
Molecular mechanism of RBM14-mediated promotion of proliferation, migration, and invasion in osteosarcoma
Highlight box
Key findings
• RNA binding motif protein 14 (RBM14) serves as an activator in osteosarcoma (OS) and exerts a major impact on boosting cellular proliferation, migration, and invasion.
• By altering the Hippo signalling system, RBM14 may be able to modulate the malignant phenotype of OS at the cellular level.
What is known and what is new?
• Research findings indicate that the expression levels of RBM14 exhibit an upregulation in OS cell lines. Suppression of RBM14 expression results in a reduction in the in vitro viability of OS cells. Conversely, overexpression of RBM14 has been observed to enhance the proliferation, migration, and metastasis of OS.
• The Hippo signalling pathway is believed to be a potential downstream pathway through which RBM14 regulates the malignant phenotype of OS.
What is the implication, and what should change now?
• RBM14 is a promising target for therapy in OS management.
• Further research is needed to develop novel therapeutic approaches targeting RBM14 and to explore its potential for treating OS patients effectively.
Introduction
Osteosarcoma (OS) is a type of malignant tumour that is very uncommon but is extremely aggressive and can spread to other parts of the body (1,2). The majority of patients diagnosed with OS are children and teenagers. Although there has been significant progress in clinical treatment, many obstacles still need to be overcome (3,4). Traditional therapeutic approaches, including surgical resection, radiation, and chemotherapy, exhibit constrained efficacy and can potentially lead to negative outcomes and relapse (5,6). Because of this, it is now more important than ever to look into developing innovative treatment options.
In the past few years, gene-targeted therapy has received a considerable amount of interest as an innovative treatment option (7,8). In contrast to conventional therapeutic approaches, gene targeting therapy offers a more specific means of intervening in the abnormal signaling pathways or altered genes seen in cancer cells. This targeted approach effectively hinders the development and propagation of tumours (9,10). The implementation of such an individualised therapeutic strategy has created a renewed sense of optimism among individuals diagnosed with OS.
Gene-targeted therapy, also referred to as gene therapy, represents a propitious approach that seeks to combat or forestall diseases by selectively targeting specific genes. Diverging from conventional therapies, which primarily focus on symptom management or protein substitution, gene-targeted therapy entails modifying the genetic material within cells to directly address the underlying cause of the disease (7,8). One methodology employed in gene-targeted therapy involves the introduction of a functional copy of a gene into the patient’s cells using viral vectors or other delivery systems. This facilitates the production of the deficient or defective protein by the therapeutic gene, thereby reinstating normal cellular function (9,10). Alternatively, gene editing techniques such as clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (CRISPR-Cas9) can be utilized to precisely manipulate the patient’s own genes, rectifying mutations or disrupting deleterious genes (11,12). In comparison to traditional therapies, gene-targeted therapy offers notable advantages, including the potential for long-term or permanent treatments that directly tackle the underlying genetic cause of the disease, as well as personalized treatments tailored to an individual’s genetic profile (13). This innovative therapeutic approach has garnered significant interest, particularly in the realm of cancer, where it exhibits efficacy in impeding tumor growth and propagation. In summary, the implementation of this personalized therapeutic strategy has instilled a renewed sense of optimism among individuals diagnosed with OS.
Transcriptome sequencing technology plays a crucial role in the investigation of gene targeted treatment (14,15). Through the examination of transcriptome data derived from samples obtained from patients, a full understanding of the variations of gene expression within tumour cells may be obtained. This analysis enables the identification of important target genes that are closely linked to the processes of carcinogenesis, progression, and resistance to therapy (16,17). This high-throughput sequencing method offers solid support for deciphering the oncogenic pathways of OS, which paves the way for a more in-depth study of the molecular properties of this type of cancerous tumour. In addition, researchers may examine changes in cellular activities and behaviours brought about by gene overexpression or knockdown by combining several approaches for gene disruption. This allows them to deduce the potential roles that these genes play in the development of tumours (18,19). The utilisation of gene perturbation experiments facilitates the investigation of the impact of genetic differences on essential cellular behaviours such as cell signalling processes, proliferative capacity, invasiveness, and metastatic potential (20,21). This approach offers invaluable knowledge into the development and advancement of OS.
In essence, the management of OS continues to face numerous hurdles, necessitating the exploration of innovative therapeutic strategies. Gene-targeted therapy emerges as a promising avenue, offering precise interventions that specifically target the aberrant signaling pathways exhibited by tumor cells. Moreover, the integration of transcriptome sequencing, gene disruption experiments, and other methodologies provides invaluable insights into the oncogenic pathways underlying OS (22,23). Indeed, recent investigations have shed light on the disrupted expression of RBM14 in a diverse array of tumor types, thereby establishing its intimate correlation with the initiation and progression of malignant diseases (24-26). This genetic sequence encodes a ribonucleoprotein, an intricate entity that has garnered recognition as a multifaceted nuclear coactivator and a profound regulator of RNA splicing (27,28). The upregulated expression of RBM14 in diverse tumor types suggests its potential involvement in the complex process of tumorigenesis. For instance, the methyltransferase like 3 (METTL3) assumes a role in facilitating the m6A hypermethylation of RBM14, consequently propelling the advancement of hepatocellular carcinoma (24). Additionally, RBM14 formed a complex with non-POU domain containing octamer binding (NONO) and impeded the NONO-mediated exon 6 skipping of discs large MAGUK scaffold protein 1 (DLG1), thereby promoting the proliferation of gallbladder cancer cells (29). However, the functional role of RBM14 in OS remains unclear.
