
Modeling of new markers for the diagnosis and prognosis of pancreatic cancer based on the transition from inflammation to cancer
Highlight box
Key findings
• Acute pancreatitis (AP) and chronic pancreatitis (CP) co-related genes are significantly correlated with pancreatic adenocarcinoma (PAAD). TINAG, DDC, SPDEF, and APOBEC1 could serve as predictors of PAAD. The risk model developed in this study could be used to predict the prognosis of PAAD patients.
What is known, and what is new?
• Genes involved in AP/CP might be important for PAAD development.
• This report identified the genes involved in AP/CP/PAAD transformation.
What is the implication, and what should change now?
• AP could transform into PAAD in certain patients.
Introduction
The incidence of pancreatic cancer continues to increase both in China and abroad (1-3). Pancreatic adenocarcinoma (PAAD) is asymptomatic in its early stage, and the prognosis of PAAD patients is poor. Currently, there is a lack of specific diagnostic biomarkers for PAAD (4,5). Thus, advances need to be made in PAAD diagnosis or prognosis to improve PAAD patient survival.
It has been continuously demonstrated that long-term chronic pancreatitis (CP) is an important risk factor of PAAD. Patients with CP have a high risk of developing PAAD (6,7). Acinar cells, which secret digestive enzymes, are thought to be very important in the inflammation of pancreatitis, causing ductal metaplasia, a precursor to pancreatic carcinogenesis. Inflammatory molecules also promote tumor growth through epithelial and mesenchymal secretion. These findings provide evidence that anti-inflammation agents could be a preventive and/or therapeutic agent for PAAD (8-11). Conversely, CP can develop from acute pancreatitis (AP). The evolution of AP-CP-PAAD is a prominent stage in PAAD onset and development, and the co-expression of AP-CP genes may serve as potential new diagnostic and prognostic markers for early PAAD (12).
Bioinformatics is an emerging discipline that mainly includes biology, mathematics, and computer science, and is being rapidly developed by the Human Genome Project (13,14). The vast amount of biomedical data has helped to elucidate the relative biological knowledge and convoy biological information more comprehensively and effectively. With the continuous accumulation of biological data and the launch of strategic planning for precision medicine, bioinformatics has become increasingly important and crucial to the current development of related fields (15).
In this article, based on the AP-CP-PAAD transformation and experimental validation studies using cancer-related databases, we investigated and validated new predictors for early PAAD diagnosis and prognosis. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1365/rc).
Methods
Data resource
Expression profile microarrays (GSE109227 and GSE41418) were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds/) for AP and CP mice, respectively, which are a genome-wide messenger RNA (mRNA) expression data (16). The GSE109227 data set contained six samples of AP tissue and five samples of normal pancreatic tissue. The GSE41418 data set contained six tissue samples of CP tissue and six samples of normal pancreatic tissue. Transcriptome sequencing (RNA-sequencing) data and matched clinical information data were obtained from The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov) for four normal controls and 150 PAAD cases with fragments per kilobase per million mapped fragments (FPKM)-style data. Non-pancreatic ductal adenocarcinoma patients (27 cases) (17) and patients with no survival information (one case) were excluded from the survival analysis (18). A total of 182 cancer patient samples with complete survival prognosis data were screened from the International Cancer Genome Consortium (ICGC) database; all the available data from the database were used to maximize the power and generalizability of the results. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Differential expression analysis and Venn analysis of shared common differentially expressed genes (co-DEGs)
An online differential analysis was performed of the GEO database data using GEO2R with the threshold set at P<0.05 to identify the significantly differently expression genes in AP and CP. The shared co-DEGs in the GSE109227 and GSE41418 data sets were also analyzed by the online database VENNY 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/). The list of genes to be analyzed was uploaded to the database to display the Venn diagram and the list of related shared genes (19).
Analysis of variance and one-way Cox regression analysis
Data on the survival time and survival status of PAAD patients were extracted; patients with a survival time of 0 days (one case) were removed. The survival time and status data were combined with the DEG data to form a matrix, and a differential analysis was first performed of the co-DEGs of the TCGA-PAAD data set. The threshold was set to P<0.05. Using the “survival” package of R software, a one-way Cox regression analysis was performed, and a P value <0.05 indicated that the DEG was related to the prognosis of PAAD patients. After the Venn analysis, the target genes were screened for association with both PAAD occurrence and prognosis. The “pheatmap” package was used to map a heat map of the target genes. A target gene forest map was then drawn using the survival package (20,21).
Prognostic risk-score model construction
Least absolute shrinkage and selection operator (LASSO) Cox regression analysis
The TCGA-PAAD cohort was analyzed by LASSO regression using L1 regularization statistics. Based on the results of the one-way Cox regression analysis, the prognosis-related genes with P values <0.05 were further analyzed using the LASSO regression algorithm of the “glmnet” R package, and the prognosis-related genes were further screened by a subsequent analysis based on the parameter Lambda values (22).
