
The rs9909659G/A polymorphisms of the STAT3 gene provide prognostic information in acute myeloid leukemia
Introduction
Acute myelogenous leukemia (AML) is a heterogeneous clone disorder of hematopoietic stem cells, with an increased incidence in adults; the median age at presentation is about 70 years old (1). Cytarabine combined with anthracycline has long been considered the standard induction regimen, with a complete remission (CR) rate of about 70% (2). However, the clinical efficacy and prognosis vary in AML patients. Recent studies have demonstrated that genetic factors are a major prognostic factor of AML (3-5).
Single nucleotide polymorphisms (SNPs), which can be interpreted as DNA sequence polymorphisms attributed to a single nucleotide mutation, are the most frequently occurring human genetic variations (6). SNPs with rich sites, representativeness, and genetic stability contribute to phenotypic diversity, drug tolerance, different susceptibility to diseases, and differences in response to environmental factors. As a result, SNPs are popularly used as molecular markers to facilitate the identification of genetic factors responsible for diseases, early diagnosis, prediction of occurrence, treatment outcome, prognosis, and potential targeted molecular therapy (7-9). Some genes have been reported to be associated with the chemotherapy response in AML patients (10-12). However, the ideal candidate gene predicting the chemotherapy response remains unknown.
The Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway mediates signaling by cytokines, which control survival, proliferation, and differentiation of several cell types. STATs are activated by phosphorylation of a tyrosine residue located in the C-terminus transactivation domain. Following JAK-mediated phosphorylation, STAT proteins dimerize and translocate to the nucleus, where they regulate gene expression (13). The more prominent factor in this signaling pathway is signal transducer and activator of transcription 3 (STAT3) for cancer and leukemia, and the activation pathway of STAT3 is shown in Figure 1. The interleukin-6 family of cytokines function as ligands bound to corresponding cytokine receptors are the most well-known traditional activators of STAT3. Bromberg’s group confirmed that the mutants of STAT3, A661, and N663 are constitutively active and transform cells (14). Therefore, constitutive STAT3 activation promotes cellular transformation and plays an important role in human tumors (15). The STAT3 transcription factor is constitutively activated in human AML cell lines and may contribute to the autonomous proliferation of AML blasts (16). Steensma’s group also considered that STAT signaling pathways are activated in most AML cases (17). The role of JAK/STAT mutations in cancer has only recently begun to be explored, and mutations in components of this pathway are possibly present in a wide variety of cancers.
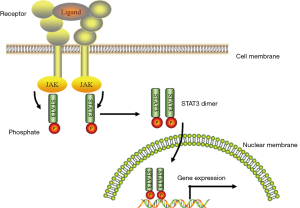
In this study, we aimed to confirm the STAT3 genotype distributions of AML patients in a Chinese population and further evaluate the association between STAT3 polymorphisms and treatment outcomes of AML.
Methods
Patients
A total of 152 patients with newly diagnosed AML according to FAB criteria at the Drum Tower Hospital in Jiangsu Province between May 2011 and September 2015 were enrolled in the study. This study was approved by the ethics committee of the hospital and performed in accordance with the Declaration of Helsinki. All patients provided written informed consent before enrollment. AML patients who were diagnosed with any other cancer types or other hematological malignancies were eliminated from the study. The M3 subtype (acute promyelocytic leukemia) and patients who received allogeneic hematopoietic stem cell transplantation (HSCT) or autologous HSCT were excluded. Cytogenetic risk groups were divided according to the Medical Research Council cytogenetic classification system (18). The presence of 5/del(Jeny5q), 7/del(Jeny7q), abn3q, complex aberrations (≥3 independent aberrations), t(Jeny9;22), and t(Jeny6;9) were identified as unfavorable karyotypes, whereas t(Jeny8;21) and inv (18) were classified as favorable karyotypes. The remaining patients, that is, those with normal karyotype and all other karyotypic aberrations, comprised the intermediate risk group. Gene mutation risk groups were divided according to the NCCN gene mutation classification system (19). NPM1 and CEBPA were identified as favorable gene mutations, whereas FLT3/ITD and C-KIT were classified as unfavorable gene mutations. The patients were then divided into two groups according to age >45 vs. ≤45; white blood cell (WBC) counts ≥50×109/L vs. <50×109/L; and level of serum lactate dehydrogenase (LDH) ≥1,000 vs. <1,000 IU/L.
