
Comprehensive proteomic profiling of lung adenocarcinoma: development and validation of an innovative prognostic model
Highlight box
Key findings
• We established an efficacious protein model designed for the prognosis prediction of lung adenocarcinoma (LUAD) patients and uncovered a CD38 expression paradox. Our utilization of clinical samples for validation bolstered the practicality of this study.
What is known and what is new?
• In clinical diagnostics, markers like CYFRA21-1, CEA, ProGRP, NSE, and SCCA are commonly employed for disease detection. Despite their widespread use, these markers lack high specificity and often provide only suggestive rather than definitive information.
• The objective of this study was to identify proteins that could potentially forecast the prognosis of lung cancer patients, focusing specifically on the proteome of LUAD patients.
What is the implication, and what should change now?
• Our investigation indicates that proteomic models demonstrate robust predictive efficacy in prognosticating outcomes for individuals with lung adenocarcinoma. Furthermore, our findings suggest a paradoxical expression of CD38 within solid tumors. Subsequent to this, it is imperative to substantiate the model’s applicability through an expanded analysis of clinical samples. Additionally, there is a pressing need to delve into the biological functions of CD38 in the context of solid tumors.
Introduction
Bronchogenic lung cancer, originating primarily from bronchial mucosa or glands, is generally referred to as lung cancer and presents a significant risk to human health. As demonstrated in a 2020 global cancer statistics study (1), lung cancer accounted for 2,206,771 new cases, comprising 11.4% of total cancer incidences, and positioned second following breast cancer. Moreover, the death toll reached 1,796,144, representing 18.0% of total cancer fatalities and securing the first rank. China, home to 20% of the global population, witnessed lung cancer incidence and mortality rates accounting for 35.6% and 37.6% respectively, of the world’s totals. The country’s 5-year lung cancer survival rate stands at a mere 15.6% due to late-stage diagnosis. Lung cancer manifests predominantly as small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), with the latter constituting the majority of cases (80–85%). NSCLC primarily includes lung adenocarcinoma (LUAD) and lung squamous carcinoma (LUSC), with LUAD emerging as the most common subtype. Early diagnosis is typically challenging; thus, patients often present with advanced-stage disease by the time symptoms manifest, culminating in poor prognosis. Presently, lung cancer screening heavily relies on chest X-rays and computed tomography (CT) scans. Diagnostic imaging largely depends on clinicians’ disease comprehension and image interpretation skills. Furthermore, the dearth of specificity in early-stage lung cancer imaging further complicates early detection.
The recent advancements in cancer-related biomarker research have ushered in a new era for lung cancer detection and treatment. Particularly noteworthy are the developments in immunotherapy, initiated with studies on the clinical effectiveness of pembrolizumab, nivolumab, and atezolizumab (2-5) as second-line therapy for lung cancer. These studies led to the Food and Drug Administration (FDA)’s endorsement of immune checkpoint inhibitors as a secondary treatment for lung cancer. Following the promising results from the Keynote024 study (6), the FDA approved pembrolizumab as a primary treatment for patients with programmed death-ligand 1 (PD-L1) expression levels above 50% in lung cancer, highlighting the beneficial impact of single-agent immunotherapy for these patients. Regarding diagnosis and disease detection, markers such as cytokeratin 19 fragment (CYFRA21-1), carcinoembryonic antigen (CEA), gastrin-releasing peptide precursor (ProGRP), neuron-specific enolase (NSE), and squamous cell carcinoma antigen (SCCA) are frequently used in clinical settings. However, none of these markers exhibit high specificity and often only have suggestive value. Therefore, the quest for a marker closely related to lung cancer continues to be a pressing research direction.
The objective of this study was to identify proteins that could potentially forecast the prognosis of lung cancer patients, focusing specifically on the proteome of LUAD patients. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1940/rc).
Methods
Data collection
The Cancer Genome Atlas (TCGA) program data portal (https://portal.gdc.cancer.gov/) was used to access the RNA sequencing and protein expression data of LUAD, as well as the accompanying clinical and pathological data. We collected 59 normal specimens and 541 tumor specimens for a total of 600 specimens. We then integrated the corresponding information. We created training and test sets from all samples at random, and we utilized the training set to build a LAUD protein prognostic model and the test set to assess our model. From each of the five LUAD patients at the Affiliated Hospital of Hangzhou Normal University, paraffin sections were obtained from two different sites, with each site providing a pair of sections including both tumor tissues and adjacent non-tumor tissues. This procedure, approved by the hospital’s research ethics review, resulted in a total of 10 pairs of paraffin sections. The study was a retrospective study, and the samples used were from the Department of Pathology, which received approval for exemption from informed consent. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Affiliated Hospital of Hangzhou Normal University [2023(E2)-KS-036] and the requirement for individual consent for this retrospective analysis was waived.