Model construction
Based on the regression coefficients obtained from the gene expression and multifactorial Cox regression analysis, a linear risk assessment model related to survival was constructed, and the coefficient value of each gene was obtained using the expression values of the genes in the model in each sample, the survival time, and the status data analysis results. The following formula was used to calculate the risk score for each pancreatic cancer patient in TCGA and the ICGC data sets, respectively: risk score = β1 × mRNA1EXP + β2 × mRNA2EXP + … + βn × mRNAnEXP. The risk model was constructed using 1000-times testing. A principal component analysis with t-distribution and randomized nearest neighbor embedding (t-SNE) was performed using the “Rtsne” R package to analyze the differentiation between the high-risk and low-risk groups of TCGA and the ICGC data sets, respectively, and the results were visualized by the “ggplot2” package of R software. Next, to assess whether the model could be used as a prognostic factor independent of other clinical features, the “survival” package of R software was used to perform univariate and multi-variate independent prognostic analyses, the results were visualized by drawing forest plots, and the “limma” R package was used for the risk difference analysis (23,24).
Clinical correlation analysis
The prognostic risk characteristics were analyzed by the “ggpubr” package of R software with patient age, gender, clinical grade or pathological stage, and the R package was used to plot each clinical characteristic. The box plots of clinical characteristics and patient risk scores were created using the R package.
Experiment-related materials and methods
Primary antibodies
Anti-tubulointerstitial nephritis antigen (TINAG) antibody was obtained from Proteintech (Wuhan, China). Anti-dopa decarboxylase (DDC), kelch like family member 36 (KLHL36), apolipoprotein B messenger RNA editing enzyme catalytic subunit 1 (APOBEC1), SAM pointed domain containing ETS transcription factor (SPDEF) antibodies were from Sangon Biotech (Shanghai) Co., Ltd.
Small-interfering RNA sequences
These sequences were synthesized by GenePharma (Shanghai, China) (Table 1).
Table 1
| Gene name | Company | Sequence (5'-3') |
|---|---|---|
| TINAG-Homo-115 | GenePharma | Positive: GGACCGGAUAUAAGAUCUUTT |
| Trans: AAGAUCUUAUAUCCGGUCCTT | ||
| TINAG-Homo-540 | GenePharma | Positive: GGACAGCAAUGGAAAUGUUTT |
| Trans: AACAUUUCCAUUGCUGUCCTT | ||
| TINAG-Homo-795 | GenePharma | Positive: GGAUGGACUCAUGGCCCAUTT |
| Trans: AUGGGCCAUGAGUCCAUCCTT | ||
| DDC-Homo-311 | GenePharma | Positive: GGACAUCAUCAACGACGUUTT |
| Trans: AACGUCGUUGAUGAUGUCCTT | ||
| DDC-Homo-720 | GenePharma | Positive: GCACACUCCUCAGUGGAAATT |
| Trans: UUUCCACUGAGGAGUGUGCTT | ||
| DDC-Homo-895 | GenePharma | Positive: GCUCCUUUGACAAUCUCUUTT |
| Trans: AAGAGAUUGUCAAAGGAGCTT | ||
| KLHL36-Homo-191 | GenePharma | Positive: GCCAUACAAGAUCAGCGAATT |
| Trans: UUCGCUGAUCUUGUAUGGCTT | ||
| KLHL36-Homo-358 | GenePharma | Positive: GCGACUACUUCAACUCCAUTT |
| Trans: AUGGAGUUGAAGUAGUCGCTT | ||
| KLHL36-Homo-1235 | GenePharma | Positive: GGAUGCGGCCUCCAAUCUUTT |
| Trans: AAGAUUGGAGGCCGCAUCCTT | ||
| APOBEC1-Homo-99 | GenePharma | Positive: GGAGUUUGACGUCUUCUAUTT |
| Trans: AUAGAAGACGUCAAACUCCTT | ||
| APOBEC1-Homo-142 | GenePharma | Positive: GCCUGUCUGCUCUACGAAATT |
| Trans: UUUCGUAGAGCAGACAGGCTT | ||
| APOBEC1-Homo-318 | GenePharma | Positive: GGAAUGCUCCCAGGCUAUUTT |
| Trans: AAUAGCCUGGGAGCAUUCCTT | ||
| SPDEF-Homo-839 | GenePharma | Positive: GCCUGCAAGCUGCUCAACATT |
| Trans: UGUUGAGCAGCUUGCAGGCTT | ||
| SPDEF-Homo-1227 | GenePharma | Positive: GCCGCUUCAUUAGGUGGCUTT |
| Trans: AGCCACCUAAUGAAGCGGCTT | ||
| TMEM94-Homo-1337 | GenePharma | Positive: GCUGUCUCCUCUCAGGAAATT |
| Trans: UUUCCUGAGAGGAGACAGCTT | ||
| TMEM94-Homo-1506 | GenePharma | Positive: GCCCAGAGACUGUACUGUUTT |
| Trans: AACAGUACAGUCUCUGGGCTT | ||
| TMEM94-Homo-1980 | GenePharma | Positive: GCCUCAAUGUGCUGCUGAATT |
| Trans: UUCAGCAGCACAUUGAGGCTT |
Immunohistochemistry staining
The PAAD tissue microarrays were purchased from the Shanghai Core Ultra Biological Co., Ltd. (item no. HPanA020PG01), and the number of valid cancer tissue cases for the TINAG, DDC, SPDEF, APOBEC1, and KLHL36 detection chips was 20. The tissue samples were incubated with the anti-APOBEC1 (Shanghai Biotechnology Co., Ltd.) and anti-KLHL36 (Shanghai Biotechnology Co., Ltd.) polyclonal antibodies. Finally, the protein expression levels were scored under the microscope by multiple uninformed pathologists. The intensity and area of staining were categorized into the following four grades: 0 (negative), 1 (weakly positive), 2 (moderately positive), and 3 (strongly positive).
