the expression levels of cuproptosis-related regulatory genes in HCC tissues from the prognostic signature. We first examined the expression of these genes in 30 HCC tissue samples and the corresponding adjacent tissues by qRT-PCR, western blotting, and IHC. As expected, the results of the qRT-PCR experiments were consistent with the differential analysis of the databases, and it was found that LIPT1, DLAT, and CDKN2A were all expressed at higher transcript levels in the HCC tissues than in the adjacent tissues (Figure 8A). Their protein expressions were detected by Western blot and IHC, and the data indicated that the protein expressions of LIPT1, DLAT and CDKN2A were all significantly increased in HCC tissues (Figure 8B,8C).
Discussion
The occurrence and development of HCC is a multiple stages and factors mediated and complex process characterized by high incidence, postoperative recurrence, and poor prognosis (27). Abnormal proliferative signal transduction, growth cycle disorders, genomic instability, epigenetic changes, and other complex interactions lead to the rapid proliferation and metastasis of HCC cells. Resistance to cell death is also a significant feature of HCC cells, which is an important reason for the failure of treatment regimens or the development of drug resistance (28,29). In the realm of individualized precision medicine, the selective targeted induction of cancer cell death may be an effective therapeutic strategy. Among the targetable cell death pathways are those of apoptosis, necroptosis, pyroptosis, and ferroptosis (30-33). In recent years, the importance of copper metabolism in tumor evolution has gradually become more prominent. Disturbances in copper metabolism can promote tumorigenesis by activating tumor proliferation-related signaling pathways, regulating tumor microangiogenesis, and remodeling the matrix and the inflammatory microenvironment (34). The latest research has revealed that copper ions can induce cell death even when other cell death modalities were inhibited, indicating that this novel copper-dependent mechanism of cell death is different from the known pathways mentioned above; thus, this process has been defined as cuproptosis (14). The targeted induction of cuproptosis in a variety of tumors has become a potentially effective method for tumor treatment with better prospects for clinical application. How the regulatory genes associated with cell cuproptosis interact in HCC and whether they are related to the survival time of patients remain unknown. In this study, we performed a comprehensive analysis of the expression, prognostic value, and immuno-microenvironment of ten genes that regulate cuproptosis in HCC.
First, the landscape of genetic variation in the HCC genome based on the TCGA database for these cuproptosis-related regulators was summarized. Further analysis of the TCGA-LIHC transcriptome and HPA database revealed that the proteins and mRNAs of these cuproptosis-related regulatory genes were differentially expressed between the normal and liver cancer tissues. Among them, CDKN2A showed more obvious changes in the epigenetic and expression levels of the genes, which is also consistent with the conclusions obtained in many previous studies (35,36). Next, we performed an unsupervised clustering analysis of differentially expressed regulatory genes to generate two molecular subtypes, revealing the differences between different subtypes in prognosis, clinicopathological characteristics, genomic mutations, expression profile of cuproptosis-related regulatory genes, TME, and enriched pathways in patients. Prognostic analysis revealed that cluster 1 survived longer than cluster 2, and the two molecular subtypes differed significantly in tumor stage. It is well known that the TP53 gene encodes the tumor protein p53, a tumor suppressor that inhibits cell division and proliferation; TP53 mutations may cause increased genomic instability (37). Interestingly, we found that HCC patients in cluster 2 exhibited more TP53 mutations, suggesting that a poor prognosis may be associated with a higher rate of TP53 mutations in this group; however, this requires more data and further experimental confirmation. Analysis of the possible signaling pathways involved in the two molecular subtypes using GSEA software revealed that both subtypes were enriched in many metabolism-related pathways, most of which were related to copper metabolism (38,39). Accumulating evidence suggests that immune cells in the TME play a vital role in tumorigenesis (40). These innate immune cells include macrophages, neutrophils, DCs, congenital lymphocytes, myelogenous inhibitory cells, and NK cells, which may have antitumor or tumor-promoting functions. Therefore, basic and clinical research on tumor immunotherapy has become a popular research focus where significant progress has been made. In particular, immune checkpoint blockade has been successfully used in the treatment of various tumors (41). Recent research has revealed a close relationship between copper metabolism and tumor immunity. We therefore performed ssGSEA to assess the abundance of immune cells in two subsets of cuproptosis-related regulatory genes, while predicting their potential to respond to