
CD112 is an epithelial-to-mesenchymal transition-related and immunological biomarker in pan-cancer
Introduction
CD112, known as poliovirus receptor-related 2 (PVRL2) or nectin-2, is a member of the nectin family that is predominantly found in epithelial adhesion junctions (1). Some kinds of cells, including neurons, epithelial cells, fibroblasts, and endothelial cells, express CD112 (2). Previous studies indicate that CD112 plays its immunological roles via interacting with multiple receptors (3). Shibuya and Tahara-Hanaoka have confirmed that CD112 can react with two co-inhibitory ligands CD112 receptor (CD112R) and T cell immunoreceptor with Ig and ITIM domains (TIGIT) to impede T cell proliferation (4,5). Compared with CD112R, TIGIT shows a weak affinity for CD112 (6). Blockade of either TIGIT or CD112R solely promotes cytokine production and cell division in CD4 positive (CD4+) T cells slightly. CD112R inhibition and TIGIT inhibition, however, result in a sharply increase in CD4+ T cell diffusion (7). In particular, CD112R blockade enhances cytotoxicity and CD8 positive (CD8+) T cells expansion (8). Furthermore, CD112R and programmed cell death protein 1 (PD-1) blockade have less of an impact on T cell activation than anti-CD112R and anti-TIGIT combined (7,8).
In recent years, there is an accumulating body of literature on the function of CD112 in solid tumors, and Oshima et al. discovered that CD112 acted in tumor cell adhesion, proliferation, and lymph node metastasis in ovarian cancer (OV) (9). Following research has indicated a link between increased CD112 manifestation and an unfavorable prognosis and aggressiveness in bladder urothelial carcinoma (BLCA) (7,10) and acute myelocytic leukemia (AML) (11). Murter et al. found that melanoma and colon cancer tumor development stopped in CD112R defective mice models, indicating that the CD112R-CD112 pathway is essential for regulating the destruction of tumor cells (12).
Even though immune checkpoint inhibitors (ICIs) have been proven in treating a number of types of cancer effectively in recent years, many cancer patients do not respond to them with acquired resistance (13). Effector T cells may be depleted and dysfunctional in the tumor microenvironment (TME) under these circumstances (14,15), and T cells with dysfunctional surface receptors, including cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) and PD-1, have more co-inhibitory receptors. These facts suggest that the identification of new immune checkpoint pathways would enhance immunotherapy’s response rate and increase its efficacy. CD112 is one of the ligands for the immune checkpoint TIGIT, and the interruption of the interaction between TIGIT and CD112 represents a current strategy in cancer immunotherapy (16). Therefore, CD112 may serve as a potential target for ICIs.
Epithelial-to-mesenchymal transition (EMT) is a dynamic cellular process where epithelial cells, characterized by cell-cell adhesion and apical-basal polarity, turn into mesenchymal cells that exhibit increased motility and invasive properties. This transition involves the downregulation of epithelial markers such as E-cadherin and the upregulation of mesenchymal markers such as N-cadherin and vimentin (17). CD112, a cell adhesion molecule implicated in diverse cellular processes, may interact with these pathways, potentially influencing the EMT process and cancer progression. However, the correlation between CD112 and EMT pathway has not been reported.
Though CD112 has been investigated in multiple tumors (1,18), however, its clinical implications and immunological roles in most other tumors remain to be elucidated. Our findings corroborate that CD112 is an EMT-related pan-cancer prognostic biomarker linked to the immunological microenvironment and offer a novel avenue for further exploration of the mechanism of CD112 in tumor immunity. We present this article in accordance with the REMARK and MDAR reporting checklists (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2258/rc).
Methods
Survival and prognostic analysis
The Cancer Genome Atlas (TCGA) database provided clinical details on the various cancers. The association between CD112 expression and the overall survival (OS) and progression-free interval (PFI) of patients, which were displayed as Kaplan-Meier (KM) curves, was examined using the KM plotter database (http://kmplot.com/analysis/), and the P value is determined using univariate survival analysis. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
The expression of CD112 in malignancies
The EdgeR software is applied to analyze the variations in CD112 expression levels between tumor and normal tissues. The R application ggplot is employed to generate violin plots. A representative immunohistochemistry (IHC) image for CD112 was taken from https://www.proteinatlas.org/, The Human Protein Atlas (HPA). The expression levels of the CD112 gene in cancer patients and healthy people were downloaded from UALCAN (http://ualcan.path.uab.edu/analysis.html).