Cell proliferation assays
The cells under the indicated treatments were inoculated in 96-well plates (5,000 cells/well). The cells were plastered, and incubated for 12, 24, 48, and 72 hours. The old medium was discarded and replaced with 100 µL of new medium, or Cell Counting Kit-8 (CCK-8) reagent (10 µL/well) was added and mixed well. The medium was then transferred to the cell incubator for 2 hours. The absorbance of the cells was measured at 450 nm with an enzyme marker.
Transwell migration assays
Transwell migration chambers were used. The cells were cultured in the upper chamber with complete medium in the lower chamber. The chambers were transferred to the incubator and removed after 24 hours. The cells were fixed and then underwent crystal violet staining. Cell migration in each well was observed under an inverted microscope, counted, and then statistically analyzed.
Pathway analysis of TINAG, DDC, SPDEF, and APOBEC1
The TINAG, DDC, SPDEF, and APOBEC1 genes were screened by TCGA-PAAD-RNA-sequencing genomic screening through the online website LinkedOmics database (https://www.linkedomics.org/login.php) to obtain the relevant co-expression gene clusters. The relevant co-expression gene clusters were then subjected to a Gene Set Enrichment Analysis-Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis via the online WebGestalt database (https://www.webgestalt.org/), and the pathway atlas was obtained.
Statistical analysis
All the bioinformatics analyses were performed in R software (version 3.6.1). The remaining statistical analyses were performed using SPSS software (version 25.0) and GraphPad Prism software (version 9.0). The following P values were considered statistically significant: *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001.
Results
Co-expression analysis of DEGs in AP and CP
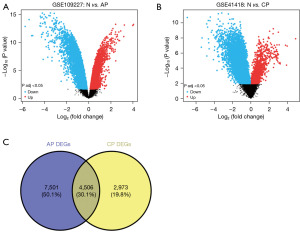
The GSE109227 data set, which contained six mouse AP tissue samples and five matched normal control tissue samples, was selected first. The DEGs were screened using GEO2R and are presented in a volcano plot (Figure 1A). A total of 12,007 AP-DEGs were identified in the AP data set. Similarly, the GSE41418 data set, which contained six mouse CP tissue samples and six matched normal control tissue samples, was next selected to screen the DEGs in CP (Figure 1B). A total of 7,479 CP-DEGs were identified in the CP data set. Next, to identify the DEGs common to AP and CP, we analyzed the intersecting genes between the AP-DEGs and CP-DEGs (i.e., the co-DEGs) through the online database VENNY 2.1. In total, 4,506 co-DEGs were identified (Figure 1C). Additionally, 3,453 co-DEGs were identified in TCGA data set.
Differential analysis of the co-DEGs in TCGA-PAAD
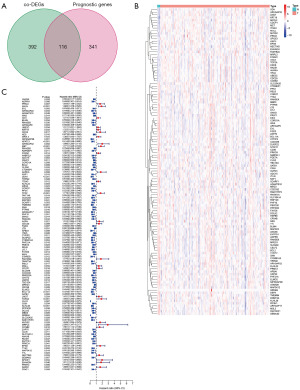
Among the 3,453 co-DEGs, 508 co-DEGs were found to be differentially expressed in PAAD, while 457 co-DEGs were correlated with PAAD prognosis. Specifically, trefoil factor 2 (TFF2), TINAG, trefoil factor 1 (TFF1), aquaporin 5 (AQP5), SPDEF, anterior gradient protein 2 (AGR2), APOBEC1, kallikrein-related peptidase 6 (KLK6), DDC, mucin 13 (MUC13), claudin 18 (CLDN18), annexin A10 (ANXA10), and tetraspanin 1 (TSPAN1) presented with the largest fold expression changes in PAAD (i.e., a log fold change >3) (Table 2). The two groups of genes were then subjected to Venn analysis. We found that 116 genes (which we refer to as target gene clusters) were closely associated with both PAAD development and prognosis (Figure 2A). The expression of the target gene clusters in PAAD is shown in a heat map (Figure 2B). A forest plot of these genes in PAAD is also presented. In total, 28 up-regulated genes and 88 down-regulated genes were found (Figure 2C).