immunotherapy based on the abundance of immune checkpoint expression in tumor tissues from patients with different subtypes and the TIDE database. As expected, there were significant differences in the degree of infiltration of multiple immune cells between the two subtypes, suggesting that cuproptosis probably plays an important role in the heterogeneity of the immune response between tumors. In contrast, much higher expression levels of immune checkpoints were observed in patients with HCC in cluster 2, implying that even though these patients have a poor prognosis, they may benefit more from some immunotherapies. We also studied the association of the two molecular subtypes with immunotherapy response based on the TIDE algorithm, which can effectively predict treatment response to immune checkpoint blockade (42). These results were consistent with the above immunotherapy-related analyses, and HCC patients in cluster 2 were more sensitive to immunotherapy. These results are similar to the conclusion based on transcriptomics and metabolomics that the metabolic and transcriptional profiles are different in patients who respond differently to immunotherapy with checkpoint inhibitors (43). Notably, the recent clinical trials have found that the combination of atezolizumab and bevacizumab can simultaneously target the two key pathogenic hallmarks of HCC immune escape and angiogenesis to achieve a higher objective tumor response rate and a longer OS (44-46). This combined treatment strategy has gradually become the first-line treatment therapy for advanced HCC (47). Therefore, we speculate that HCC patients in cluster 2 may also obtain better prognosis by combining tyrosine kinase inhibitors targeting angiogenesis such as bevacizumab on the basis of immunotherapy. In addition, the robustness of utilizing the GDSC database and the R package “pRRophetic” in predicting the response to chemotherapy has been demonstrated in different clinical trials (48). We calculated the half maximal inhibitory concentration (IC50) values for multiple targeted agents to assess the sensitivity of patients and found that both molecular subtypes had their respective sensitive targeted agents, which may indicate that cuproptosis-related regulatory genes play a potential role in it. Clinical trials have reported that Axitinib, ATRA and veliparib are effective or well tolerated in the treatment of patients with HCC (49-51). In addition, luminespib, ponatinib, Akt inhibitor VIII, AICAR and saracatinib have also been proved to inhibit the malignant phenotype of HCC cells in vitro or in vivo experiments (52-56). Collectively, these findings may provide more suitable personalized treatment options for HCC patients. However, the role of cuproptosis-related regulatory genes in these treatment allocations needs to be further clarified. We look forward to the addition of clinical trials and basic researches in follow-up studies to confirm our findings.
To further analyze the prognostic potential of genes involved in the regulation of survival-related cuproptosis, after screening by univariate Cox regression analysis and narrowing the scope by LASSO regression analysis, the risk scores of the three genes (LIPT1, DLAT, and CDKN2A) derived from their multivariate Cox regression coefficients formed a multigene signature, which was analyzed through the training and testing cohorts. Patients with HCC were divided into low- and high-risk groups, and the prognosis of patients in the low-risk group was better. Previous studies have shown that the pyroptosis related gene signature, autophagy related gene signature, ferroptosis related gene signature, N6-methyladenosine (m6A) related gene signature, and peroxisome proliferators-activated receptors (PPAR) related multigene signature predict 3-year OS of HCC with AUCs of 0.645, 0.650, 0.635, 0.667, and 0.685, respectively (57-61), which are similar to our study. In addition to the good predictive performance for HCC prognosis, the cuproptosis-related genes molecular classification and risk signature constructed in our study have more advantages. For example, it can distinguish immune checkpoint genes into high- and low expression group, and found that many of the genes mutated at high frequency in HCC differ in their mutation spectrum among different subtypes of patients, while predicting multiple targeted chemotherapy drug sensitivity differences. It was validated using the TCGA-LIHC entire cohort and ICGC-LIRI cohort. Univariate and multivariate Cox regression analyses were used to screen independent prognostic factors, and a nomogram was constructed for independent prognostic factors. The ROC curve showed that this signature exhibited good performance in predicting the prognostic outcomes of patients with HCC. In conclusion, our signature may be a useful clinical tool to assist doctors in effectively evaluating patient prognosis.
Finally, we detected the expression of ten cuproptosis-related regulatory genes in HCC tissues by qRT-PCR, western blotting, and IHC, and the results were consistent with the results of previous bioinformatic analyses of databases, indicating that the prognostic signature may be a promising tool for predicting the survival outcomes of HCC patients.