Construction of LUAD prognosis related protein signature
Initially, we conducted univariate COX proportional regression analysis with a P value threshold of 0.05 to identify proteins potentially linked to the prognosis of LUAD in the training set. The objective was to investigate the association between these proteins and overall survival (OS) in patients with LUAD. The outcomes of this analysis are presented in the form of a forest plot. The volcano plot illustrates the differential expression of prognosis-related proteins in both normal and tumor tissues, with a significance threshold set at a P value of less than 0.05. Subsequently, we conducted a least absolute shrinkage and selection operator (LASSO) analysis (7,8) using the “glmnet” R package to further refine the selection of prognosis-related proteins, aiming to identify the most crucial ones. Ten-fold cross-validation was conducted on the proteins that had been previously screened in order to ascertain the optimal penalty parameter λ and establish the most suitable model. Subsequently, multivariate COX proportional regression analysis was performed using a stepwise regression approach by the “step” function, which further refined the selection of variables. This stepwise analysis allowed for the construction of a prognostic protein signature model for LUAD. The resultant prognostic signature was then utilized to compute the risk score for each sample, employing the following equation:
This was achieved by multiplying the regression coefficients (Coef) with the expression levels (xk) of the respective proteins. Following this, we calculated the risk score for each specimen, spanning both our training and test sets. Based on this computation, samples are then stratified into groups of high and low risk. This segregation is guided by the median risk score.
Assessment of the prognostic model
Upon model construction completion, our goal was to examine its functionality in numerous aspects, thus confirming its possible use in clinical settings. We assessed the model’s capacity to differentiate between high and low risk cohorts through principal component analysis (PCA) on all proteins and modeled proteins with the assistance of the “scatterplot3d” R package. For further verification, Kaplan-Meier (KM) analysis was conducted on both risk cohorts across all datasets, the training set, and the test set. The analysis was supplemented by progression-free survival (PFS) examination in these risk cohorts, statistical significance being underscored by P values beneath 0.05. As we proceeded, our assessment incorporated receiver operating characteristic (ROC) curve analysis to investigate the predictive potential of both the protein model and clinicopathological factors for patient survival, as well as the model’s forecasting ability for 1-, 2-, and 3-year patient survival. This predictive capacity was quantified through the area under curve (AUC), computed using the “timeROC” R package. The “survival” and “survminer” R packages were also featured in this process. Conclusively, we probed the risk scores, survival condition, and expression metrics of six prognostic proteins across all datasets, the training set, and the test set. Patients were classified into groups of high and low expression hinging on protein expression differences. The survival rates of these clusters were then scrutinized via KM survival curve analysis.
Comprehensive clinical analysis and construction of the nomogram
To verify the model’s practicality across a variety of patient demographics, we segregated the patients into subgroups according to various clinicopathological data, including age, gender, TNM stages (Tumor, Node, Metastasis stages), and overall stage. This classification allowed us to perform in-depth analyses, one of which involved scrutinizing OS in high- and low-cohorts via KM survival curve analysis. With the assistance of “corrplot” and “circlize” R packages, we inspected the differential expression of each protein represented in the model across these different subgroups, and visually presented the results through boxplots. Subsequently, both univariate and multivariate COX regression analyses were conducted to evaluate the independent prognostic power of our model and the prognostic value inherent in various clinicopathological factors. With the intention of directly benefiting clinical practice, we devised a nomogram that amalgamated both risk scores and clinicopathological factors. This tool was intended to predict the survival of LUAD patients at 1-, 3-, and 5-year intervals. The precision of these nomograms in forecasting survival was further confirmed through calibration curves.
Gene set enrichment analysis (GSEA) and construction of the protein coexpression network
To probe the potential regulatory mechanisms differentiating the cohorts of high and low risk, we employed the “c2.cp.kegg.symbols.gmt” and “c5.go.symbols.gmt” gene sets as our background gene sets. The “clusterProfiler” package enabled us to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses on patients within the high- and low-risk cohorts. This approach allowed us to investigate the potential biological functions and signaling pathways that may bear relevance to patients in various risk cohorts. We set a P value of less than 0.05 as the screening threshold, and the top five most significant pathways were retained and visualized for further scrutiny. Moreover, we harnessed the “corrplot” and “circlize” packages to examine interactions between the six proteins represented in our model, with results visually presented in a comprehensive circos plot. Continuing our exploration, we evaluated the co-expression relationships between model proteins and other proteins using the “ggplot2” and “ggalluvial” packages, representing the findings in a detailed Sankey diagram.