Chemotherapy regimens and therapeutic effect evaluation
All patients were treated with standard induction chemotherapy (45 mg/m2/day i.v. daunorubicin for 1–3 days and 100 mg/m2/day i.v. cytarabine for 1–7 days). Consolidation therapies were performed based on two more cycles of high-dose cytarabine (2–3 g/m2/day i.v. for 3 consecutive h for 1–3 days). CR was defined according to the criteria of the International Working Group (19). Partial remission (PR) indicated that at least one of the standards on clinical manifestation, blood analysis, and bone marrow was not met; less than 20% of blast cells were observed in the bone marrow. Non-remission (NR) indicated that all standards of CR on clinical manifestation, blood analysis, and bone marrow were not met; more than 20% of blast cells were observed in the bone marrow. Early death was defined as death within 8 weeks from the start of the first induction therapy course. Leukemia-free survival (LFS) was defined as the interval from the date of confirmed CR to the date of confirmed relapse from any causes, whereas event-free survival (EFS) was defined as the interval from the date of treatment to the date of a confirmed relapse, persistence of the disease, or death from any causes. Overall survival (OS) was defined as the length of time from the date of diagnosis to the date of death from any causes or last follow-up.
DNA extraction and polymorphism genotyping
We selected two SNPs within the STAT3 gene that have been reported to be associated with cancer development and outcome (20-22). These SNPs coding polymorphisms for amino acid substitutions likely affect the resulting protein structure and function. They occur at a relatively high frequency, thereby affecting a relatively large portion of the general population.
Genomic DNA was isolated from peripheral blood leukocytes by the phenol-chloroform extraction method. SNP genotyping was performed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry according to the manufacturer’s instructions. The STAT3 genotypes were inspected, and the results were tested for departures from the Hardy-Weinberg equilibrium separately for cases by SNP analyzer software.
Statistical analysis
Demographic and clinical variables of different genotypes were evaluated using the Pearson Chi-squared test or Fisher’s exact test, as appropriate. The Kaplan-Meier product-limit method was used to determine OS. The differences were assessed by the log-rank test. The prognostic impact of different variables on survival (LFS and EFS) was determined by multivariate Cox proportional hazards model. All data were statistically analyzed using a commercially available statistical software package (SPSS 19.0; IBM Corp., Armonk, NY, USA). All tests were bilateral, and P<0.05 was considered statistically significant. Odds ratios with 95% confidence intervals (CIs) were also calculated.
Results
Clinical characteristics and distribution of genotypes
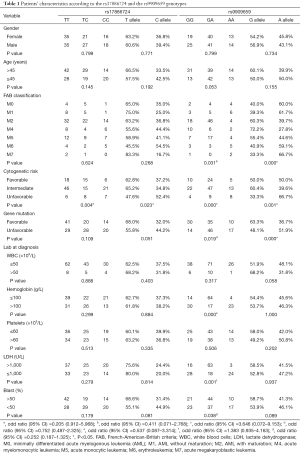
As shown in Table 1, patient characteristics, such as gender, age, FAB classification, leukocytosis at the time of presentation, WBC and platelet counts, and levels of hemoglobin and LDH, were not significantly different among the groups with different STAT3 genotypes in rs17886724. The frequency of the TC/CC genotype of rs17886724 was higher in the unfavorable group than that of the T/T genotypes (P=0.004). Meanwhile, the C allele in rs17886724 was more frequently classified into the unfavorable group (P=0.023). Age and gender classifications were similar in different genotypes of rs9909659. Moreover, patients with M7, low hemoglobin, unfavorable cytogenetic factor, unfavorable gene mutation factor, and high LDH were associated with the GA/AA genotype in rs9909659 (P=0.031, P=0.000, P=0.000, P=0.019, and P=0.001, respectively).
Full table
The STAT3 genotype distributions are displayed in detail in Table 1. The frequencies of the TT, TC, and CC genotypes in rs17886724 were 46.05%, 31.58%, and 28.37%, respectively, whereas those of the GG, GA, and AA genotypes in rs9909659 were 28.95%, 53.29%, and 17.76%, respectively. Linkage disequilibrium (LD) analysis showed no LD between rs17886724 and rs9909659 in the STAT3 SNPs.
Treatment outcomes according to STAT3 genotypes
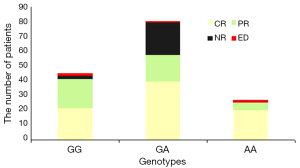
All 152 patients received standard remission induction chemotherapy. Responses of patients to treatment were evaluated after every chemotherapy cycle. Responses were subdivided into four groups, namely, CR, PR, NR, and early death. Differences in the chemotherapy response based on the STAT3 genotypes are shown in Table 2. The polymorphic genotypes of rs17886724T/C were not significantly different between AML patients who had good response and those who had poor response to standard chemotherapy (P>0.05), but the TT genotype in rs17886724 showed a higher CR rate after the first chemotherapy cycle (P=0.037). As reflected in the statistical results, different responses to chemotherapy in AML patients were associated to STAT3 rs9909659 genotypes. Figure 2 shows that the rate of AML patients with the rs9909659 G/G genotype achieving good response was higher than those with the G/A and A/A genotypes (P=0.038).