Correlation analysis of CD112 with the immune microenvironment
The tumor purity was examined using the R software program estimate. Using immune and stromal scores based on the Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) can simplify the evaluation of immunological and stromal components in malignancies. This technique computes immune and stromal scores by examining specific gene expression traits of immune and stromal cells in order to predict the invasion of non-tumor cells. Data were also acquired from the Tumor Immune Evaluation Resource (TIMER) database to investigate the association between CD112 expression and 21 markers of immune cell subsets.
Immune checkpoint and neoantigen
Each tumor sample’s neoantigen count is calculated independently using ScanNeo, and the relationship between the expression of CD112 and neoantigen count is examined. Additionally, the correlations of expression between CD112 and immune checkpoint genes that are shared are investigated. The expression of each of these immune checkpoint genes is separated, and a connection between it and CD112 is computed.
Mismatch repairs (MMRs) and methyltransferase
MMR proteins are an intracellular MMR system that causes high levels of somatic mutations when critical gene activities are lost and DNA replication mistakes cannot be corrected (19). The relationship between mutations and gene expression was investigated in this work using the five MMR genes MutL homolog 1, MutS homolog 2, MutS homolog 6, postmeiotic segregation increased 2, and epithelial cell adhesion molecule (EPCAM). Gene expression was altered without the DNA sequence changing as a result of the chemical modification of DNA known as DNA methylation.
Gene set enrichment analysis (GSEA)
GSEA makes use of gene-set data to find significant correlations between gene sets and the degree to which they are expressed in particular functional gene sets. This method was used to investigate the connections between cellular components, molecular processes, and gene expression. A lot of information about gene function and molecular networks may be found in the Kyoto Encyclopedia of Genes and Genomes (KEGG). We used GSEA to evaluate the enrichment of genes in the high CD112 expression group utilizing the KEGG and HALLMARK pathways (http://www.gsea-msigdb.org/gsea/index.jsp).
Cell lines and cell culture
The normal intestinal epithelial cells (NCM460; RRID:CVCL_0460) and human colorectal cancer (CRC) cell lines SW1116 (RRID:CVCL_0544), DLD1 (RRID:CVCL_KV93), SW480 (RRID:CVCL_0546), HCT116 (RRID:CVCL_XC97), and are all purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). In full Dulbecco’s modified Eagle medium (DMEM) media with 10% fetal bovine serum (FBS) (Biological Industries), NCM460, SW480, and DLD1 cells were grown. Complete Roswell Park Memorial Institute (RPMI) medium was used to grow SW1116. Complete McCoy’s 5 A medium was used to cultivate HCT116. Every 1 to 2 days, the culture media were updated.
Real-time quantitative polymerase chain reaction (RT-qPCR)
The RNA was extracted from CRC cell lines using TRIzol (Invitrogen, Carlsbad, CA, USA), and cDNA was produced using SuperScript® III Reverse Transcriptase (Invitrogen). Then, using an ABI 7500 quantitative PCR apparatus, RT-qPCR is performed (TaKaRa, Dalian, China). Relative the expression of CD112 was calculated by the 2−ΔΔCT method. Primer sequences were synthesized by Qixiang Biotech Company (Nantong, China) and were listed below: CD112-forward (F): ACTCAGGTGTCAGGGACCCT; CD112-reverse (R): TCCACTTTGCAGGTGACCGT.