Tumor immune environment and the sensitivity of immunotherapy
In our endeavor to discern the variations in the tumor immune environment of patients in different risk cohorts, we exploited the CIBERSORT algorithm (9) to analyze the proportion of 22 immune cell infiltrates in LUAD samples obtained from the TCGA database. For a nuanced examination of the differences in the proportions of immune cell infiltration between high- and low-risk cohorts, we employed the “limma” package. Visualization of these disparities was accomplished with the “ggpubr” and “fmsb” packages, resulting in box plots and radar plots. Furthermore, we employed The Cancer Immunome Atlas (TCIA) (https://tcia.at/home) database to evaluate the sensitivity of LUAD patients to immunotherapy. We initiated by downloading the pertinent data for LUAD patients from the TCIA database, which included information related to programmed cell death 1 (PD-1) and CTLA4 immune checkpoints, as well as the immunophenoscore (IPS). Subsequent to this, we conducted an analysis of the disparity in IPS between high- and low-risk cohorts. The IPS functioned as an immunogenicity measure, with a higher IPS indicative of stronger immunogenicity. This consequently suggested a more favorable immunotherapy response.
Consensus clustering analysis
Aiming to gain a comprehensive understanding of the clinical characteristics of patients with LUAD, we conducted an unsupervised consensus clustering analysis using the ConsensusClusterPlus package. This procedure allowed us to categorize LUAD patients into distinct subtypes. Following this, we conducted a survival analysis among the various subtypes to investigate any potential disparities. Additionally, we delved into whether different clinicopathological indicators, such as age, gender, and TNM stage, varied significantly between the subtypes. To present these findings more effectively, we employed the “ggplot2” package, converting the results into visually understandable histograms.
Validation of prognostic protein expression levels
We obtained immunohistochemical images of the model proteins in both lung cancer tissue and normal lung tissue from the Human Protein Atlas (HPA) database (https://www.proteinatlas.org/) to facilitate a comparison of the expression levels of the model proteins. The selection of images, representing protein expression in LUAD, was based on clear staining patterns and relevance to LUAD pathology. We prioritized high-resolution images with consistent protein expression patterns, ensuring an accurate and representative depiction of the protein expression in lung cancer tissues. Subsequently, we identified the “optimal protein” (exhibiting the most significant difference) for validation, basing our selection on the results of differential expression of the model proteins in subgroups according to clinicopathological factors. With the help of the “plyr”, “reshape2”, and “ggpubr” packages, we conducted a pan-cancer analysis of these “optimal proteins” in human tumors. A differential expression analysis of the “optimal protein” in various tumors was conducted, and then the results were arranged according to the expression of the “optimal protein” in different tumors, with the findings represented in a box plot. As a subsequent step, to further validate the “optimal protein”, we implemented immunofluorescence staining on formalin-fixed paraffin-embedded (FFPE) samples of lung cancer tissues and paraneoplastic tissues from five LUAD patients at the Affiliated Hospital of Hangzhou Normal University. In this investigation, the antibody against CD38 (Santa Cruz Biotechnology, Dallas, TX, USA; Cat# sc-18858, RRID:AB_627050) was utilized. We quantified the immunofluorescence intensity using Image J software to conduct a semi-quantitative analysis of protein expression, thereafter examining whether the protein exhibited differential expression in lung cancer tissues and paracancer tissues employing a t-test.
Statistical analysis
Statistical analysis was carried out using R software (version 4.0.2). For all tests, a P value of less than 0.05 was considered statistically significant. The normality of data distribution was assessed using Shapiro-Wilk test. Differences between groups were analyzed using the Student’s t-test for normally distributed data and the Mann-Whitney U test for non-normally distributed data. Chi-square or Fisher’s exact test were used for categorical data analysis. Survival analysis was performed using KM method and differences between survival curves were evaluated using the log-rank test. Multivariate survival analysis was carried out using Cox proportional hazards model. The proportional hazards assumption was tested using Schoenfeld residuals. ROC curve analysis was used to evaluate the prognostic accuracy of the protein signature, with the area under the curve (AUC) indicating predictive performance. For the analysis of protein expression levels and immunotherapy response, Pearson or Spearman correlation analysis was employed depending on data distribution. GSEA was performed to identify significant pathways associated with risk scores, and the Benjamini-Hochberg method was used for adjusting P values for multiple comparisons.
Results
Construction of the protein risk signature and evaluation of the prognostic model
At the commencement of our investigation, we harvested RNA sequencing data, protein expression data, and corresponding clinicopathological data from 600 LUAD samples via the TCGA database. An amalgamation of these datasets resulted in a collection of 357 samples with comprehensive data, forming the cornerstone of our study. We randomly partitioned these 357 samples into two equitable sets: the training set, encompassing 179 cases, and the test set, inclusive of 178 cases. This partitioning was instrumental in facilitating subsequent model construction. We began with univariate regression to probe the association of proteins with OS, identifying ten significant proteins (Table 1). These were depicted in a forest plot (Figure 1A), where the x-axis showed hazard ratios on a log scale, and the y-axis listed proteins. The length of the lines represented 95% confidence intervals. A volcano plot (Figure 1B) illustrated differential protein expression: the x-axis showed log2 hazard ratios and the y-axis showed the negative log10 P values. Red points indicated high-risk proteins, green points indicated low risk, and black points were nonsignificant. We further refined the selection of prognostic proteins using LASSO regression (Figure 1C,1D). This process, guided by the optimal lambda value obtained through cross-validation, identified eight key proteins (NDUFB4, CD49B, UQCRC2, SMAD1, CD134, FOXM1, CD38, KAP1) associated with prognosis. Subsequently, six of these proteins (CD49B, UQCRC2, SMAD1, FOXM1, CD38, KAP1) were selected via multivariate regression proportional analysis to construct the final prognostic model (Table 2). Risk scores for each sample were calculated by multiplying each protein’s expression level in the model by its corresponding regression coefficient, and then summing up these products. Ultimately, all samples were bifurcated into high- and low-risk cohorts based on the median value of the risk scores from the training set samples.