Full table
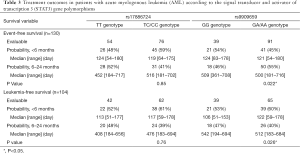
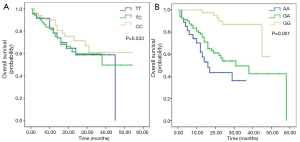
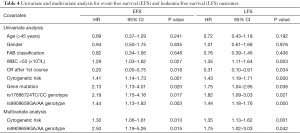
The median follow-up time was 14 months (range, 1–24 months). As shown in Table 3, the <6 months rates of EFS for patients who had a median EFS duration of 124 and 119 days with the TT and TC/CC genotypes in rs17886724 were 48% and 57%, respectively, whereas the <6 months rates of EFS for patients who had a median EFS duration of 124 and 121 days with the GG and GA/AA genotypes in rs9909659 were 53% and 43%, respectively. The <6 and 6–24 months rates of LFS for patients who had a median LFS duration of 113 and 117 days with the TT and TC/CC genotypes in rs17886724 were 56% and 61%, respectively, whereas the <6 and 6–12 months rates of LFS for patients who had a median LFS duration of 106 and 122 days with the GG and GA/AA genotypes in rs9909659 were 53% and 60%, respectively. However, the genotype frequencies of rs17886724T/C and rs9909659G/A were not significantly different between patients with EFS <6 months and EFS ≥6 months, as well as between LFS <6 months and LFS ≥6 months. The Kaplan-Meier method and log-rank test showed that patients with the GA/AA genotype in rs9909659G/A had significantly worse OS than those with the GG genotype (P=0.001) (Figure 3A). For other SNPs, patients with the TC/CC genotype of rs17886724T/C did not have significantly worse OS than those with the TT genotype (P=0.553) (Figure 3B). The survival analyses by Cox regression model are shown in Table 4. On univariate analysis, WBC >50×109/L (P=0.027, P=0.003), CR after 1st Course (P=0.018, P=0.034), cytogenetic risk (P=0.001, P=0.000), gene mutation (P=0.020, P=0.036), rs17886724TC/CC genotype (P=0.017, P=0.021), and rs9909659GA/AA genotype (P=0.003, P=0.000) were significantly associated with EFS and PFS. However, on multivariate Cox regression analysis, only cytogenetic risk (P=0.013, P=0.001, respectively) and rs9909659GA/AA genotype (P=0.015, P=0.042, respectively) remained as an independent significant predictor for EFS and PFS.
Full table
Full table
Discussion
In this retrospective study, we explored the associations between STAT3 gene polymorphisms and treatment outcome in patients with AML in a Chinese population. Our analysis highlights the relevance of the gene variant in Ara-C-based chemotherapy response. We found that the genotypes of STAT3 rs17886724 and rs9909659 were associated with the clinical characteristics of patient with AML. The AML patients with the rs9909659GA/AA genotype were not sensitive to standard chemotherapy.
Constitutive activation of STAT proteins, especially STAT3 and STAT5, has been detected in a variety of human cancers (23,24). Furthermore, various oncoproteins have been shown to directly cause STAT activation (25,26). These proteins include the leukemogenic fusion oncoprotein tyrosine kinases, such as breakpoint cluster region (Bcr)-Abelson (Abl), TEL-JAK2, and TEL-platelet-derived growth factor-beta receptor. Thus, aberrant STAT activation may place a central role in cancer development. A testable hypothesis is that heterochromatin formation constitutes a tumor suppression mechanism, and heterochromatin destabilization is a crucial change in the global chromatin structure that promotes oncogene-induced tumorigenesis. As one of the most extensively studied STAT members, STAT3 signaling has proven to be important in the pathogenesis of classical Hodgkin’s lymphoma (27). Several groups confirmed that STAT3 protein has a primary oncogene of nasal-type natural killer cell lymphoma, and STAT3 is a malignancy factor in cutaneous T-cell lymphoma (28,29). The occurrence of STAT3 activation in AML is not plausible (30). Recent evidence revealed that STAT3 plays a vital role in mediating drug resistance to targeted cancer therapies and chemotherapies (31). STAT3 gene polymorphisms are important in diagnosing various diseases and evaluating responses of patients to therapies. Lei et al. (32) suggested that rs1905339 (A > G) in the STAT3 region is associated with an increased breast cancer risk (P=0.000, CI: 1.03–1.08). Permuth-Wey et al. reported that STAT3 genetic polymorphisms are associated with unfavorable response to platinum-based therapy in patients with advanced serous epithelial ovarian cancer (33). SNPs of STAT3 were identified to be independent predictors for OS of lung cancer patients in a Han Chinese population (34). However, only few studies have investigated the association of STAT3 genetic polymorphisms with chemotherapy response in AML patients.