To bridge this knowledge gap, our study aims to explore the role of RBM14 in the development and progression of OS, shedding light on the underlying mechanisms specific to this disease. We analyzed RBM14 expression profiles in cancerous tissues and cell populations using data from diverse databases such as The Cancer Genome Atlas (TCGA) project, The Genotype-Tissue Expression (GTEx) Project, Gene Expression Omnibus (GEO) database, and cancer cell line encyclopedia (CCLE) data. Additionally, functional gain and loss experiments was conducted using cellular models to evaluate the impact of RBM14 on OS cell proliferation, migration, and invasiveness. In parallel, the Genetic Perturbation Similarity Analysis Database (GPSAdb) has generated 6,096 gene sets from 3,048 gene perturbation RNA-seq datasets, categorized into up-regulated and down-regulated gene sets. By leveraging gene set enrichment analysis (GSEA) with these meticulously curated gene sets, we have identified genes that exhibit similar downstream effects when RBM14 expression is perturbed, indicating their potential as downstream targets of RBM14 in modulating malignant characteristics in OS.
These investigations will establish a conceptual framework for the development of innovative therapeutic strategies in the management of OS. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2070/rc).
Methods
Expression analysis and data processing
RBM14 expression data were collected from several sources for transcriptional analysis and data processing. The transcription data from TCGA, GTEx, and CCLE, along with pertinent medical specimens, were acquired from The University of California Santa Cruz Xena (UCSC Xena) platform (30). The OS tissue and cell data were gathered from the GEO database, and whereas a gene perturbation dataset was acquired from the GPSAdb (31). The data processing as well as analysis were performed with R software, specifically version 4.2.2.
Survival analysis
The Univariate Cox proportional hazard regression was utilised to evaluate the individual risk ratios (hazard ratio) for overall survival time (OS-time). The survival analysis was conducted using the R packages “survival” and “survminer”. The association between RBM14 expression and OS-time was assessed using the Kaplan-Meier method, enabling the graphical representation of survival curves. Furthermore, the log-rank test was employed to ascertain the statistical significance of the observed association.
Cell lines and culture conditions
In our laboratory, the human OS cell lines U2OS (CL-0236, Pricella Life Science & Technology Co., Ltd., Wuhan, China) and MG63 (CL-0157, Pricella Life Science & Technology Co., Ltd.) were consistently cultivated for cell culture purposes. U2OS cells were cultivated in McCoy’s 5A medium, while MG63 cells were cultured in minimum essential medium (MEM) medium supplemented with non-essential amino acids (NEAA). Both media were supplemented with 10% fetal bovine serum (FBS), 100 units per millilitre (mL) of penicillin, and 100 milligrammes per millilitre (mg/mL) of streptomycin. The cells were subjected to incubation at a temperature of 37 ℃ in an environment with controlled humidity and a carbon dioxide concentration of 5%.
Cell transfection
The RBM14 overexpression plasmid (pcDNA3.1-RBM14) was obtained from Shanghai Hanheng Biotechnology Co., Ltd. (Shanghai, China) for the purpose of conducting gene overexpression studies. The siRNA molecule designed to specifically target RBM14 was obtained from Shanghai Hanheng Biotechnology Co., Ltd. The first si-RBM14 sequence was 5'-GCATTCTGGCCATAGAGCTCGTATT-3', while the second si-RBM14 sequence was 5'-AAATCTTCATTTTGCCGCCGCAACC-3'. Furthermore, a siRNA with a scrambled sequence of 5'-TGCTGACTCCAAAGCTCTG-3' was also included. The cell transfection method was conducted using Lipofectamine 3000 transfection reagent (Cat No. L3000015, Invitrogen, Thermo Fisher Scientific, Inc., Shanghai, China) following the guidelines provided by the manufacturer. The cells that had been collected were used for conducting further studies 48 hours after transfection.
Western blot analysis
In order to conduct the Western blot analysis, we first extracted the proteins with radioimmunoprecipitation assay (RIPA) buffer and then used the bicinchoninic acid (BCA) assay to determine their concentration. Next, an equivalent quantity of protein was partitioned on sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently deposited onto polyvinylidene difluoride (PVDF) membranes (Millipore). Subsequently, the membranes were blocked using a 5% fat-free milk solution and subjected to examination using primary antibodies, including RBM14 (Proteintech, Cat No. 10196-1-AP) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH, Proteintech, Cat No. 60004-1-Ig). This was then followed by washing the membranes in a solution with Tris-buffered saline including Tween-20. The membrane’s preparation was assisted by the use of specific horseradish peroxidase-conjugated secondary antibodies prior to treatment with an enhanced chemiluminescence (ECL) kit manufactured by Millipore. The antibodies that were employed in the study were obtained from Wuhan Sanying Biotechnology Co., Ltd. (Wuhan, China).
Cell proliferation analysis
In order to assess cell proliferation, cells were seeded at a density of 1×104 cells per well in a 96-well plate and subsequently cultivated under controlled conditions of 5% CO2 at 37 ℃. Cell proliferation was assessed at certain time points (24, 48, 72, and 96 hours after seeding) using a Cell Counting Kit 8 (CCK8) assay kit, following the guidelines provided by the manufacturer. To ensure the reliability of our findings, we performed a total of five separate trials for each group and subsequently graphed the average values obtained from these tests.