Table 1
| Protein | HR | HR.95L | HR.95H | P value |
|---|---|---|---|---|
| NDUFB4 | 0.197 | 0.040 | 0.966 | 0.05 |
| TRAP1 | 1.605 | 1.017 | 2.531 | 0.04 |
| CYCLINB1 | 1.355 | 1.030 | 1.784 | 0.03 |
| CD49B | 1.856 | 1.043 | 3.304 | 0.04 |
| UQCRC2 | 3.469 | 1.059 | 11.367 | 0.04 |
| SMAD1 | 3.290 | 1.129 | 9.590 | 0.03 |
| CD134 | 0.146 | 0.028 | 0.778 | 0.02 |
| FOXM1 | 2.049 | 1.188 | 3.534 | 0.01 |
| CD38 | 0.351 | 0.151 | 0.816 | 0.02 |
| KAP1 | 4.437 | 1.703 | 11.563 | 0.002 |
HR, hazard ratio; L, low; H, high.

Table 2
| Protein | Coef |
|---|---|
| CD49B | 0.64292591 |
| UQCRC2 | 1.219037602 |
| SMAD1 | 0.998231812 |
| FOXM1 | 0.553722336 |
| CD38 | −0.639864924 |
| KAP1 | 1.035762646 |
Coef, coefficients.
After developing the model, we conducted a comprehensive evaluation of its performance. In Figure 2A, PCA on all proteins showed overlapping clusters with no clear separation of risk cohorts, with axes representing the principal components. Figure 2B’s PCA, on model proteins, delineated distinct high- and low-risk cohorts. KM curves (Figure 2C for the entire dataset, Figure 2D for the test set, and Figure 2E for the training set) showed the survival probability over time on the y-axis, revealing a statistically significant difference in OS between the high-risk (red line) and low-risk (blue line) cohorts. For PFS, Figure 2F indicated no significant disparity in survival probability between the risk cohorts. ROC curve analysis demonstrated that the risk model’s AUC values were higher compared to other clinicopathological factors, as illustrated in Figure 3A. Specifically, the AUC values for the risk model at 1, 3, and 5 years were 0.685, 0.618, and 0.635, respectively, as shown in Figure 3B. This highlighted the superior predictive accuracy of the risk model over traditional clinicopathological factors. Investigation into patient risk score and survival time across the entire dataset, the training set, and the test set indicated an increasing mortality rate in proportion to escalating risk scores. Moreover, heatmaps demonstrating protein-related expression highlighted CD49B, UQCRC2, SMAD1, FOXM1, and KAP1 as being highly expressed in the high-risk cohort, whilst CD38 exhibited high expression in the low-risk cohort (Figure 3C-3E). Analysis of protein expression and patient survival curves manifested longer OS in patients with low expression of CD49B, UQCRC2, FOXM1, and KAP1, as opposed to their high expression counterparts. In contrast, high levels of CD38 expression indicated longer OS compared to its low expression counterparts. No noteworthy relationship was discovered between SMAD1 expression and OS (Figure 3F).


Comprehensive clinical analysis
The patient cohort was further segmented into several subgroups in accordance with age, gender, TNM-stages, and stage. Subsequent analysis of KM survival curves within these diversified subgroups (Figure 4A) revealed a longer OS for the low-risk cohort relative to the high-risk counterpart. However, within the T3–4 patient subgroup, no significant difference in OS was noted between the two risk cohorts. A deep-dive into the relationship between model proteins and clinicopathological factors revealed unique patterns of protein expression. The x-axis in Figure 4B categorized clinical factors such as age, gender, and TNM-stages, while the y-axis measured the expression levels of model proteins. CD38 showed differential expression in the age and TNM-stage subgroups; CD49B in the gender and T-stage subgroups; FOXM1 in the gender subgroup; KAP1 in the M-stage and stage subgroups; and the risk score in the T-stage subgroup also indicated differential expression. The univariate and multivariate COX regression analysis, depicted in Figure 5A,5B, established the risk score as an independent prognostic indicator, as indicated by P values less than 0.05 in both analyses. The x-axis represented the hazard ratio for clinical variables such as age, gender, and TNM-stages, with the y-axis listing these variables. The boxes represented the hazard ratios with their confidence intervals, and the dashed line denoted a hazard ratio of one. We assembled nomograms that incorporated both risk scores and clinicopathological factors for LUAD patients. These nomograms, as depicted in Figure 5C, provided a visual representation where points were assigned for each factor, including M stage, T stage, N stage, overall stage, gender, age, risk scores, and, to predict the OS at 1, 3, and 5 years. The calibration plot in Figure 5D was scrutinized to assess the accuracy of our prediction model. The alignment of the plotted points with the diagonal line represented the model’s reliability. Points closer to the diagonal line indicated more accurate predictions, and our model demonstrated superior prognostic accuracy at 1 and 3 years, as evidenced by the plotted points’ proximity to the 45-degree line, compared to the 5-year predictions.