Our study had some limitations; the first was that our prognostic signature of cuproptosis-related genes was constructed and validated using retrospective data from public databases. In the future, we will use our prospective multicenter clinical data for further validation. Second, there are few studies on the regulation of cuproptosis-related regulatory genes in HCC; therefore, more experimental findings are needed in the future to refine the molecular classification and risk signature. Additionally, in vitro and in vivo experiments are needed to elucidate the specific mechanism of cuproptosis-related regulatory genes in the development of liver carcinogenesis and immunotherapy.
Conclusions
In conclusion, our results indicated that cuproptosis is strongly associated with HCC. We identified two cuproptosis-associated subtypes of HCC and further evaluated these subtypes for differences in immune networks and signaling pathways, providing additional insights into the relationship between cuproptosis and immunity in HCC. Furthermore, a prognostic signature of cuproptosis-related regulatory genes was constructed and proved to have significant predictive value. The promising prognostic accuracy of this signature may facilitate individualized prognosis management and therapeutic intervention.
Acknowledgments
We acknowledge TCGA database and ICGC database for providing their platforms and contributors for uploading their meaningful datasets.
Funding: This work was supported by funds from
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1876/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1876/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1876/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1876/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Second Affiliated Hospital of Nanchang University (No. Review [2021] No. (084)). Informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hepatocellular carcinoma. Nat Rev Dis Primers 2021;7:7. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019;156:477-491.e1. [Crossref] [PubMed]
- Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589-604. [Crossref] [PubMed]
- Anwanwan D, Singh SK, Singh S, et al. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer 2020;1873:188314. [Crossref] [PubMed]
- Xu LX, He MH, Dai ZH, et al. Genomic and transcriptional heterogeneity of multifocal hepatocellular carcinoma. Ann Oncol 2019;30:990-7. [Crossref] [PubMed]
- Cabral LKD, Tiribelli C, Sukowati CHC. Sorafenib Resistance in Hepatocellular Carcinoma: The Relevance of Genetic Heterogeneity. Cancers (Basel) 2020;12:1576. [Crossref] [PubMed]
- Ge EJ, Bush AI, Casini A, et al. Connecting copper and cancer: from transition metal signalling to metalloplasia. Nat Rev Cancer 2022;22:102-13. [Crossref] [PubMed]
- Shanbhag VC, Gudekar N, Jasmer K, et al. Copper metabolism as a unique vulnerability in cancer. Biochim Biophys Acta Mol Cell Res 2021;1868:118893. [Crossref] [PubMed]
- Kahlson MA, Dixon SJ. Copper-induced cell death. Science 2022;375:1231-2. [Crossref] [PubMed]
- Garza NM, Swaminathan AB, Maremanda KP, et al. Mitochondrial copper in human genetic disorders. Trends Endocrinol Metab 2023;34:21-33. [Crossref] [PubMed]
- Gao W, Huang Z, Duan J, et al. Elesclomol induces copper-dependent ferroptosis in colorectal cancer cells via degradation of ATP7A. Mol Oncol 2021;15:3527-44. [Crossref] [PubMed]
- Li Y. Copper homeostasis: Emerging target for cancer treatment. IUBMB Life 2020;72:1900-8. [Crossref] [PubMed]
- Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022;375:1254-61. [Crossref] [PubMed]
- Li Y, Du Y, Zhou Y, et al. Iron and copper: critical executioners of ferroptosis, cuproptosis and other forms of cell death. Cell Commun Signal 2023;21:327. [Crossref] [PubMed]
- Zheng P, Zhou C, Lu L, et al. Elesclomol: a copper ionophore targeting mitochondrial metabolism for cancer therapy. J Exp Clin Cancer Res 2022;41:271. [Crossref] [PubMed]
- Li Y, Yang J, Zhang Q, et al. Copper ionophore elesclomol selectively targets GNAQ/11-mutant uveal melanoma. Oncogene 2022;41:3539-53. [Crossref] [PubMed]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572-3. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Therneau T. survival: Survival analysis, including penalised likelihood. 2010.
- Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010;33:1-22. [Crossref] [PubMed]
- Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med 2013;32:5381-97. [Crossref] [PubMed]
- Yan J, Lei J, Chen L, et al. Human Leukocyte Antigen F Locus Adjacent Transcript 10 Overexpression Disturbs WISP1 Protein and mRNA Expression to Promote Hepatocellular Carcinoma Progression. Hepatology 2018;68:2268-84. [Crossref] [PubMed]
- Sadeghi Rad H, Monkman J, Warkiani ME, et al. Understanding the tumor microenvironment for effective immunotherapy. Med Res Rev 2021;41:1474-98. [Crossref] [PubMed]
- Bader JE, Voss K, Rathmell JC. Targeting Metabolism to Improve the Tumor Microenvironment for Cancer Immunotherapy. Mol Cell 2020;78:1019-33. [Crossref] [PubMed]
- Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018;24:1550-8. [Crossref] [PubMed]
- Finn RS, Zhu AX. Evolution of Systemic Therapy for Hepatocellular Carcinoma. Hepatology 2021;73:150-7. [Crossref] [PubMed]
- Llovet JM, Montal R, Sia D, et al. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol 2018;15:599-616. [Crossref] [PubMed]
- Ladd AD, Duarte S, Sahin I, et al. Mechanisms of drug resistance in HCC. Hepatology 2023; Epub ahead of print. [Crossref] [PubMed]
- Tan Y, Chen Q, Li X, et al. Pyroptosis: a new paradigm of cell death for fighting against cancer. J Exp Clin Cancer Res 2021;40:153. [Crossref] [PubMed]
- Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol 2020;17:395-417. [Crossref] [PubMed]
- Gong Y, Fan Z, Luo G, et al. The role of necroptosis in cancer biology and therapy. Mol Cancer 2019;18:100. [Crossref] [PubMed]
- Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting Ferroptosis to Iron Out Cancer. Cancer Cell 2019;35:830-49. [Crossref] [PubMed]
- Michniewicz F, Saletta F, Rouaen JRC, et al. Copper: An Intracellular Achilles' Heel Allowing the Targeting of Epigenetics, Kinase Pathways, and Cell Metabolism in Cancer Therapeutics. ChemMedChem 2021;16:2315-29. [Crossref] [PubMed]
- Zhou Y, Wang XB, Qiu XP, et al. CDKN2A promoter methylation and hepatocellular carcinoma risk: A meta-analysis. Clin Res Hepatol Gastroenterol 2018;42:529-41. [Crossref] [PubMed]
- Sun J, Li J, Guo Z, et al. Overexpression of Pyruvate Dehydrogenase E1α Subunit Inhibits Warburg Effect and Induces Cell Apoptosis Through Mitochondria-Mediated Pathway in Hepatocellular Carcinoma. Oncol Res 2019;27:407-14. [Crossref] [PubMed]
- Danovi S. TP53-dependent genomic instability. Nat Genet 2022;54:1584. [Crossref] [PubMed]
- Blades B, Ayton S, Hung YH, et al. Copper and lipid metabolism: A reciprocal relationship. Biochim Biophys Acta Gen Subj 2021;1865:129979. [Crossref] [PubMed]
- Zhong CC, Zhao T, Hogstrand C, et al. Copper (Cu) induced changes of lipid metabolism through oxidative stress-mediated autophagy and Nrf2/PPARγ pathways. J Nutr Biochem. 2022;100:108883. [Crossref] [PubMed]
- Lei X, Lei Y, Li JK, et al. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett 2020;470:126-33. [Crossref] [PubMed]
- Sharma P, Siddiqui BA, Anandhan S, et al. The Next Decade of Immune Checkpoint Therapy. Cancer Discov 2021;11:838-57. [Crossref] [PubMed]
- Wang YC, Wang X, Yu J, et al. Targeting monoamine oxidase A-regulated tumor-associated macrophage polarization for cancer immunotherapy. Nat Commun 2021;12:3530. [Crossref] [PubMed]
- Triozzi PL, Stirling ER, Song Q, et al. Circulating Immune Bioenergetic, Metabolic, and Genetic Signatures Predict Melanoma Patients' Response to Anti-PD-1 Immune Checkpoint Blockade. Clin Cancer Res 2022;28:1192-202. [Crossref] [PubMed]
- Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N Engl J Med 2020;382:1894-905. [Crossref] [PubMed]