Table 2
| Gene | Control | PAAD | logFC | P value |
|---|---|---|---|---|
| TFF2 | 6.83664 | 267.8866 | 5.292191 | 0.002974 |
| TINAG | 0.033313 | 1.003763 | 4.913173 | 0.005472 |
| TFF1 | 40.79442 | 802.0375 | 4.297226 | 0.007221 |
| AQP5 | 2.230661 | 41.41324 | 4.214549 | 0.002468 |
| SPDEF | 1.084305 | 18.34634 | 4.08065 | 0.001386 |
| AGR2 | 13.91381 | 211.2185 | 3.924146 | 0.001386 |
| APOBEC1 | 0.426995 | 6.257341 | 3.873257 | 0.008128 |
| KLK6 | 2.854156 | 40.6557 | 3.832321 | 0.009135 |
| DDC | 0.24065 | 2.951894 | 3.616632 | 0.007471 |
| MUC13 | 5.292654 | 60.24678 | 3.508821 | 0.002562 |
| CLDN18 | 11.69363 | 126.2321 | 3.432284 | 0.021434 |
| ANXA10 | 7.037711 | 67.75781 | 3.267209 | 0.016284 |
| TSPAN1 | 17.2421 | 142.9409 | 3.051412 | 0.002204 |
PAAD, pancreatic adenocarcinoma; co-DEGs, common differentially expressed genes; FC, fold change.
Risk-score model construction
PAAD patient data downloaded from TCGA or the ICGC databases were used as training and validation sets. A LASSO regression analysis was conducted on the above 116 target gene groups, and the following 19 key prognosis-related genes were identified: enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase (EHHADH), MET Proto-Oncogene, Receptor Tyrosine Kinase (MET), DNA methyltransferase 3 alpha (DNMT3A), Rho GTPase activating protein 17 (ARHGAP17), tubulin tyrosine ligase (TTL), EGFR pathway substrate 8, signaling adaptor (EPS8), BCL11 transcription factor A (BCL11A), KLHL36, differentially expressed in FDCP 8 homolog (DEF8), RELB proto-oncogene, NF-kB subunit (RELB), crystallin alpha B (CRYAB), transmembrane protein 94 (TMEM94), cell division cycle 20 (CDC20), solute carrier family 16 member 14 (SLC16A14), cyclin B2 (CCNB2), nectin cell adhesion molecule 3 (NECTIN3), anillin, actin binding protein (ANLN), amyloid beta precursor protein binding family A member 1 (APBA1), and DNA topoisomerase II alpha (TOP2A) (Figure 3A,3B and Table 3). The following risk-score equation based on the expression of the 19 key genes was formulated: 0.08945 × EHHADH + 0.24165 × MET + (−0.12536) × DNMT3A + (−0.54232) × ARHGAP17 + (−0.1063) × TTL + 0.244083 × EPS8 + (−0.03683) × BCL11A + (−0.46656) × KLHL36 + (−0.00994) × DEF8 + (−0.29732) × RELB + (−0.0521) × CRYAB + (−0.4635) × TMEM94 + 0.118142 × CDC20 + (−0.03301) × SLC16A14 + 0.170268 × CCNB2 + 0.001 × NECTIN3 + 0.075642 × ANLN + (−0.62613) × APBA1 + 0.096801 × TOP2A.