GSEA and construction of the protein coexpression network
Our exploration into the biological mechanisms and regulatory pathways implicated in varying risk cohorts utilized GO and KEGG analyses. The results indicated significant enrichment in five key pathways for genes of patients in the high-risk cohort: cell cycle, Parkinson’s disease, pyrimidine metabolism, ribosome, and spliceosome. These genes were also associated with biological processes such as appendage development, morphogenesis, and epidermis development; cellular components like the ribosomal subunit; and molecular functions such as the structural constituent of the ribosome (Figure 6A). Conversely, genes of patients in the low-risk cohort were notably enriched in distinct pathways: allograft rejection, asthma, intestinal immune network for immunoglobulin A (IgA) production, primary immunodeficiency, and systemic lupus erythematosus. These genes were linked to cellular components such as the immunoglobulin complex, circulating immunoglobulin complex, and T cell receptor complex; alongside biological processes such as antigen binding and immunoglobulin receptor binding (Figure 6B). Subsequent to this, a co-expression analysis was performed between the model proteins, and the outcomes are depicted in Figure 6C. The circos plot illustrated the strength and direction of the co-expression relationships among model proteins, with red lines indicating positive correlation and green lines indicating negative correlation. The intensity of the color on each line reflected the magnitude of the correlation coefficient, demonstrating the degree of co-expression between the proteins. To shed further light on the interplay among these proteins, we analyzed the regulatory relationships between the model proteins and other proteins, as represented in the Sankey diagram (Figure 6D). The Sankey diagram detailed the complex interactions and the flow of regulatory influence from model proteins to various other proteins implicated in LUAD, highlighting the pathways and potential mechanisms that these proteins may have influenced.
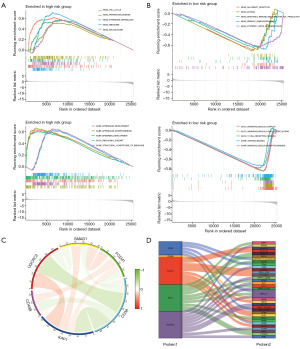
Exploration of tumor immune microenvironment and response to immunotherapy
To examine the extent of immune cell infiltration in various risk cohorts, we deployed the CIBERSORT algorithm. The outcome of this analysis revealed differing degrees of Plasma cell infiltration within the high- and low-risk cohorts, with more pronounced infiltration in the low-risk cohort (Figure 7A,7B). However, the infiltration levels of other immune cells did not exhibit any significant disparities between the two risk cohorts. Further, we conducted an immunotherapy sensitivity analysis using the TICA database for patients in the high- and low-risk cohorts. This analysis disclosed a higher Immune Potential Score (IPS) in the low-risk cohort than in the high-risk counterpart in the presence of positive immune checkpoints (PD-1 and/or CTLA4), suggesting an enhanced immune response within the low-risk cohort (Figure 7C).

Consensus clustering analysis
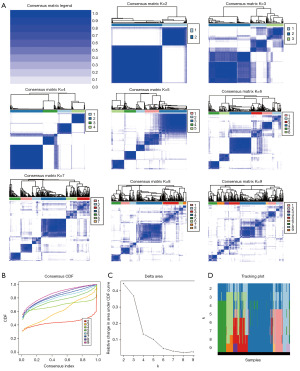
The application of unsupervised consensus clustering analysis to the prognostic proteins identified an optimal value of k=2 for the LUAD samples, as evidenced by the consensus matrix (Figure 8A), the consensus cumulative distribution function (CDF) (Figure 8B), the delta area plot (Figure 8C), and the tracking plot (Figure 8D). Subsequently, we partitioned the sample into two distinct subtypes: subtype 1 and subtype 2. Probing survival rates across these subtypes uncovered that subtype 2 demonstrated significantly enhanced OS (Figure 9A). Additionally, when examining the clinical correlations within the varying clusters, we discerned pronounced discrepancies in N-stage and T-stage amid the two subtypes (Figure 9B).