- Galle PR, Finn RS, Qin S, et al. Patient-reported outcomes with atezolizumab plus bevacizumab versus sorafenib in patients with unresectable hepatocellular carcinoma (IMbrave150): an open-label, randomised, phase 3 trial. Lancet Oncol 2021;22:991-1001. [Crossref] [PubMed]
- Cheng AL, Qin S, Ikeda M, et al. Updated efficacy and safety data from IMbrave150: Atezolizumab plus bevacizumab vs. sorafenib for unresectable hepatocellular carcinoma. J Hepatol 2022;76:862-73. [Crossref] [PubMed]
- Stefanini B, Ielasi L, Chen R, et al. TKIs in combination with immunotherapy for hepatocellular carcinoma. Expert Rev Anticancer Ther 2023;23:279-91. [Crossref] [PubMed]
- Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41:D955-61. [Crossref] [PubMed]
- Zhang B, Zhang X, Zhou T, et al. Clinical observation of liver cancer patients treated with axitinib and cabozantinib after failed sorafenib treatment: a case report and literature review. Cancer Biol Ther 2015;16:215-8. [Crossref] [PubMed]
- Sun J, Mao F, Liu C, et al. Combined FOLFOX4 with all-trans retinoic acid versus FOLFOX4 with placebo in treatment of advanced hepatocellular carcinoma with extrahepatic metastasis: a randomized, double-blind comparative study. Signal Transduct Target Ther 2023;8:368. [Crossref] [PubMed]
- Gabrielson A, Tesfaye AA, Marshall JL, et al. Phase II study of temozolomide and veliparib combination therapy for sorafenib-refractory advanced hepatocellular carcinoma. Cancer Chemother Pharmacol 2015;76:1073-9. [Crossref] [PubMed]
- Buontempo F, Ersahin T, Missiroli S, et al. Inhibition of Akt signaling in hepatoma cells induces apoptotic cell death independent of Akt activation status. Invest New Drugs 2011;29:1303-13. [Crossref] [PubMed]
- Liao X, Song G, Xu Z, et al. Oxaliplatin resistance is enhanced by saracatinib via upregulation Wnt-ABCG1 signaling in hepatocellular carcinoma. BMC Cancer 2020;20:31. [Crossref] [PubMed]
- Gao J, Xiong R, Xiong D, et al. The Adenosine Monophosphate (AMP) Analog, 5-Aminoimidazole-4-Carboxamide Ribonucleotide (AICAR) Inhibits Hepatosteatosis and Liver Tumorigenesis in a High-Fat Diet Murine Model Treated with Diethylnitrosamine (DEN). Med Sci Monit 2018;24:8533-43. [Crossref] [PubMed]
- Liu C, Mu X, Wang X, et al. Ponatinib Inhibits Proliferation and Induces Apoptosis of Liver Cancer Cells, but Its Efficacy Is Compromised by Its Activation on PDK1/Akt/mTOR Signaling. Molecules 2019;24:1363. [Crossref] [PubMed]
- Augello G, Emma MR, Cusimano A, et al. Targeting HSP90 with the small molecule inhibitor AUY922 (luminespib) as a treatment strategy against hepatocellular carcinoma. Int J Cancer 2019;144:2613-24. [Crossref] [PubMed]
- Fu XW, Song CQ. Identification and Validation of Pyroptosis-Related Gene Signature to Predict Prognosis and Reveal Immune Infiltration in Hepatocellular Carcinoma. Front Cell Dev Biol 2021;9:748039. [Crossref] [PubMed]
- Ye W, Shi Z, Zhou Y, et al. Autophagy-Related Signatures as Prognostic Indicators for Hepatocellular Carcinoma. Front Oncol 2022;12:654449. [Crossref] [PubMed]
- Luo L, Yao X, Xiang J, et al. Identification of ferroptosis-related genes for overall survival prediction in hepatocellular carcinoma. Sci Rep 2022;12:10007. [Crossref] [PubMed]
- Kong W, Li X, Xu H, et al. Development and validation of a m(6)A-related gene signature for predicting the prognosis of hepatocellular carcinoma. Biomark Med 2020;14:1217-28. [Crossref] [PubMed]
- Xu W, Chen Z, Liu G, et al. Identification of a Potential PPAR-Related Multigene Signature Predicting Prognosis of Patients with Hepatocellular Carcinoma. PPAR Res 2021;2021:6642939. [Crossref] [PubMed]