According to the present research, no association was found between STAT3 genotypes and responses to chemotherapy in AML patients with rs17886724, as well as clinical characteristics among different genotypes in rs17886724. Simultaneously, patients with hemoglobin levels less than 100 g/L exhibited LDH of more than 1,000. Unfavorable cytogenetic factors were associated with the GA/AA genotype in rs9909659, which may be associated with poor response to chemotherapy and poor OS. These potential factors in the rs9909659GA/AA genotype could be related to the poor prognosis of AML. No significant associations were found among EFS duration, LFS duration, and different genotypes in rs9909659, which may be due to the limited observation time and samples.
In conclusion, AML patients with the GA/AA genotype in rs9909659 were not sensitive to standard chemotherapy, which could also be a predictor for OS in AML patients. The observation duration and number of patients should be increased in future work. Further studies should be conducted to overcome the limitations of this study, such as the relatively small number of patients, small number of target genotypes, and follow-up time. Replication studies would help determine the potential mechanism of STAT3 SNPs, as well as develop novel therapeutic target strategies to improve the chemotherapy response and prognosis.
Acknowledgments
We would like to thank all the volunteers who took part in this study.
Funding: This work was supported by the National Natural Science Foundation of China (81400162, 81570174), the Natural Science Foundation of Jiangsu Province (BK20140100), and the Technique Development Foundation of Nan Jing (Outstanding Youth Foundation, JQX15004).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.07.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the ethics committee of the hospital and performed in accordance with the Declaration of Helsinki (as revised in 2013). All patients provided written informed consent before enrollment.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Estey E, Döhner H. Acute myeloid leukaemia. Lancet 2006;368:1894-907. [Crossref] [PubMed]
- Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet 2013;381:484-95. [Crossref] [PubMed]
- Singh H, Werner L, Deangelo D, et al. Clinical outcome of patients with acute promyelocytic leukemia and FLT3 mutations. Am J Hematol 2010;85:956-7. [Crossref] [PubMed]
- Flach J, Dicker F, Schnittger S, et al. An accumulation of cytogenetic and molecular genetic events characterizes the progression from MDS to secondary AML: an analysis of 38 paired samples analyzed by cytogenetics, molecular mutation analysis and SNP microarray profiling. Leukemia 2011;25:713-8. [Crossref] [PubMed]
- Varn FS, Andrews EH, Cheng C. Systematic analysis of hematopoietic gene expression profiles for prognostic prediction in acute myeloid leukemia. Sci Rep 2015;5:16987. [Crossref] [PubMed]
- Pang GS, Wang J, Wang Z, et al. Predicting potentially functional SNPs in drug-response genes. Pharmacogenomics 2009;10:639-53. [Crossref] [PubMed]
- Diabetes Genetics Initiative of Broad Institute of Harvard and MIT. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331-6. [Crossref] [PubMed]
- Fang HM, Tian G, Zhou LJ, et al. FGFR4 genetic polymorphisms determine the chemotherapy response of Chinese patients with non-small cell lung cancer. Acta Pharmacol Sin 2013;34:549-54. [Crossref] [PubMed]
- Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet 2015;47:856-60. [Crossref] [PubMed]
- Zhe N, Wang J, Chen S, et al. Heme oxygenase-1 plays a crucial role in chemoresistance in acute myeloid leukemia. Hematology 2015;20:384-91. [Crossref] [PubMed]
- van den Heuvel-Eibrink MM, van der Holt B, Burnett AK, et al. CD34-related coexpression of MDR1 and BCRP indicates a clinically resistant phenotype in patients with acute myeloid leukemia (AML) of older age. Ann Hematol 2007;86:329-37. [Crossref] [PubMed]
- Damiani D, Tiribelli M, Franzoni A, et al. BAALC overexpression retains its negative prognostic role across all cytogenetic risk groups in acute myeloid leukemia patients. Am J Hematol 2013;88:848-52. [Crossref] [PubMed]