Validation of CD38 expression level
Our comprehensive pan-cancer analysis, as illustrated in Figure 10A, displayed significant differential expression of CD38 between normal and tumor tissues across various cancer types, notably in LUAD, BRCA, CHOL, COAD, HNSC, KICH, KIRC, LIHC, LUSC, PCPG, PRAD, READ, and THCA. In Figure 10B, the relative expression levels of CD38 are depicted, showcasing the variability in its expression among different tumor types. Subsequently, through the utilization of immunohistochemical images from the HPA database, we confirmed differential expression of CD38 and other model proteins in LUAD tissues as opposed to normal lung tissues, with a significant upregulation in the former (Figure 10C). Our prior analysis singled out CD38 as having the most prominent expression across the diverse subgroups. With this discovery, we designated CD38 as the “prime protein” for subsequent validation. Immunofluorescence staining was performed on 10 pairs of lung cancer and adjacent non-tumor tissues (Figure 10D), followed by semi-quantitative analysis (Figure 10E). The results clearly indicated that CD38 is highly expressed in lung cancer tissues compared to adjacent non-tumor tissues.

Discussion
The landscape of lung cancer research is heavily populated by investigations centered on identifying relevant biomarkers due to their potential utility in early screening, diagnosis, efficacy evaluation, and prognosis prediction. One promising direction was the exploration of autoantibodies against lung cancer as such markers. Massion et al. (10) reported a substantial improvement in their CT screening risk model’s diagnostic potency for lung cancer upon integrating antibodies to p53, NY-ESO-1, CAGE, GBU4-5, SOX2, HuRt, melanoma antigen, and MAGE-A4. The modified model yielded a specificity exceeding 92% and a positive predictive value surpassing 70%. Similarly, the study by Dai et al. (11) demonstrated that the sensitivity for detecting NSCLC increased to 84% when the detection of anti-ENO1 was combined with two other tumor protein biomarkers (CEA and CYFRA 21-1). Another study (12) capitalized on complement fragment 4d (C4d) to differentiate benign from malignant lung nodules. However, the restricted sensitivity of these autoantibodies curtails their utility in early screening. Alternative strategies for biomarker discovery, such as microRNA, circulating tumor DNA, circulating tumor cells, exosomes, and DNA methylation, have been exploited (13-17), albeit they suffer from the drawbacks of the testing processes being inconvenient and expensive, thus limiting their translation into clinical practice. While the pursuit of lung cancer biomarkers at the RNA and DNA levels continues to attract attention, it is crucial to remember that proteins primarily mediate biological functions. Proteins such as CEA, NSE, CYFRA21-1, ProGRP, and SCCA serve as primary lung cancer biomarkers in clinical settings. NSE and ProGRP are principally employed for SCLC’s early diagnosis, efficacy monitoring, and prognostic assessment (18-20), while CEA, CYFRA21-1, and SCCA are predominantly used for NSCLC (21-23). CYFRA21-1 and SCCA hint at a higher likelihood of LUSC, and a close correlation exists between CEA and LUAD. Nevertheless, the credibility of CEA is compromised by various factors that can induce its elevation, including different adenocarcinoma types (24) and certain benign diseases (25). Hence, the pressing requirement for identifying additional protein biomarkers closely tied to LUAD, the most impactful subtype of lung cancer, is clear. Catering to this need, our research aims to scrutinize the database for proteins with a strong association with LUAD (CD49B, UQCRC2, SMAD1, FOXM1, CD38, and KAP1-6) and build a model to predict the prognosis of LUAD patients.
Leveraging the LASSO-COX algorithm, we developed a prognostic signature, subsequently determining the risk score for every sample. These samples were then segregated into two risk cohorts using the median value. Subsequently, we employed PCA analysis, ROC curve analysis, and KM curve survival analysis to assess the performance of our protein model. The PCA analysis suggested the protein model’s effectiveness in categorizing samples into the two risk cohorts. The ROC curve further attested to the model’s superior predictive ability for LUAD patient prognosis compared to other factors. Additionally, survival analysis demonstrated a more favorable prognosis in the low-risk cohort than in the high-risk cohort, although no significant variation in PFS between the two categories was noted. Patient PFS evaluation presents a multifaceted clinical challenge as it often mandates a comprehensive assessment by clinicians considering patient status, clinical examination metrics, and imaging results (26). Interestingly, our study revealed a direct correlation between risk scores and patient mortality, signifying an incremental increase in mortality with higher risk scores. Upon conducting clinical correlation analysis, we found a significant divergence in prognosis among the high- and low-risk cohorts across different clinicopathologic factor subgroups. This suggests the potential broad applicability of our protein model to LUAD patients. Further, COX regression analysis revealed the risk score’s potential as an independent prognostic indicator. We subsequently incorporated clinicopathologic factors to develop a nomogram, aiming to provide a practical tool for clinicians in predicting LUAD patient survival.