Western blot analysis
The radioimmunoprecipitation assay (RIPA) buffer which includes a combination of phenylmethylsulfonyl fluoride (PMSF), phosphatase inhibitors, and protease inhibitors were applied to cleave the clinical tissues and CRC cells. Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the same quantity of protein was transferred to a polyvinylidene fluoride (PVDF) membrane. To stop non-specific binding, the membrane was blocked with 5% skim milk for 2 hours at the ambient temperature. The following primary antibodies were subsequently applied to the membranes and incubated for an additional overnight period at 4 ℃: CD112 (1:1,000, Abcam, Cambridge, UK), N-cadherin (1:2,000, Proteintech, Wuhan, China), E-cadherin (1:2,000, Proteintech), and matrix metalloproteinase-9 (MMP-9; 1:2,000, Proteintech), vimentin (1:2,000, Proteintech), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:10,000, Proteintech). The secondary antibody (goat anti-mouse or goat anti-rabbit enzyme-labeled) was given by the Proteintech Company (1:10,000).
Cell counting kit-8 (CCK-8) assay
Cell proliferation was detected using the CCK-8 method (Dojindo, Kumamoto, Japan). 48 hours after siRNA transfection, cells were plated at a density of 3,000 cells per well and injected in 96 wells in 100 mL of medium. Ten µL of CCK-8 was added to each well at 0, 24, 48, 72, and 96 hours, and the wells underwent incubation for 2 hours at 37 ℃. At 450 nm, the optical density (OD) values were calculated.
CD112 interference
Negative control small interfering (si)RNA (si-NC) and siRNA against CD112 are purchased from GenePharma Ltd. (Shanghai, China). Using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA), transient transfection is performed as directed by the manufacturer. SW1116 and DLD1 cells were transfected with si-NC or si-CD112 after growing to 40–60% confluence in six-well plates and having undergone two dishes in phosphate-buffered saline (PBS). The siRNA sequences are listed below: CD112-790-F: GACGAGGGCAACUACACUUTT, R: AAGUGUAGUUGCCCUCGUCTT; CD112-1000-F: GCCAAAGAGACUCAGGUGUTT, R: ACACCUGAGUCUCUUUGGCTT.
Colony formation assay
Six-well plates were used to plate cells (500 cells/well) following transfection. Colonies were visible in culture after 10 to 15 days. After being fixed with 4% paraformaldehyde for 30 minutes, cell colonies were stained with 0.5% crystal violet for 5 minutes. Extra staining solution was then washed away with PBS.
5-ethynyl-2-deoxyuridine (EdU) assay
The transfected cells were then fixed using a 4% polyvinyl alcohol solution after being grown in media containing EdU. The permeabilization process Using 0.5% Triton X-100 solution, cells were made permeable so that antibodies may identify and bind EdU. Anti-EdU antibodies were used in cells to create EdU-DNA-antibody complexes. The cells were then viewed under the fluorescent microscope (Olympus, Japan) after being stained with a secondary antibody that is labeled with luciferase.
Cell migration assay
Transwell plates (8.0-mm in diameter) were used to evaluate cell migration (Corning, NY, USA). A total of 5×104 CRC cells were resuspended in 200 µL of serum-free media in the upper chamber and 500 µL of complete medium containing 20% FBS in the lower chamber at 48 hours after transfection. The cells were cultured for 48 hours. The Image J program was used to count the number of cells that made it past the membrane in three randomly chosen fields.
Statistical analysis
The Student’s t-test was used to analyze gene expression. The survival study made use of the Cox proportional risk regression model, the KM curve, and the log-rank test. Tumor mutation burden (TMB) and microsatellite instability (MSI) were analyzed simultaneously using the Spearman test. The aforementioned research studies, which used the R package (version 4.0.3) and the R language software packages (ggplot2, clusterProfiler, GSVA, and enrichPlot), were deemed statistically significant if P<0.05.