5-ethynyl-2'-deoxyuridine (EdU) assay
In order to complete the EdU assay, we utilised the EdU assay kit that was provided to us by RiboBio Company (Guangzhou, China). The cells were seeded in a 96-well plate at a density of 2×104 cells per well and afterward treated with EdU at a concentration of 50 µM for a duration of two hours. Following a 30-minute fixation of the cells in a 4% paraformaldehyde solution (Leagene, Beijing, China), a permeabilization step was performed using a 0.3% Triton X-100 solution (Jiayan Biotech, Shanghai, China) for a duration of 10 minutes. The cells were subsequently exposed to glycine (2 mg/mL, Sangon Biotech, Shanghai, China) for a duration of five minutes, after which they were incubated with the Apollo reaction mixture for a period of 30 minutes. Following the application of Hoechst33342 (Invitrogen) for a duration of 30 minutes, the nuclei were subjected to microscopic examination.
Cell migration and invasion assays
We used Transwell inserts with a pore size of 8 µm (provided by Guangzhou Jet Bio-Filtration Co., Ltd., Guangzhou, China) and BD BioCoat Matrigel invasion chambers (provided by BD Biosciences in Bedford, MA, USA) for the cell migration and invasion tests, respectively. After the transfection procedure, we extracted cells and suspended them in serum-free media. Then, we seeded roughly one log10 cell (for the migration assay) or 1.5 log10 cells (for the invasion assay) into the appropriate inserts. After that, cells were kept in an incubating chamber at 37 ℃ for twenty-four hours with FBS acting as a chemoattractant.
Following the time span of incubation, any cells that had migrated or invaded the tissue were captured on camera after being fixed in methanol containing 20% and stained with crystal violet containing 0.1% from Invitrogen. After that, we used a microscope to determine the total number of cells that had migrated to a new location or invaded an existing one. The ratio of the number of migrated or invaded cells found in a designated sample to the number of cells found in a control sample was used to determine the cell migration and invasion rate.
Differential gene expression analysis of RNA-seq
TRIzol was used to separate and purify the sample’s total RNA once it had been separated. The NanoDrop ND-1000 was utilised in order to ascertain the total RNA’s quantity as well as its level of purity. The Bioanalyzer 2100 was utilised in order to perform an analysis on the RNA and validate its integrity. After fragmentation and synthesis of cDNA with the use of hexamer primers, qualified RNA samples were enriched in eukaryotic mRNA using magnetic beads. This was followed by the enrichment of the samples. After that, the double-stranded cDNA was cleaned up with the help of some magnetic beads called AMPure XP. After that, end repair, A-tailing, and adapter ligation were carried out, and AMPure XP magnetic beads were utilised in order to determine the appropriate fragment size. Finally, using Base Calling, the raw image data files that were collected from PE150 sequencing on the Illumina HiSeq 2500 platform were converted to FASTQ files.
A number of quality control tests were carried out with the assistance of FastQC just before the data analysis was carried out. The readings were processed by Trimmomatic in order to get rid of bases of poor quality, adaptor contamination, and ambiguous nucleotides. After the reads were trimmed, they were matched to a reference genome with the assistance of alignment tools such as HISAT2. After that, duplicate reads were identified with Picard Tools and eliminated from the data set. In order to acquire read counts at the gene level, the tool featureCounts was utilised. These read counts represent the total amount of reads that were mapped to each gene in the reference annotation. In order to take into consideration the changes in library size and RNA composition, the read counts were normalised with TMM or one of the other normalisation methods. In the final step, a differential gene expression analysis using DESeq2 (32) was performed on the read count matrix after it had been normalised.
GSEA
In order to carry out GSEA (33), we first sorted the differentially expressed genes from the differential gene expression groups into decreasing order based on the logfold change (logFC) values. After that, we ran an enrichment analysis by making use of the GSEA function that was included in the “clusterProfile” R package (34,35). The GPSAdb was consulted in order to get perturbed gene sets.
Functional and pathway enrichment analysis
Gene Ontology (GO) keywords and enrichment analyses have been carried with the help of Metascape (http://metascape.org) (36) in order to determine the different functional and pathway classes contained within the Hallmark gene sets. The statistical thresholds that were utilised consisted of a minimum count of 3, a significance level of P less than 0.01, and an enrichment factor that was more than 1.5.
Protein-protein interaction enrichment analysis
The following databases were utilised in the process of conducting protein-protein interaction enrichment analysis: STRING (37), BioGrid (38), OmniPath, and InWeb_IM (39). The analysis was done on each gene list that was provided. In particular, this research took into account only the physical interactions that took place within STRING and BioGrid that received a physical score of 0.132 or higher. Proteins that are capable of forming physical interactions with at least one other member of the gene list that was provided are included in the network that was produced as a result. The Molecular Complex Detection (MCODE) algorithm (40) has been employed to determine which components of the network are tightly coupled when the network contains between 3 and 500 proteins. This aids in the discovery of clusters of proteins that, on the basis of their physical interactions, are strongly related to one another.
Statistical analysis
R (version 4.2.2) and GraphPad Prism 8.0 were utilised throughout the statistical analysis process. The results of our study were reported as the mean accompanied by the standard deviation (SD). All experiments were performed in triplicate (technical repeats). In order to analyse the differences between two sets of normally distributed data, we utilised independent Student’s t-tests. Chi-squared tests were utilised for the purpose of comparison in situations in which the data did not have a normal distribution. Every test of the hypothesis was conducted using a two-pronged approach, and a P value of less than 0.05 was determined to be statistically significant.