- Darnell JE Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994;264:1415-21. [Crossref] [PubMed]
- Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell 1999;98:295-303. [Crossref] [PubMed]
- Haura EB, Turkson J, Jove R. Mechanisms of disease: Insights into the emerging role of signal transducers and activators of transcription in cancer. Nat Clin Pract Oncol 2005;2:315-24. [Crossref] [PubMed]
- Spiekermann K, Biethahn S, Wilde S, et al. Constitutive activation of STAT transcription factors in acute myelogenous leukemia. Eur J Haematol 2001;67:63-71. [Crossref] [PubMed]
- Steensma DP, McClure RF, Karp JE, et al. JAK2 V617F is a rare finding in de novo acute myeloid leukemia, but STAT3 activation is common and remains unexplained. Leukemia 2006;20:971-8. [Crossref] [PubMed]
- Grimwade D, Hills RK, Moorman AV, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010;116:354-65. [Crossref] [PubMed]
- Cheson BD, Bennett JM, Kopecky KJ, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol 2003;21:4642-9. [Crossref] [PubMed]
- Zhong Y, Wu J, Chen B, et al. Investigation and analysis of single nucleotide polymorphisms in Janus kinase/signal transducer and activator of transcription genes with leukemia. Leuk Lymphoma 2012;53:1216-21. [Crossref] [PubMed]
- Zhong Y, Feng J, Chen B, et al. Signal transducer and activator of transcription 3 (STAT3) gene polymorphisms are associated with treatment outcomes in acute myeloid leukemia. Int J Lab Hematol 2012;34:383-9. [Crossref] [PubMed]
- Connelly TM, Koltun WA, Berg AS, et al. A single nucleotide polymorphism in the STAT5 gene favors colonic as opposed to small-bowel inflammation in Crohn's disease. Dis Colon Rectum 2013;56:1068-74. [Crossref] [PubMed]
- Bromberg J. Stat proteins and oncogenesis. J Clin Invest 2002;109:1139-42. [Crossref] [PubMed]
- Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer 2004;4:97-105. [Crossref] [PubMed]
- Benekli M, Baer MR, Baumann H, et al. Signal transducer and activator of transcription proteins in leukemias. Blood 2003;101:2940-54. [Crossref] [PubMed]
- Sternberg DW, Gilliland DG. The role of signal transducer and activator of transcription factors in leukemogenesis. J Clin Oncol 2004;22:361-71. [Crossref] [PubMed]
- Holtick U, Vockerodt M, Pinkert D, et al. STAT3 is essential for Hodgkin lymphoma cell proliferation and is a target of tyrphostin AG17 which confers sensitization for apoptosis. Leukemia 2005;19:936-44. [Crossref] [PubMed]
- Coppo P, Gouilleux-Gruart V, Huang Y, et al. STAT3 transcription factor is constitutively activated and is oncogenic in nasal-type NK/T-cell lymphoma. Leukemia 2009;23:1667-78. [Crossref] [PubMed]
- Chen YW, Guo T, Shen L, et al. Receptor-type tyrosine-protein phosphatase κ directly targets STAT3 activation for tumor suppression in nasal NK/T-cell lymphoma. Blood 2015;125:1589-600. [Crossref] [PubMed]
- Sommer VH, Clemmensen OJ, Nielsen O, et al. In vivo activation of STAT3 in cutaneous T-cell lymphoma. Evidence for an antiapoptotic function of STAT3. Leukemia 2004;18:1288-95. [Crossref] [PubMed]
- Zhao C, Li H, Lin HJ, et al. Feedback Activation of STAT3 as a Cancer Drug-Resistance Mechanism. Trends Pharmacol Sci 2016;37:47-61. [Crossref] [PubMed]
- Lei J, Rudolph A, Moysich KB, et al. Genetic variation in the immunosuppression pathway genes and breast cancer susceptibility: a pooled analysis of 42,510 cases and 40,577 controls from the Breast Cancer Association Consortium. Hum Genet 2016;135:137-54. [Crossref] [PubMed]
- Permuth-Wey J, Fulp WJ, Reid BM, et al. STAT3 polymorphisms may predict an unfavorable response to first-line platinum-based therapy for women with advanced serous epithelial ovarian cancer. Int J Cancer 2016;138:612-9. [Crossref] [PubMed]
- He F, Yang R, Li XY, et al. Single nucleotide polymorphisms of the NF-κB and STAT3 signaling pathway genes predict lung cancer prognosis in a Chinese Han population. Cancer Genet 2015;208:310-8. [Crossref] [PubMed]