Results
CD112 is abnormally expressed in tumor tissue
The CD112 expression differences of 27 tumors from the GTEx database and TCGA database are shown in Figure 1A, and the results indicated that in adrenocortical carcinoma (ACC), breast invasive carcinoma (BRCA), OV, BLCA, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), rectum adenocarcinoma (READ), colon adenocarcinoma (COAD), cholangiocarcinoma (CHOL), esophageal carcinoma (ESCA), brain lower grade glioma (LGG), glioblastoma multiforme (GBM), lung adenocarcinoma (LUAD), uterine corpus endometrial carcinoma (UCEC), liver hepatocellular carcinoma (LIHC), prostate adenocarcinoma (PRAD), thyroid carcinoma (THCA), pancreatic adenocarcinoma (PAAD), stomach adenocarcinoma (STAD), and uterine carcinosarcoma (UCS), CD112 expression was much higher, indicating that most malignancies have unusually high levels of this protein when compared to normal tissue. In addition, in COAD (N=327, P<0.001), LIHC (N=421, P<0.001), BRCA (N=1,211, P<0.001), and STAD (N=449, P<0.001) patients, most patients’ genes expressed CD112 at higher levels (Figure 1B-1E). Representative IHC photos from the HPA database support these findings (Figure 1F-1I). Next, we selected six pairs of CRC and paracancerous tissue specimens to determine CD112 expression by western blotting which revealed that CD112 was expressed in cancer tissues from CRC patients substantially (Figure 1J). Moreover, PCR and western blot assays suggested that CD112 expressed highly in intestinal cancer cells compared with NCM460 (Figure 1K,1L).
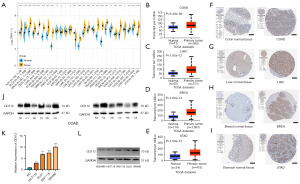
High CD112 expression is correlated with unfavorable outcomes across tumors
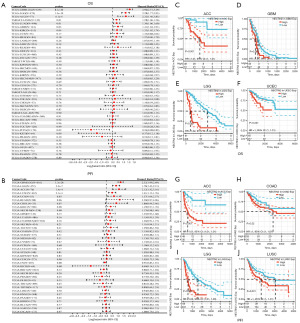
By employing TCGA RNA sequencing (RNA-seq), we evaluated the prognostic implications of CD112 in 44 TCGA cancer types. Utilizing the clinical data from UCSC Xena (Figure 2A), we found that worse OS is related to increased CD112 expression in glioblastoma and low-grade glioma (GBMLGG) [hazard ratio (HR) =2.96; 95% confidence interval (CI): 2.51–3.49; P=8.1e−40], LGG (HR =2.28; 95% CI: 1.77–2.93; P=1.3e−10), and ACC (HR =2.32; 95% CI: 1.40–3.83; P=6.1e−4). Conversely, greater levels of CD112 expression are associated with superior prognosis in pheochromocytoma and paraganglioma (PCPG) (HR =0.26; 95% CI: 0.08–0.82; P=0.02). Furthermore, a forest plot of the 38 different TCGA tumor PFI and gene expression is presented in Figure 2B. Similar to the OS analysis, the higher expression of CD112 is correlated with a poorer PFI significantly in the GBMLGG (HR =2.32; 95% CI: 1.99–2.69; P=1.3e−28), LGG (HR =1.78; 95% CI: 1.43–2.21; P=2.3e−7), ACC (HR =2.12; 95% CI: 1.42–3.16; P=1.41e−4), and lung squamous cell carcinoma (LUSC) (HR =1.41; 95% CI: 1.13–1.76; P=2.1e−3). In contrast, the lower PFI in kidney chromophobe (KICH) is associated with lower CD112 expression (HR =0.40; 95% CI: 0.14–1.15; P=0.08). Subsequent OS analysis of the data revealed that CD112 is a risk factor in ACC (N=79, P<0.001), GBM (N=160, P<0.001), LGG (N=509, P<0.001), and UCEC (N=542, P<0.001) (Figure 2C-2F). Furthermore, the KM survival analysis revealed that the expression of CD112 adversely impacted the PFI of patients in ACC (N=79, P<0.001), COAD (N=453, P=0.02), LGG (N=509, P<0.001), and LUSC (N=496, P=0.001) (Figure 2G-2J). These results implied that the expression of CD112 is significantly linked to a poor prognosis in ACC, LGG, and UCEC. In conclusion, high CD112 expression is related to poor prognosis in pan-cancer.