Results
RBM14 is a potential prognostic marker and therapeutic target in bone sarcoma
In order to investigate how RBM14 is expressed in various cancers, we searched the TCGA database for 33 distinct cancers and analysed the results. The findings demonstrated that in contrast to the standard control group, RBM14 was increased in several types of cancer; this indicates that RBM14 may function as a factor that is detrimental for cancer patients who have the abovementioned cancers (Figure 1A). Following that, we carried out survival analysis and univariate regression analysis on these samples of cancer to compare their outcomes. According to the findings, the level of RBM14 expression did not appear to have a meaningful correlation with the general survival rate of the majority of cancer individuals. For instance, in the assessment of OS-time, RBM14 demonstrated noteworthy associations with the prognoses of four distinct cancer types, namely adenoid cystic carcinoma (ACC), liver hepatocellular carcinoma (LIHC), skin cutaneous melanoma (SKCM), and pheochromocytoma and paraganglioma (PCPG). However, among those four kinds of cancer, RBM14 exhibited a lower level of expression in ACC in comparison with standard samples (Figure 1B). As a consequence of the study described above, we came to the conclusion that RBM14 has been shown to be both an unfavorable prognostic factor and a viable therapeutic target for a subset of malignancies.
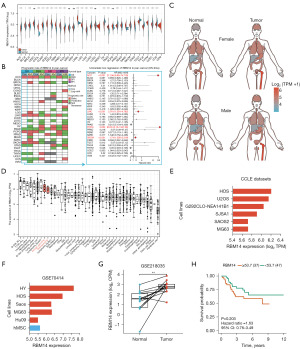
After comprehensive exploration of RBM14 expression in various cancer tissues and cell lines, we proceeded to integrate data from the TCGA and GTEx databases. This integration allowed us to analyze RBM14 expression levels in different organs of the human body under both normal and cancerous conditions. Remarkably, our research findings consistently demonstrated that RBM14 exhibited the highest expression levels in bone tumor tissues, regardless of gender, when compared to normal samples. This observation strongly suggests a significant association between RBM14 and bone tumors (Figure 1C). Additionally, leveraging the CCLE database, we examined RBM14 expression in 40 cancer cell lines and found that its expression was notably elevated in bone tumors compared to other types of tumors (Figure 1D).
Based on these compelling findings, we determined OS as the primary focus of our subsequent study. We conducted further analysis of RBM14 expression in OS cell lines. Within the CCLE dataset, we observed heightened expression of RBM14 in HOS and U2-OS cell lines, whereas SAOS2 and MG63 cell lines exhibited lower expression levels (Figure 1E). Furthermore, utilizing the GEO database, we thoroughly examined RBM14 expression in both OS tissues and cell lines. Comparative analysis with the standard control group demonstrated a substantial upregulation of RBM14 expression in both OS cell lines and tissues (Figure 1F,1G), in stark contrast to the expression levels observed in the normal control group. These findings strongly suggest the involvement of RBM14 as a tumor-promoting factor in OS, thereby emphasizing its potential role in the progression of this disease. In addition, we investigated whether or not there was a correlation among the magnitude of RBM14 expression and the amount of time patients with OS survived using the KM (Kaplan-Meier) and COX analyses (Figure 1H). According to the outcomes, individuals who exhibited low levels of RBM14 expression had significantly higher rates of survival than those who exhibited high levels of RBM14 expression (P>0.05). On the other hand, these findings might call for additional validation using a larger sample size. In conclusion, our research has shown that RBM14 has the potential to be both a therapeutic target and a determinant in determining the prognosis of OS.
Overexpression of RBM14 promotes proliferation, migration, and invasion abilities of OS cells
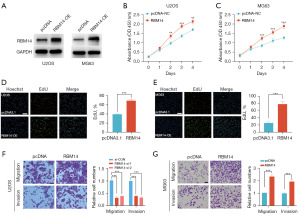
In order to provide evidence that RBM14 plays a role in OS, we conducted investigations in which RBM14 was overexpressed in U2OS and MG63 cell lines (Figure 2A, Figure S1A-S1D). After that, we determined the rate of cell division by using the CCK8 and the Edu assay. When compared to the group acting as a control, the results of our investigations showed that RBM14 overexpression greatly increased the proliferation of U2OS and MG63 cells; this demonstrates RBM14’s ability to enhance OS proliferation (Figure 2B-2E). The following step consisted of analysing the impact that RBM14 had on the migratory and invading capabilities of OS cells using Transwell assays. According to the findings of our research, RBM14 overexpression led to a significant rise in the total number of cells capable of migration and invasion in both the U2OS and MG63 cell lines when compared to the controls (Figure 2F,2G). As a result of these observations, we hypothesize that elevating RBM14 levels in OS cells can significantly improve their capacity for motility and infiltration. In the end, the findings from our experiments demonstrate that RBM14 is able to boost OS cells’ capabilities of proliferation, migration, and invasion, highlighting the potential significance of this protein in the pathogenesis of OS.
Suppression of RBM14 inhibits the proliferation, migration, and invasive abilities of OS cells
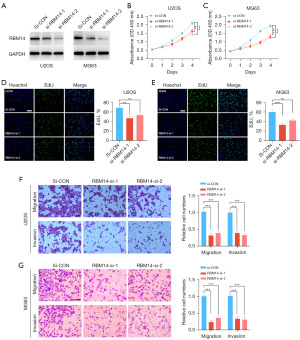
We conducted knockdown experiments of RBM14 in OS cell lines (U2OS and MG63) and evaluated cell proliferation ability using CCK8 and EDU assays in order to better examine the role of RBM14 in controlling the malignant phenotype of OS. This was done so that we could learn more about the role that RBM14 plays in regulating the malignant phenotype of OS. The findings demonstrated that inhibiting RBM14 with a knockdown (Figure 3A, Figure S1E-S1H) had a substantial impact on the ability of U2OS and MG63 cells to proliferate in comparison to the control group (Figure 3B-3E). Using the Transwell migration and invasion tests, the following step consisted of determining how the RBM14 knockdown affected the OS cells’ ability to migrate and invade new tissue. The findings showed that inhibiting RBM14 in U2OS and MG63 cells considerably reduced their capacity for migration and invasion (Figure 3F,3G). Because of this, the findings of our research indicate that inhibiting RBM14 can dramatically reduce the ability of OS cells to migrate and invade nearby tissues. In conclusion, we demonstrated that RBM14 plays a role in enhancing OS cell proliferation, motility, and invasion capabilities through tests involving both RBM14 overexpression and RBM14 knockdown. These innovative findings give a theoretical framework for further exploring the functional processes of RBM14 in OS and utilising it as a therapeutic target.