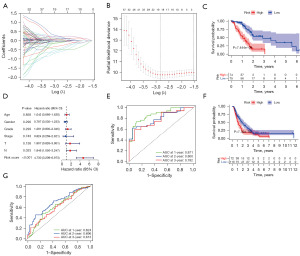
Table 3
| Gene symbol | Description | Coefficient |
|---|---|---|
| EHHADH | Enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase | 0.08945 |
| MET | MET Proto-Oncogene, Receptor Tyrosine Kinase | 0.24165 |
| DNMT3A | DNA methyltransferase 3 alpha | −0.12536 |
| ARHGAP17 | Rho GTPase activating protein 17 | −0.54232 |
| TTL | Tubulin tyrosine ligase | −0.1063 |
| EPS8 | EGFR pathway substrate 8, signaling adaptor | 0.244083 |
| BCL11A | BCL11 transcription factor A | −0.03683 |
| KLHL36 | Kelch like family member 36 | −0.46656 |
| DEF8 | Differentially expressed in FDCP 8 homolog | −0.00994 |
| RELB | RELB proto-oncogene, NF-kB subunit | −0.29732 |
| CRYAB | Crystallin alpha B | −0.0521 |
| TMEM94 | Transmembrane protein 94 | −0.4635 |
| CDC20 | Cell division cycle 20 | 0.118142 |
| SLC16A14 | Solute carrier family 16 member 14 | −0.03301 |
| CCNB2 | Cyclin B2 | 0.170268 |
| NECTIN3 | Nectin cell adhesion molecule 3 | 0.001 |
| ANLN | Anillin, actin binding protein | 0.075642 |
| APBA1 | Amyloid beta precursor protein binding family A member 1 | −0.62613 |
| TOP2A | DNA topoisomerase II alpha | 0.096801 |
Risk scores were calculated for each PAAD patient in both TCGA and the ICGC data sets, and the patients were divided into high- and low-risk groups. The study found a significant difference in overall survival between the high- and low-risk groups for TCGA-PAAD (Figure 3A-3C). The high-risk group had a worse prognosis and shorter survival time than the low-risk group. The univariate and multifactor independent prognostic analyses showed that the N stage and risk-score value were independent risk factors for PAAD prognostic (Figure 3D). A receiver operating characteristic (ROC) curves analysis was conducted to further investigate the diagnostic efficiency of the risk-score model in TCGA data set. The areas under the curve (AUCs) for 1-, 2- and 3-year survival were 0.871, 0.8, and 0.782, respectively, indicating that the model was good at predicting PAAD patient survival (Figure 3E).
The risk score was validated in the ICGC-PAAD data set. Consistent with TCGA results, the high-risk group had a worse prognosis and shorter survival time than the low-risk group (P<0.05, Figure 3F). A ROC curve analysis was conducted to further investigate the diagnostic efficiency of the risk-score model in the ICGC data set. The AUCs for 1-, 2- and 3-year survival were 0.624, 0.696, and 0.613, respectively (Figure 3G). Taken together, the results indicate that the risk score can be used to predict PAAD patient survival.
Immunohistochemical validation
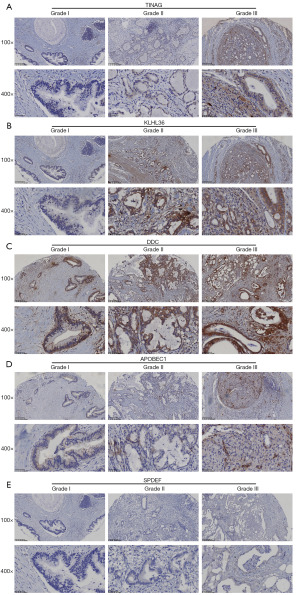
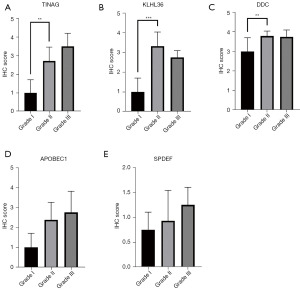
The expression of the genes related to PAAD occurrence, including TINAG, DDC, SPDEF, and APOBEC1, were tested in clinic samples. The TINAG, DDC, SPDEF, and APOBEC1 proteins increased as the tumor pathological grade increased. However, only TINAG and DDC were significantly differentially expressed in grades I and II (P<0.05) (Figures 4,5). In addition, 19 key genes in the prognosis risk model were also tested in the clinic samples. As observed in the risk model, KLHL36 was negatively associated with a poor prognosis; however, our results showed that KLHL36 protein expression was significantly more increased in grade II than grade I patients (P<0.05) (Figures 4,5).
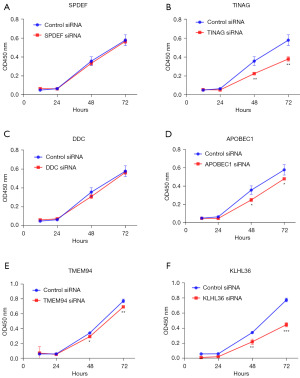
Effects of SPDEF, TINAG, DDC, APOBEC1, TMEM94, and KLHL36 on PAAD cell growth
SPDEF, TINAG, DDC, APOBEC1, TMEM94, and KLHL36 were knocked down, and the proliferation of PAAD cells was investigated by CCK-8 cell assays (Figure 6). The results showed that the proliferation of PAAD cells was significantly inhibited following the knock down of TINAG, APOBEC1, TMEM94, and KLHL36 (at 48 and 72 hours) (Figure 6B,6D-6F). However, the knock down of SPDEF and DDC did not affect the proliferation of PAAD cells (Figure 6A,6C). These results suggest that TINAG, APOBEC1, TMEM94, and KLHL36 inhibit the proliferation of PAAD cells.
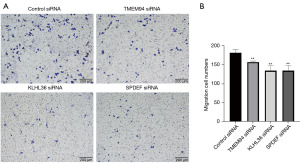
Effects of SPDEF, TINAG, DDC, APOBEC1, TMEM94, and KLHL36 on PAAD cell migration
To verify the effect of the above genes on PAAD cell migration, we performed transwell assays. The results showed that the down-regulation of SPDEF, TMEM94, and KLHL36 significantly inhibited PAAD cell migration (Figure 7). However, the down-regulation of TINAG, DDC, and APOBEC1 did not affect the migratory ability of PAAD cells.