Correlation between CD112 and immune infiltration
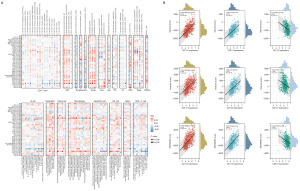
Next, we obtained the correlation data of CD112 in 33 distinct cancer types and 21 independent immune invasion cells from the TIMER database and examined the interaction between gene expression and the scores of these immune cells in order to look into the connections between immune invasion and CD112 expression in multiple kinds of malignancies (Figure 3A). In contrast to CD4+ T cell, T cell, B cell, and CD8+ T cell, our results showed a substantial negative connection between CD112 expression and cancer-associated fibroblast (CAF), endo, myeloid-derived suppressor cell (MDSC), neutrophil, and monocyte. This suggested that CD112 contributes substantially to the immune infiltration. Subsequently, through tumor purity analysis, we discovered a positive correlation between CD112 expression and both GBMLGG (N=656, R=0.61, P=2.2e−69) and LGG (N=504, R=0.55, P=6.3e−41) within the StormalScore. Conversely, a negative association was found with PRAD (N=495, R=−0.32, P=2.2e−13). In the context of ImmuneScore, CD112 expression displayed a positive correlation with GBMLGG (N=656, R=0.54, P=2.5e−51) and LGG (N=504, R=0.49, P=5.9e−32), but a negative link with THCA (N=503, R=−0.26, P=5.2e−9). Similarly, within the ESTIMATEScore, CD112 expression was positively associated with GBMLGG (N=656, R=0.59, P=8.3e−62) and LGG (N=504, R=0.53, P=1.7e−37), while negatively correlated with PRAD (N=495, R=−0.29, P=2.2e−11) (Figure 3B). Overall, our results demonstrate that CD112 may have a role in the malignancies’ immune infiltration.
CD112 expression in pan-cancer is associated with TMB and MSI
TMB is a promising indicator of immunotherapy response for a variety of cancer types (20). We found that CD112 is positively connected with TMB in THYM (P=1.8e−17), PAAD (P=4.4e−5), LUAD (P<0.001), LGG (P=8.5e−5), STAD (P=0.01), head and neck squamous cell carcinoma (HNSC) (P=0.03), ESCA (P=1.3e−5), BLCA (P=1.8e−5), and COAD (P=0.003), and negatively associated with THCA (P=1.5e−6), BRCA (P=2.1e−12), and CESC (P=0.02) (Figure 4A).
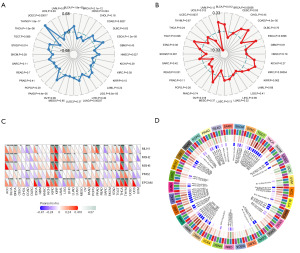
In tumor tissue, faulty DNA MMR function leads to MSI (21). Fifteen percent of CRC silence MSI by epigenetic MLH1 silencing or a germline mutation in one of the MMR genes (22). Then we next assessed the connections between CD112 and MSI, and the consequences suggested that in BLCA (P=0.01), testicular germ cell tumors (TGCT) (P=0.03), OV (P=0.02), kidney renal clear cell carcinoma (KIRC) (P<0.001), ESCA (P=0.01), and COAD (P=4.3e−5), CD112 is associated with MSI positively. In contrast, CD112 is negatively related to UCS (P=0.01), UCEC (P=0.004), READ (P=0.03), and BRCA (P<0.001) (Figure 4B).
MMR genes and the expression of DNA methyltransferase
MMR genes are essential for preserving genomic integrity, when important genes in this mechanism stop working, errors appear in DNA replication and results in greater somatic mutation rates and cancer (23,24). Next, in order to learn more about CD112’s role in the development of cancer, we looked at the relationship between its expression and the genes involved in MMR in pan-cancer. As illustrated in Figure 4C, it indicated that CD112 expression is positively related to MMR gene mutation levels in 11 cancers, including LIHC, BLCA, ACC, kidney renal papillary cell carcinoma (KIRP), THCA, OV, LUAD, PRAD, KIRC, PCPG, and UCEC. Conversely, in COAD and sarcoma (SARC), CD112 expression exhibited a negative relationship with MMR gene mutation levels.