GSEA identifies RBM14-regulated upregulated and downregulated genes and gene sets
We conducted RNA-seq sequencing on U2OS cells, both overexpressing and with knockdown of RBM14, aiming to delve deeper into the genetic and molecular mechanisms through which RBM14 governs the malignant characteristics of OS (table available at https://cdn.amegroups.cn/static/public/tcr-23-2070-1.xlsx). In our quest to unravel the downstream regulatory genes influenced by RBM14, we employed these RNA-seq data as our input. By employing GSEA in conjunction with the aforementioned input data and the extensive collection of 6,096 gene sets from GPSAdb, we were able to identify genes that, upon knockdown or knockout, exhibited similar effects on downstream genes as observed in our input data. These genes hold significant potential as prospective downstream target genes of RBM14 (Figure 4).
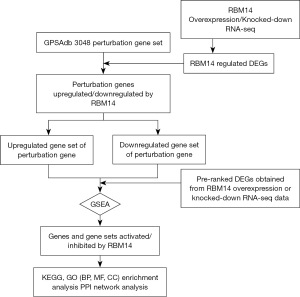
Initially, we identified RBM14 as the regulator of 619 upregulated genes (logFC >0, adj.P value 0.05) and 925 downregulated genes (logFC <0, adj.P value 0.05) (Figure 5A,5B). Additionally, we obtained RNA-seq sequencing data for 3048 perturbed genes from the GPSAdb, which included 1458 perturbed genes. Through differential analysis of these perturbed genes, we discovered 158 downregulated perturbed genes and 48 upregulated perturbed genes that were under the control of RBM14. Based on this differential analysis, we established a collection of upregulated genes known as the Perturbation Gene Upsets, as well as a set of downregulated genes referred to as the Perturbation Gene Downsets, both of which constitute the Perturbation Gene Regulatory Network.
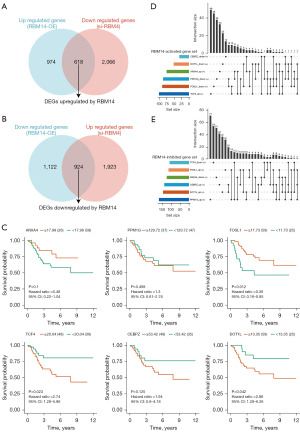
Subsequently, we conducted a GSEA using these gene sets to explore the genes whose expression levels were affected by RBM14 knockdown or overexpression. This investigation allowed us to identify disrupted genes and gene sets that RBM14 has the potential to activate or inhibit (Table 1). Specifically, within the dataset of RBM14 upregulated perturbed genes, we defined a gene set as being active under RBM14 regulation if the Perturbation Gene Upsets were triggered upon RBM14 overexpression and suppressed upon RBM14 knockdown. In other words, the Perturbation Gene Upsets were activated by RBM14 overexpression and inhibited by RBM14 knockdown.
Table 1
| Perturbation genes regulated by RBM14 | Up or down geneset regulated by perturbation gene | GSEA results* | Genes and gene sets activated/inhibited by RBM14 | |
|---|---|---|---|---|
| Overexpression RBM14 | Knocked-down RBM14 | |||
| Upregulated perturbation gene | Perturbation gene upsets | NES >0 | NES <0 | Activated |
| Perturbation gene downsets | NES <0 | NES >0 | Inhibited | |
| Downregulated perturbation gene | Perturbation gene upsets | NES <0 | NES >0 | Inhibited |
| Perturbation gene downsets | NES >0 | NES <0 | Activated | |
GSEA results*: all GSEA enriched gene sets meet adj.P value <0.05. GSEA, gene set enrichment analysis; RBM14, RNA binding motif protein 14; NES, normalized enrichment score.
Similarly, in the context of the Perturbation Gene Downsets, we considered a gene set to be suppressed under RBM14 regulation if it was repressed following RBM14 overexpression but activated after RBM14 knockdown. Likewise, within the dataset of RBM14 downregulated perturbed genes, we defined a gene set as being suppressed under RBM14 regulation if the Perturbation Gene Upsets were inhibited by RBM14 overexpression and activated by RBM14 knockdown.
Finally, we found two genes (TCF4 and ANXA4) that were affected by RBM14’s activation, as well as six gene sets that were affected by RBM14’s activation: TCF4_upset, ANXA4_upset, CEBPZ_downset, DOT1L_downset, PPM1G_downset, and FOSL1_downset. In addition, we found that RBM14 decreased the expression of 4 disrupted genes (CEBPZ, DOT1L, PPM1G, and FOSL1) and 6 perturbed gene sets (TCF4_downset, ANXA4_downset, CEBPZ_upset, DOT1L_upset, PPM1G_upset, and FOSL1_upset). After then, we analysed the survival of the RBM14-regulated genes that had been altered before in relation to OS (Figure 5C). TCF4, which is a gene that is triggered by RBM14, was found to have a strong link with the prognosis of OS patients once the findings were analysed. FPSL1 and DOT1L revealed substantial connections with the prognosis of OS patients. On the other hand, the RBM14-suppressed gene collection contained genes that showed no such associations. An upset plot (Figure 5D,5E) was used to visualize the total amount of genes that were shared by both these two affected gene sets.