In our research, we conducted an analysis to understand the differential expression of model proteins across various subgroups. Intriguingly, CD38 outperformed as the “optimal protein”, thereby prompting a detailed examination of its expression. Notably, among the model proteins, CD49B, UQCRC2, SMAD1, FOXM1, and KAP1-6 exhibited heightened expression in the high-risk cohort, whereas CD38 was predominantly expressed in the low-risk cohort. The prognosis for the CD38 high-expression cohort outperformed that of its low-expression counterpart, suggesting CD38’s role as a “protective factor”. Conversely, the other proteins, exhibiting superior prognosis in the low-expression group, are suggested to function as “risk factors”. Intriguingly, this outcome contradicts our subsequent validation analysis, thus highlighting a “contradiction”. Both the HPA database’s immunohistochemistry results and the immunofluorescence staining of clinical samples revealed elevated CD38 expression in tumors, implying its role as a tumor promoter. Current research consensus acknowledges CD38’s ability to dampen the immune response, with anti-CD38 therapeutics employed in multiple myeloma treatment. Within solid tumors, CD38 is commonly perceived to enable tumor cells to evade immunotherapy via immune response suppression (27). However, the regulatory function of CD38 within tumor cells remains elusive. Some studies have reported high CD38 expression in lung cancer, indicating its role as an oncogenic factor (28,29). These findings align with the results of immunofluorescence staining and semi-quantitative analysis in our study. The other study, however, revealed lower CD38 expression in lung cancer, suggesting an inhibitory role (30). These results resonate with the pan-cancer analysis in our study, demonstrating lower CD38 expression in LUAD, LIHC, BRCA, COAD, CHOL, KICH, LUSC, PCPG, PRAD, READ, and THCA. Zucali et al. showed no significant antitumor activity associated with CD38 inhibition (31), and Ng et al. indicated improved prognosis in HCC patients exhibiting high CD38 expression (32). Consequently, we propose that this “contradiction” may arise from the existence of protein interactions or potential regulatory pathways involving CD38 in lung cancer and other solid tumors. Further exploration is warranted to uncover the underlying mechanisms.
In addition, we conducted gene enrichment analysis among patients categorized into high- and low-risk cohorts. Interestingly, the genetic makeup of high-risk cohort patients appeared to be substantially enriched in pathways associated with metabolism. This observation led us to postulate that tumors in the high-risk cohort exhibit greater proliferation, invasiveness, and migratory abilities, thus indicating poorer prognosis. Conversely, the genetic profiles of patients within the low-risk cohort were primarily enriched in immune-related pathways, suggesting a superior immune infiltration status for this cohort. Subsequent analysis of immune cell infiltration within the tumor microenvironment revealed that only plasma cells demonstrated significant infiltration in the low-risk cohort. Nevertheless, the infiltration levels of other immune cells did not differ significantly between the groups, preventing an assessment of immune infiltration abundance via the risk scores. With CD38 showcasing heightened expression in the low-risk cohort, the potential suppression of the immune response could explain this outcome. Analysis of immunotherapy sensitivity indicated a potentially greater therapeutic benefit for the low-risk cohort patients. Further, we conducted consensus clustering analysis on our samples to investigate the prognostic and clinical correlations across various subtypes.
Admittedly, this study bears certain limitations. Our findings, derived primarily from public databases, and validated with a limited number of clinical samples, somewhat undermine the robustness of our model. In future research, we aim to corroborate our model utilizing a larger set of clinical samples. Concurrently, we plan to delve into biomolecular experimentation to unravel the enigma surrounding the “contradiction” of CD38. Further exploration into potential regulatory pathways of CD38 in solid tumors will also be a central focus.
Conclusions
In summary, we established an efficacious protein model designed for the prognosis prediction of LUAD patients. By integrating it with clinicopathologic data, we devised a nomogram, thereby introducing novel tools for clinical application. Our utilization of clinical samples for validation bolstered the practicality of this study. Ultimately, it is our aspiration that this research will serve as a foundation for future explorations into the role of proteins in lung cancer pathogenesis.