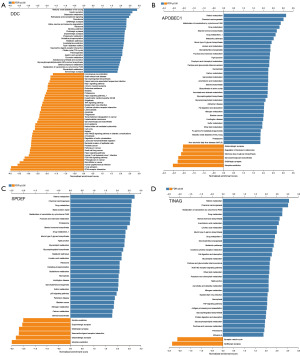
Pathway analysis for TINAG, DDC, SPDEF, and APOBEC1
Based on the above results, TINAG, DDC, SPDEF, and APOBEC1 appear to be very important in PAAD occurrence. Thus, we analyzed the pathways in which these genes were involved. The results showed that in pancreatic cancer, the TINAG, DDC, SPDEF, and APOBEC1 genes may be involved in the following pathways: neuroactive ligand-receptor interactions, cytokine-receptor interactions, natural killer cell-mediated cytotoxicity, cancer pathways, cancer proteoglycans, the extracellular matrix (ECM)-receptor interaction, the tumor necrosis factor (TNF) signaling pathway, the repressor activator protein 1 (RAP1) signaling pathway, the Hippo signaling pathway, focal adhesion, tight junction, mannose type O-glycan biosynthesis, etc. (Figure 8). Meanwhile, we also found that some pathways were related to AP/CP, including apoptosis, the protein 53 (p53) signaling pathway, the phosphatidylinositol 3-kinase (PI3K)-Akt signaling pathway, the cell cycle, the cytokine-cytokine receptor interaction, glutathione metabolism, the regulation of actin cytoskeleton, leukocyte transendothelial migration, the metabolism of xenobiotics by cytochrome P450, necroptosis, linoleic acid metabolism, glycerophospholipid metabolism peroxisome, porphyrin and chlorophyll metabolism, and proteasome (Figure 8).
Discussion
In this study, we used data sets from mice with AP or CP from the GEO database. AP and CP could be clearly determined in mouse that were not so easy in the clinic (25). However, the use of mouse models also had some disadvantages. Notably, there are differences in gene expression between humans and mice in both protein-coding and non-coding genes (26).
In this study, we found that TFF2, TINAG, TFF1, AQP5, SPDEF, AGR2, APOBEC1, KLK6, DDC, MUC13, CLDN18, ANXA10, and TSPAN1 may be potential early diagnostic markers for PAAD. Among them, TFF2, TFF1, AGR2, AQP5, KLK6, MUC13, CLDN18, ANXA10, and TSPAN1 have been found to be expressed in PAAD (27-34). These genes have been shown to affect the occurrence, development, and metastasis of PAAD through multiple pathways and regulatory networks (28). Consistent with previous research (27-34), we found that TFF2, TFF1, AGR2, AQP5, KLK6, MUC13, CLDN18, ANXA10, and TSPAN1 mRNAs were highly expressed in PAAD. Thus, a significant portion of our screened target genes were found to be associated with PAAD, which provides supporting evidence of the feasibility of our analytical approach. Notably, the AP-CP-PAAD mechanism plays a key role in the development of PAAD.
The associations between TINAG, DDC, SPDEF, and APOBEC1 and PAAD had not previously been examined. The bioinformatics analysis also found that the fold changes of the genes in PAAD were larger than those in AP or CP. Together, the immunohistochemistry and cellular assay results suggest that TINAG is the potential new early diagnostic marker for PAAD.
According to the prognostic risk-score model consisting of 19 genes, MET and ANLN were positively associated with a poor prognosis in PAAD, while DNMT3A, KLHL36, DEF8, and TMEM94 were negatively associated with a poor prognosis in PAAD. The hepatocyte growth factor (HGF)/MET signaling pathway promotes the motility, histomorphogenesis and mesenchymal-epithelial transition of epithelial cells in the body (35), and also plays a role in the development and progression of many mesenchymal-derived tumors (36). DNMT3A promotes proliferation by activating the signal transducer and activator of transcription 3 (STAT3) signaling pathway and inhibiting apoptosis in PAAD (37). Bioinformatics analysis results have identified DEF8 and MET in a risk scoring system for predicting the prognosis of patients with resectable PAAD (38). The experiments were not validated; however, the reliability of this model was confirmed by previous finding. ANLN was included in another risk-score model for overall survival prediction (39). All these finding support the conclusion reached by this study.