Functional enrichment and protein network analysis reveals the potential regulatory molecular mechanisms of RBM14
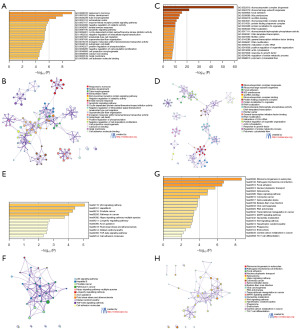
In order to gain a deeper understanding of the molecular pathways that are controlled by RBM14, we carried out GO and Kyoto Encyclopaedia of Genes and Genomes (KEGG) enrichment studies on the altered gene sets that had either been activated or inhibited. On the RBM14-activated gene sets that were disrupted, we carried out GO enrichment analysis (which included Biological Process, Molecular Function, and Cellular Component) as well as protein-protein interaction network analysis, as can be shown in Figure 6A,6B. The most significantly enriched GO terms were “response to hormone”, “kidney development”, “tube morphogenesis”, and “extracellular matrix”, followed by “enzyme-linked receptor protein signalling pathway”. In addition, “p53 signalling pathway”, “Hepatitis B”, “Prostate cancer”, “Pathways in cancer”, and “Hippo signalling pathway-multiple species” were the top five enriched terms in the results of the KEGG pathway analysis (Figure 6C,6D). After that, we repeated the previous step with the RBM14-inhibited, perturbed gene sets and got the same results. The terms “ribonucleoprotein complex biogenesis”, “ribosomal large subunit biogenesis”, “focal adhesion”, “90S preribosome”, and “snoRNA binding” were the top five enriched GO terms (Figure 6E,6F). “Ribosome biogenesis in eukaryotes”, “Pathogenic Escherichia coli infection”, “Focal adhesion”, “Nucleocytoplasmic transport”, and “Spliceosome” were some of the enriched KEGG terms (Figure 6G,6H). It is interesting to note that while conducting the enrichment analysis described above, we came to the conclusion that the Hippo signalling pathway is enriched in the RBM14 activated or inhibited perturbed gene sets. This lends credence to the idea that there is a close connection between this pathway and the regulation molecular processes of RBM14.
Discussion
RBM14 is an RNA binding protein that plays a significant part in the development and progression of several different types of cancer. It does this by controlling the production of downstream genes of interest and modifying erratically stimulated signaling pathways, which in turn influences the proliferation and metastasis of tumour cells (26). On the contrary hand, it is not yet known whether RBM14 demonstrates a similarly important role in OS. In light of this, we decided to carry out an exhaustive investigation of the biological processes and activities of RBM14 in OS. According to the results of our investigation, RBM14 is significantly expressed in OS tissues and cells and correlated with the likelihood of survival of people who have OS. The overproduction of RBM14 in OS cells has been shown to considerably boost OS cell proliferation, migration, and invasion, whilst the knockdown of RBM14 in OS cells has been demonstrated to significantly reduce these processes. Based on these data, RBM14 may provide an intriguing target for the treatment of OS because it appears to play an important part in the disease’s progression. It is possible that advances in the clinical management of OS will become possible as a result of further research into the molecular regulatory mechanisms of RBM14 in OS and the development of therapeutic techniques targeting RBM14.
Initial investigations suggest that RBM14 plays a role in the regulation of a variety of physiological and pathological processes. RBM14, for example, is an example of a new inhibitor of the assembly of the centrosomal protein complex. Through the activity of the STIL/CPAP complex, the loss of RBM14 in human cells can induce ectopic development of the centrosomal protein complex, which can lead to genomic instability and cancer (41). According to the findings of another study, RBM14 is an essential factor in the maturation process of oocytes. When RBM14 is knocked down, the oocyte’s -tubulin can become too acetylated, which can alter the spindle’s shape and the alignment of the chromosomes. This can lead to spindle abnormalities and chromosomal abnormalities throughout the maturation process of the egg. According to the findings of this investigation, RBM14 could serve both as a possible biomarker of oocyte quality and as a new therapeutic target for treating females whose oocytes do not mature properly (42). In addition to this, RBM14 is an essential factor in the embryonic stem cells’ (ESCs) capacity to preserve their pluripotency and initiate differentiation at an early stage. Mouse embryonic stem cells (mESCs) have their gene expression profile changed when RBM14 is depleted. This leads to the downregulation of genes associated with pluripotency as well as genes implicated in the Wnt and TGF-signalling pathways, which in turn affects the pluripotency and differentiation of mESCs (43). Based on these findings, it appears that a defect in RBM14 plays an important part in both physiological and pathological processes, and that it is a significant target in the biological processes that were previously discussed. However, our understanding of the role that RBM14 plays in tumours is still in its infancy at this point.
Recent findings point to the possibility that RBM14 plays a role in the DNA damage response (DDR). Defects in the DDR are linked to a number of illnesses, including cancer and neurological conditions. According to the findings of this research, RBM14 has the ability to efficiently recruit XRCC4 and XLF to chromatin in the event that DNA has been damaged. This has the impact of controlling non-homologous end joining (NHEJ) and influencing DNA damage repair (44). Further investigation has revealed that the radioresistance and recurrence of glioblastoma multiforme (GBM) tumours are caused, at least in part, by the preferential activation of stemness and DDR in the tumor-initiating cells of the cancer. Controlling the DNA-PK-dependent NHEJ pathway is one method that RBM14 can facilitate the repair of DNA. These findings shed light on a previously unknown role of the RNA-binding protein RBM14, which is the regulation of DNA repair and the maintenance of cells that can initiate tumours. Targeting the pathways that are dependent on RBM14 might stop the growth of new tumours and put an end to this fatal disease (45).