Acknowledgments
We thank the TCGA (The Cancer Genome Atlas) database for generously sharing a large amount of data.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1940/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1940/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1940/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1940/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Affiliated Hospital of Hangzhou Normal University [2023(E2)-KS-036] and the requirement for individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837-46. [Crossref] [PubMed]
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Tay JK, Narasimhan B, Hastie T. Elastic Net Regularization Paths for All Generalized Linear Models. J Stat Softw 2023;106:1. [Crossref] [PubMed]
- Wang H, Lengerich BJ, Aragam B, et al. Precision Lasso: accounting for correlations and linear dependencies in high-dimensional genomic data. Bioinformatics 2019;35:1181-7. [Crossref] [PubMed]
- Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59. [Crossref] [PubMed]
- Massion PP, Healey GF, Peek LJ, et al. Autoantibody Signature Enhances the Positive Predictive Power of Computed Tomography and Nodule-Based Risk Models for Detection of Lung Cancer. J Thorac Oncol 2017;12:578-84. [Crossref] [PubMed]
- Dai L, Qu Y, Li J, et al. Serological proteome analysis approach-based identification of ENO1 as a tumor-associated antigen and its autoantibody could enhance the sensitivity of CEA and CYFRA 21-1 in the detection of non-small cell lung cancer. Oncotarget 2017;8:36664-73. [Crossref] [PubMed]
- Ajona D, Pajares MJ, Corrales L, et al. Investigation of complement activation product c4d as a diagnostic and prognostic biomarker for lung cancer. J Natl Cancer Inst 2013;105:1385-93. [Crossref] [PubMed]
- Sozzi G, Boeri M, Rossi M, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol 2014;32:768-73. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Ilie M, Hofman V, Long-Mira E, et al. "Sentinel" circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS One 2014;9:e111597. [Crossref] [PubMed]
- Zhang JT, Qin H, Man Cheung FK, et al. Plasma extracellular vesicle microRNAs for pulmonary ground-glass nodules. J Extracell Vesicles 2019;8:1663666. [Crossref] [PubMed]
- Kim Y, Lee BB, Kim D, et al. Aberrant Methylation of SLIT2 Gene in Plasma Cell-Free DNA of Non-Small Cell Lung Cancer Patients. Cancers (Basel) 2022;14:296. [Crossref] [PubMed]
- Yan P, Han Y, Tong A, et al. Prognostic value of neuron-specific enolase in patients with advanced and metastatic non-neuroendocrine non-small cell lung cancer. Biosci Rep 2021;41:BSR20210866. [Crossref] [PubMed]
- Li L, Zhang Q, Wang Y, et al. Evaluating the diagnostic and prognostic value of serum TuM2-PK, NSE, and ProGRP in small cell lung cancer. J Clin Lab Anal 2023;37:e24865. [Crossref] [PubMed]
- Sun L, Shao Q. Expression changes and clinical significance of serum neuron-specific enolase and squamous cell carcinoma antigen in lung cancer patients after radiotherapy. Clinics (Sao Paulo) 2023;78:100135. [Crossref] [PubMed]
- Bes-Scartezini F, Saad R Junior. Prognostic assessment of tumor markers in lung carcinomas. Rev Assoc Med Bras (1992) 2022;68:313-7. [PubMed]
- Guo S, Chen J, Hu P, et al. The Value of Circulating Tumor Cells and Tumor Markers Detection in Lung Cancer Diagnosis. Technol Cancer Res Treat 2023;22:15330338231166754. [Crossref] [PubMed]
- Yu D, Du K, Liu T, et al. Prognostic value of tumor markers, NSE, CA125 and SCC, in operable NSCLC Patients. Int J Mol Sci 2013;14:11145-56. [Crossref] [PubMed]
- Peltonen R, Österlund P, Lempinen M, et al. Postoperative CEA is a better prognostic marker than CA19-9, hCGβ or TATI after resection of colorectal liver metastases. Tumour Biol 2018; [Crossref] [PubMed]
- Yang Y, Xu M, Huang H, et al. Serum carcinoembryonic antigen elevation in benign lung diseases. Sci Rep 2021;11:19044. [Crossref] [PubMed]
- Jasper K, Stiles B, McDonald F, et al. Practical Management of Oligometastatic Non-Small-Cell Lung Cancer. J Clin Oncol 2022;40:635-41. [Crossref] [PubMed]
- Chen L, Diao L, Yang Y, et al. CD38-Mediated Immunosuppression as a Mechanism of Tumor Cell Escape from PD-1/PD-L1 Blockade. Cancer Discov 2018;8:1156-75. [Crossref] [PubMed]
- Bu X, Kato J, Hong JA, et al. CD38 knockout suppresses tumorigenesis in mice and clonogenic growth of human lung cancer cells. Carcinogenesis 2018;39:242-51. [Crossref] [PubMed]
- Gao L, Liu Y, Du X, et al. The intrinsic role and mechanism of tumor expressed-CD38 on lung adenocarcinoma progression. Cell Death Dis 2021;12:680. [Crossref] [PubMed]
- Karimi-Busheri F, Rasouli-Nia A, Zadorozhny V, et al. CD24+/CD38- as new prognostic marker for non-small cell lung cancer. Multidiscip Respir Med 2013;8:65. [Crossref] [PubMed]
- Zucali PA, Lin CC, Carthon BC, et al. Targeting CD38 and PD-1 with isatuximab plus cemiplimab in patients with advanced solid malignancies: results from a phase I/II open-label, multicenter study. J Immunother Cancer 2022;10:e003697. [Crossref] [PubMed]
- Ng HHM, Lee RY, Goh S, et al. Immunohistochemical scoring of CD38 in the tumor microenvironment predicts responsiveness to anti-PD-1/PD-L1 immunotherapy in hepatocellular carcinoma. J Immunother Cancer 2020;8:e000987. [Crossref] [PubMed]