DNA methylation is of an alteration of epigenetic gene that can change gene expression (25). One crucial element in cancer development is the alteration of DNA methylation state (26). To further determine the role of CD112 expression in DNA methylation, we evaluated its correlation with four DNA methyltransferases. As depicted in Figure 4D, CD112 expression levels are associated with these four kinds of DNA methyltransferases conspicuously, particularly in LIHC, LGG, KIRP, BRCA, THCA, and TGCT. In brief, these findings suggest that CD112 may mediate the repairment of DNA methylation to regulate the progression of malignancies.
Immune checkpoint genes and immune neoantigens
Cancer immunotherapies that target adaptive immunological checkpoints in recent years have markedly improved patient outcomes for metastatic and treatment-resistant cancer types (27). As shown in Figure 5A, we compiled expression data from over 40 distinct genes to examine the connections between CD112 and immune checkpoint genes. These findings demonstrate that CD112 is positively connected to immune checkpoint genes in a number of malignancies, including LGG, GBM, KIRP, PCPG, and KIRC.
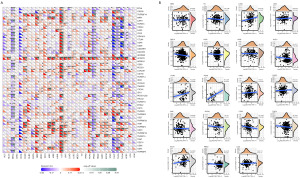
Neoantigens for cancer are antigens produced by somatic mutations found in specific malignancies (28). On account of immunogenicity and the lack of expression in healthy tissues, neoantigens are regarded as crucial targets for cancer immunotherapy (29). In 19 tumors, we assessed the association between CD112 expression and neoantigens (Figure 5B), and the results revealed that in LUAD (R=0.161, P<0.05), READ (R=0.278, P<0.05), and BLCA (R=0.209, P<0.05), CD112 expression is positively linked with neoantigen. Collectively, these results suggest the potential of CD112 as an immunodiagnostic biomarker.
Knockdown of CD112 inhibits the proliferation and invasive ability of CRC cells
To elucidate the function of CD112 in CRC, siRNA-mediated knockdown of CD112 is employed in CRC cells SW1116 and DLD1 and the efficiency is subsequently verified using RT-qPCR (Figure 6A). Subsequently, the CCK8 kit is used to assess changes in cellular activity in the negative vector group and the two si-CD112 transfected groups at different time points after transfection. Results revealed that, after 48 hours of knocking down CD112, the cell proliferation ability is significantly attenuated (Figure 6B). Colony formation assays and EdU studies showed that CD112 knockdown reduced CRC cell proliferation significantly (Figure 6C,6D). Additionally, results from the Transwell migration assay and wound healing assay showed that intestinal cancer cells’ ability to migrate was restricted when CD112 was knocked down (Figure 6E,6F). Collectively, our findings imply that CD112 may play a key role in CRC cell proliferation and migration, and that CD112 knockdown inhibits the growth of intestinal cancer cells.
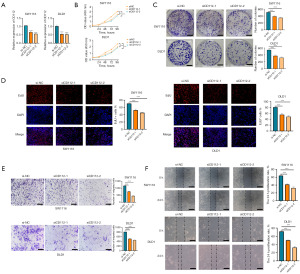
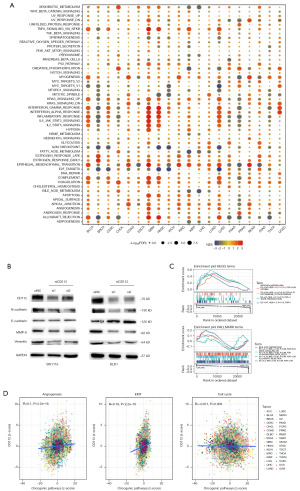
Gene enrichment of CD112 for pathway analysis
After examining the interaction between CD112 and different signaling pathways, we found an association between CD112 and EMT in several tumors (Figure 7A). The EMT is a key process in the progression of CRC and is marked by the increased expression of N-cadherin and the decreased expression of E-cadherin. Vimentin and MMP-9 are also important markers of this transition (30,31). In particular, MMP-9 is involved in the degradation of extracellular matrix, facilitating cell migration and invasion (32). Therefore, a decrease in the expression levels of N-cadherin, MMP-9, and vimentin, along with an increase in E-cadherin, suggests a suppression of EMT. Then we conducted a western blotting assay on intestinal cancer cell lines SW1116 and DLD1 after knocking down CD112. The assay results showed reduced protein expression of N-cadherin, E-cadherin, MMP-9, and vimentin, suggesting that decreased CD112 expression could inhibit EMT in CRC cells as depicted in Figure 7B. Furthermore, we utilized enrichment analysis to determine the most significant pathways associated with CD112 expression. As demonstrated in Figure 7C, the top pathways identified are nitrogen metabolism, asthma, autoimmune thyroid, and taste transduction in KEGG pathways, and bile acid metabolism, KRAS signaling, pancreas beta cells, and allograft rejection in HALLMARK pathways. Moreover, we utilized the z-score algorithm and examined the correlation between CD112 expression and pathway activity, including angiogenesis, EMT, and cell cycle. This analysis uncovered a positive correlation between CD112 and angiogenesis and EMT, reinforcing our above findings (Figure 7D).