In the study that we did on OS, we used data from sample sequencing to investigate possible target molecules and regulatory pathways of RBM14 in patients with metastatic disease. Our research suggests that RBM14 may influence the growth of malignant cells in OS via modifying the Hippo signalling system. In order to elucidate RBM14’s molecular regulation mechanism and provide a theoretical and experimental basis for targeted treatment and clinical application of RBM14 in OS, further research into the molecular mechanisms that underlie the functions that these molecules play in OS is required.
Translating the findings regarding RBM14-targeted therapies into clinical practice poses several challenges that need to be addressed. Firstly, there is a need to establish the safety and feasibility of these therapies through meticulous validation studies (46). This involves conducting rigorous research with larger sample sizes to substantiate the effectiveness and potential risks associated with RBM14-targeted treatments. However, acquiring a larger sample size can be challenging due to the inclusion of diverse patient populations. Variations in age, sex, genetic background, and disease subtypes introduce complexities in data analysis and interpretation, requiring careful consideration during the validation process. Additionally, ensuring the reproducibility of results is crucial. Independent replication studies, standardized protocols, and stringent experimental designs are necessary to validate the findings and confirm the efficacy of RBM14-targeted therapies. Reproducibility not only strengthens the reliability of the research but also enhances the generalizability of the findings to broader patient populations. Furthermore, in the case of targeting RBM14 in OS, several challenges may arise during the translation process. One challenge is the development of effective delivery methods to specifically target RBM14 in tumor cells while minimizing off-target effects. Additionally, understanding the potential resistance mechanisms that may arise during treatment is important for developing strategies to overcome or prevent resistance. In essence, tackling the obstacles inherent in the translation of research findings, encompassing aspects like delivery modalities and resistance mechanisms, alongside the exploration of novel research avenues, will undoubtedly foster the advancement of targeted therapies for OS and conceivably other malignancies.
In conclusion, RBM14 unquestionably plays a pivotal role in the development of OS, which is a pathological process. The therapeutic targeting of this protein holds tremendous potential for the effective treatment of OS and other malignancies that are connected with it. We may anticipate the development of cancer therapy strategies that are both more effective and more specific as research into RBM14 and its mechanisms of action in cancer continues to progress at a deeper level. This will be excellent news for a very large number of people, and it will also make significant contributions to the advancement of both medicine and science.
Conclusions
Our findings imply that RBM14 serves as an activator in OS and exerts a major impact on boosting cellular proliferation, migration, and invasion. In a nutshell, these are the conclusions that we have drawn from our research. By altering the Hippo signalling system, RBM14 may be able to modulate the malignant phenotype of OS at the cellular level.
These discoveries highlight the potential significance of RBM14 as an appealing therapeutic target for the treatment of OS and its management. We may be able to develop unique and highly successful therapeutic techniques for patients suffering from OS if we focus our attention on this protein.
Acknowledgments
The authors thank citexs (www.citexs.com) and Home for Researchers (www.home-for-researchers.com) for English language editing. We thank FigYa, Shipeng Guo and all members of GZDlab for the help of bioinformatic analysis and data visualization. We thank Dr. Jianming Zeng (University of Macau), and all the members of his bioinformatics team, biotrainee, for generously sharing their experience and codes.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2070/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2070/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2070/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2070/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Moukengue B, Lallier M, Marchandet L, et al. Origin and Therapies of Osteosarcoma. Cancers (Basel) 2022;14:3503. [Crossref] [PubMed]
- Whelan JS, Davis LE. Osteosarcoma, Chondrosarcoma, and Chordoma. J Clin Oncol 2018;36:188-93. [Crossref] [PubMed]
- Eaton BR, Schwarz R, Vatner R, et al. Osteosarcoma. Pediatr Blood Cancer 2021;68:e28352. [Crossref] [PubMed]
- Sheng G, Gao Y, Yang Y, et al. Osteosarcoma and Metastasis. Front Oncol 2021;11:780264. [Crossref] [PubMed]
- Meltzer PS, Helman LJ. New Horizons in the Treatment of Osteosarcoma. N Engl J Med 2021;385:2066-76. [Crossref] [PubMed]
- Tian H, Cao J, Li B, et al. Managing the immune microenvironment of osteosarcoma: the outlook for osteosarcoma treatment. Bone Res 2023;11:11. [Crossref] [PubMed]
- Leidner R, Sanjuan Silva N, Huang H, et al. Neoantigen T-Cell Receptor Gene Therapy in Pancreatic Cancer. N Engl J Med 2022;386:2112-9. [Crossref] [PubMed]