A study screening of AAD diagnostic markers from CP-related genes identified CCNB2, cell division cycle 6 (CDC6), cyclin-dependent kinase 1 (CDK1), cell division cycle 28 (CDC28), and cyclin-dependent kinase regulatory subunit 2 (CKS2) as key genes involved in the development of CP and PAAD (40). Another study conducted a microarray (Affymetrix) analysis and identified four DEGs in PAAD and CP; that is, 14-3-3sigma, S100 calcium binding protein P (S100P), S100 calcium binding protein A6 (S100A6), and integrin beta4 (ITGB4) (41). Unlike previous studies, we innovatively searched for new key genes associated with PAAD initiation, progression and metastasis from the AP-CP-PAAD perspective, thus potentially identifying key genes that play a role in the early stage of pancreatic cancer.
In the process of our research, we found a relationship between the genes related to AP, CP, and PAAD. In this study, we found that TINAG, DDC, SPDEF, and APOBEC1 showed large fold changes in tumors and can serve as new diagnostic markers for PAAD. A risk-score risk model can be used as a new tool for PAAD prognosis prediction. However, more trials and clinical studies need to be conducted.
Conclusions
TINAG, DDC, SPDEF, and APOBEC1 may serve as new early diagnostic markers for PAAD. The models constructed, which include EHHADH, MET, DNMT3A, ARHGAP17, TTL, EPS8, BCL11A, KLHL36, DEF8, RELB, CRYAB, TMEM94, CDC20, SLC16A14, CCNB2, NECTIN3, ANLN, APBA1, and TOP2A, may serve as potential diagnostic criteria for PAAD and indicators for the prognostic assessment of PAAD.
Acknowledgments
Funding: The present study was supported in part by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1365/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1365/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1365/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1365/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jia X, Du P, Wu K, et al. Pancreatic Cancer Mortality in China: Characteristics and Prediction. Pancreas 2018;47:233-7. [Crossref] [PubMed]
- Miller KD, Nogueira L, Devasia T, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin 2022;72:409-36. [Crossref] [PubMed]
- Kolbeinsson HM, Chandana S, Wright GP, et al. Pancreatic Cancer: A Review of Current Treatment and Novel Therapies. J Invest Surg 2023;36:2129884. [Crossref] [PubMed]
- National Health Commission Of The People's Republic Of China. National guidelines for diagnosis and treatment of pancreatic cancer 2022 in China (English version). Chin J Cancer Res 2022;34:238-55. [Crossref] [PubMed]
- Stoffel EM, Brand RE, Goggins M. Pancreatic Cancer: Changing Epidemiology and New Approaches to Risk Assessment, Early Detection, and Prevention. Gastroenterology 2023;164:752-65. [Crossref] [PubMed]
- Gandhi S, de la Fuente J, Murad MH, et al. Chronic Pancreatitis Is a Risk Factor for Pancreatic Cancer, and Incidence Increases With Duration of Disease: A Systematic Review and Meta-analysis. Clin Transl Gastroenterol 2022;13:e00463. [Crossref] [PubMed]
- Munigala S, Subramaniam DS, Subramaniam DP, et al. Incidence and Risk of Pancreatic Cancer in Patients with a New Diagnosis of Chronic Pancreatitis. Dig Dis Sci 2022;67:708-15. [Crossref] [PubMed]
- Pinho AV, Chantrill L, Rooman I. Chronic pancreatitis: a path to pancreatic cancer. Cancer Lett 2014;345:203-9. [Crossref] [PubMed]
- Hausmann S, Kong B, Michalski C, et al. The role of inflammation in pancreatic cancer. Adv Exp Med Biol 2014;816:129-51. [Crossref] [PubMed]
- Padoan A, Plebani M, Basso D. Inflammation and Pancreatic Cancer: Focus on Metabolism, Cytokines, and Immunity. Int J Mol Sci 2019;20:676. [Crossref] [PubMed]
- Wang L, Xie D, Wei D. Pancreatic Acinar-to-Ductal Metaplasia and Pancreatic Cancer. Methods Mol Biol 2019;1882:299-308. [Crossref] [PubMed]
- Keihanian T, Barkin JA, Souto EO. Early Detection of Pancreatic Cancer: Risk Factors and the Current State of Screening Modalities. Gastroenterol Hepatol (N Y) 2021;17:254-62. [PubMed]
- Collins FS, Patrinos A, Jordan E, et al. New goals for the U.S. Human Genome Project: 1998-2003. Science 1998;282:682-9. [Crossref] [PubMed]
- Martin-Sanchez F, Iakovidis I, Nørager S, et al. Synergy between medical informatics and bioinformatics: facilitating genomic medicine for future health care. J Biomed Inform 2004;37:30-42. [Crossref] [PubMed]