- Cavazzana M, Bushman FD, Miccio A, et al. Gene therapy targeting haematopoietic stem cells for inherited diseases: progress and challenges. Nat Rev Drug Discov 2019;18:447-62. [Crossref] [PubMed]
- Rokhzan R, Meier K. Targeted Therapy in Melanoma. N Engl J Med 2022;386:e66. [Crossref] [PubMed]
- Dale B, Cheng M, Park KS, et al. Advancing targeted protein degradation for cancer therapy. Nat Rev Cancer 2021;21:638-54. [Crossref] [PubMed]
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819-23. [Crossref] [PubMed]
- Li T, Yang Y, Qi H, et al. CRISPR/Cas9 therapeutics: progress and prospects. Signal Transduct Target Ther 2023;8:36. [Crossref] [PubMed]
- Wang HX, Li M, Lee CM, et al. CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem Rev 2017;117:9874-906. [Crossref] [PubMed]
- Jiang Z, Zhou X, Li R, et al. Whole transcriptome analysis with sequencing: methods, challenges and potential solutions. Cell Mol Life Sci 2015;72:3425-39. [Crossref] [PubMed]
- Zhang H, He L, Cai L. Transcriptome Sequencing: RNA-Seq. Methods Mol Biol 2018;1754:15-27. [Crossref] [PubMed]
- Caravagna G, Giarratano Y, Ramazzotti D, et al. Detecting repeated cancer evolution from multi-region tumor sequencing data. Nat Methods 2018;15:707-14. [Crossref] [PubMed]
- Milanez-Almeida P, Martins AJ, Germain RN, et al. Cancer prognosis with shallow tumor RNA sequencing. Nat Med 2020;26:188-92. [Crossref] [PubMed]
- Kamimoto K, Stringa B, Hoffmann CM, et al. Dissecting cell identity via network inference and in silico gene perturbation. Nature 2023;614:742-51. [Crossref] [PubMed]
- Yeo NC, Church GM. Perturbation of Gene Regulation by Genome Editing. Methods Mol Biol 2023;2594:59-68. [Crossref] [PubMed]
- Wei R, Zhang H, Cao J, et al. Sample-Specific Perturbation of Gene Interactions Identifies Pancreatic Cancer Subtypes. Int J Mol Sci 2022;23:4792. [Crossref] [PubMed]
- Chang D, Du H, Chen X, et al. A controlled random gene perturbation method identifies ARPC1B gene as a key regulator of cancer metastasis. Genes Dis 2022;10:687-9. [Crossref] [PubMed]
- Zhang W, Qi L, Liu Z, et al. Integrated multiomic analysis and high-throughput screening reveal potential gene targets and synergetic drug combinations for osteosarcoma therapy. MedComm (2020) 2023;4:e317.
- Sasa K, Saito T, Kurihara T, et al. Establishment of Rapid and Accurate Screening System for Molecular Target Therapy of Osteosarcoma. Technol Cancer Res Treat 2022;21:15330338221138217. [Crossref] [PubMed]
- Hu J, Yang L, Peng X, et al. METTL3 promotes m6A hypermethylation of RBM14 via YTHDF1 leading to the progression of hepatocellular carcinoma. Hum Cell 2022;35:1838-55. [Crossref] [PubMed]
- Tsuji K, Kawata H, Kamiakito T, et al. RNA-binding protein 14 promotes phase separation to sustain prostate specific antigen expression under androgen deprivation in human prostate cancer. J Steroid Biochem Mol Biol 2023;235:106407. [Crossref] [PubMed]
- Hu Y, Mu H, Deng Z. RBM14 as a novel epigenetic-activated tumor oncogene is implicated in the reprogramming of glycolysis in lung cancer. World J Surg Oncol 2023;21:132. [Crossref] [PubMed]
- Kai M. Roles of RNA-Binding Proteins in DNA Damage Response. Int J Mol Sci 2016;17:310. [Crossref] [PubMed]
- Mehta M, Raguraman R, Ramesh R, et al. RNA binding proteins (RBPs) and their role in DNA damage and radiation response in cancer. Adv Drug Deliv Rev 2022;191:114569. [Crossref] [PubMed]
- Yang ZY, Zhao C, Liu SL, et al. NONO promotes gallbladder cancer cell proliferation by enhancing oncogenic RNA splicing of DLG1 through interaction with IGF2BP3/RBM14. Cancer Lett 2024;587:216703. [Crossref] [PubMed]
- Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020;38:675-8. [Crossref] [PubMed]
- Guo S, Xu Z, Dong X, et al. GPSAdb: a comprehensive web resource for interactive exploration of genetic perturbation RNA-seq datasets. Nucleic Acids Res 2023;51:D964-8. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Stark C, Breitkreutz BJ, Reguly T, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 2006;34:D535-9. [Crossref] [PubMed]
- Li T, Wernersson R, Hansen RB, et al. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat Methods 2017;14:61-4. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Shiratsuchi G, Takaoka K, Ashikawa T, et al. RBM14 prevents assembly of centriolar protein complexes and maintains mitotic spindle integrity. EMBO J 2015;34:97-114. [Crossref] [PubMed]
- Qin H, Qu Y, Yuan YF, et al. RBM14 Modulates Tubulin Acetylation and Regulates Spindle Morphology During Meiotic Maturation in Mouse Oocytes. Front Cell Dev Biol 2021;9:635728. [Crossref] [PubMed]
- Chen G, Zhang D, Zhang L, et al. RBM14 is indispensable for pluripotency maintenance and mesoderm development of mouse embryonic stem cells. Biochem Biophys Res Commun 2018;501:259-65. [Crossref] [PubMed]
- Simon NE, Yuan M, Kai M. RNA-binding protein RBM14 regulates dissociation and association of non-homologous end joining proteins. Cell Cycle 2017;16:1175-80. [Crossref] [PubMed]
- Yuan M, Eberhart CG, Kai M. RNA binding protein RBM14 promotes radio-resistance in glioblastoma by regulating DNA repair and cell differentiation. Oncotarget 2014;5:2820-6. [Crossref] [PubMed]
- Bodmer W, Golubovskaya V. Cancer Immunotherapy: Where Next? Cancers (Basel) 2023;15:2358. [Crossref] [PubMed]