- Branco I, Choupina A. Bioinformatics: new tools and applications in life science and personalized medicine. Appl Microbiol Biotechnol 2021;105:937-51. [Crossref] [PubMed]
- Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol 2006;411:352-69. [Crossref] [PubMed]
- Nicolle R, Raffenne J, Paradis V, et al. Prognostic Biomarkers in Pancreatic Cancer: Avoiding Errata When Using the TCGA Dataset. Cancers (Basel) 2019;11:126. [Crossref] [PubMed]
- Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- Sun L, Dong S, Ge Y, et al. DiVenn: An Interactive and Integrated Web-Based Visualization Tool for Comparing Gene Lists. Front Genet 2019;10:421. [Crossref] [PubMed]
- Benítez-Parejo N, Rodríguez del Águila MM, Pérez-Vicente S. Survival analysis and Cox regression. Allergol Immunopathol (Madr) 2011;39:362-73. [Crossref] [PubMed]
- Koletsi D, Pandis N. Survival analysis, part 3: Cox regression. Am J Orthod Dentofacial Orthop 2017;152:722-3. [Crossref] [PubMed]
- D'Angelo GM, Rao D, Gu CC. Combining least absolute shrinkage and selection operator (LASSO) and principal-components analysis for detection of gene-gene interactions in genome-wide association studies. BMC Proc 2009;3:S62. [Crossref] [PubMed]
- Alizadeh AA, Gentles AJ, Alencar AJ, et al. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood 2011;118:1350-8. [Crossref] [PubMed]
- Emura T, Chen YH. Gene selection for survival data under dependent censoring: A copula-based approach. Stat Methods Med Res 2016;25:2840-57. [Crossref] [PubMed]
- Houdebine LM. Transgenic animal models in biomedical research. Methods Mol Biol 2007;360:163-202. [PubMed]
- Lin S, Lin Y, Nery JR, et al. Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A 2014;111:17224-9. [Crossref] [PubMed]
- Jahan R, Ganguly K, Smith LM, et al. Trefoil factor(s) and CA19.9: A promising panel for early detection of pancreatic cancer. EBioMedicine 2019;42:375-85. [Crossref] [PubMed]
- Hong X, Li ZX, Hou J, et al. Effects of ER-resident and secreted AGR2 on cell proliferation, migration, invasion, and survival in PANC-1 pancreatic cancer cells. BMC Cancer 2021;21:33. [Crossref] [PubMed]
- Silva PM, da Silva IV, Sarmento MJ, et al. Aquaporin-3 and Aquaporin-5 Facilitate Migration and Cell-Cell Adhesion in Pancreatic Cancer by Modulating Cell Biomechanical Properties. Cells 2022;11:1308. [Crossref] [PubMed]
- Zhang L, Lovell S, De Vita E, et al. A KLK6 Activity-Based Probe Reveals a Role for KLK6 Activity in Pancreatic Cancer Cell Invasion. J Am Chem Soc 2022;144:22493-504. [Crossref] [PubMed]
- Khan S, Zafar N, Khan SS, et al. Clinical significance of MUC13 in pancreatic ductal adenocarcinoma. HPB (Oxford) 2018;20:563-72. [Crossref] [PubMed]
- Wang X, Zhang CS, Dong XY, et al. Claudin 18.2 is a potential therapeutic target for zolbetuximab in pancreatic ductal adenocarcinoma. World J Gastrointest Oncol 2022;14:1252-64. [Crossref] [PubMed]
- Ishikawa A, Sakamoto N, Honma R, et al. Annexin A10 is involved in the induction of pancreatic duodenal homeobox 1 in gastric cancer tissue, cells and organoids. Oncol Rep 2020;43:581-90. [PubMed]
- Liu S, Cai Y, Changyong E, et al. Screening and Validation of Independent Predictors of Poor Survival in Pancreatic Cancer. Pathol Oncol Res 2021;27:1609868. [Crossref] [PubMed]
- Gherardi E, Birchmeier W, Birchmeier C, et al. Targeting MET in cancer: rationale and progress. Nat Rev Cancer 2012;12:89-103. [Crossref] [PubMed]
- Huang C, Liu H, Gong XL, et al. Expression of DNA methyltransferases and target microRNAs in human tissue samples related to sporadic colorectal cancer. Oncol Rep 2016;36:2705-14. [Crossref] [PubMed]
- Jing W, Song N, Liu YP, et al. DNMT3a promotes proliferation by activating the STAT3 signaling pathway and depressing apoptosis in pancreatic cancer. Cancer Manag Res 2019;11:6379-96. [Crossref] [PubMed]
- Wu C, Wu Z, Tian B. Five gene signatures were identified in the prediction of overall survival in resectable pancreatic cancer. BMC Surg 2020;20:207. [Crossref] [PubMed]
- Li Z, Hu C, Yang Z, et al. Bioinformatic Analysis of Prognostic and Immune-Related Genes in Pancreatic Cancer. Comput Math Methods Med 2021;2021:5549298. [Crossref] [PubMed]
- Li H, Hao C, Yang Q, et al. Identification of hub genes in chronic pancreatitis and analysis of association with pancreatic cancer via bioinformatic analysis. Gen Physiol Biophys 2022;41:15-30. [Crossref] [PubMed]
- Logsdon CD, Simeone DM, Binkley C, et al. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res 2003;63:2649-57. [PubMed]