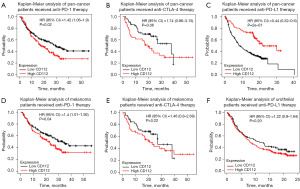
Relevance of CD112 and immunotherapy
To investigate whether CD112 acts in the context of cancer immunotherapy, our research made use of KM plotter database, which revealed that among various malignancies, those with high CD112 expression had a worse OS rate when treated with anti-PD-1 and anti-CTLA-4 treatment. Conversely, in the case of anti-programmed death ligand 1 (PD-L1) therapy, a better prognosis is observed (Figure 8A-8C). This pattern is most prominent in melanoma and urothelial, where individuals with higher CD112 expression tended to have poorer OS with anti-PD-1 and respectively anti-PD-L1 treatments (Figure 8D-8F). It demonstrates a significant association between CD112 and the outcome of immunotherapy patients.
Discussion
Immunotherapy has elicited a revolution in oncology within the past few years, yet due to the multifariousness of each patient’s anti-TME, this form of treatment is only applicable to a limited number of cancers. Consequently, biomarkers to anticipate patient response to immunotherapy have become increasingly relevant. Through this research we determined that high CD112 expression is linked with poor prognosis, both in OS and PFI, and CD112 negatively correlates with the CD4+ T cell, CD8+ T cell, and B cell in immune microenvironment. The TIMER database indicates a positive correlation between CD112 expression and MDSCs. MDSCs, a diverse group of myeloid origin cells, are known for their prominent role in suppressing immune responses, chiefly those relating to T and B cells (33,34). This immunosuppression creates a conducive environment for tumor growth and spread. The overexpression of CD112 might stimulate MDSC proliferation, thereby enhancing their immunosuppressive activities. Consequently, the immune system’s ability to mount an effective anti-tumor response is compromised. B cells, for instance, are hindered in their antibody production role, while T cells’ cytotoxic actions against tumor cells are weakened. Furthermore, MDSCs can induce regulatory T cells’ expansion, another cell type known for its immunosuppressive abilities, amplifying the effect (35). Hence, elevated expression of CD112, leading to increased MDSC activity, could be associated with malignant tumor progression and poorer prognosis due to a weakened immune response and enhanced tumor-promoting factors. Immunological checkpoints are crucial regulatory nodes within the immune system, intricately controlling the balance between immune activation and tolerance to self (36). They function by inhibiting overactive immune responses, preventing autoimmunity and maintaining immunological homeostasis (37,38). However, cancer cells exploit these checkpoints, gaining the ability to evade immune recognition and destruction, thus facilitating tumor growth and progression (39). In the context of our results, we investigated the correlation between CD112 expression and multiple immune checkpoint genes. Remarkably, our findings indicate that CD112 expression is positively associated with immune checkpoint genes in several malignancies, including LGG, GBM, KIRP, PCPG, and KIRC. This suggests that CD112 may potentially play a critical role in the immune evasion strategies employed by these tumor types. Neoantigens are highly immunogenic, specific, and abnormal proteins (40) for gene point mutation (41), deletion mutation, gene fusion, and so on (42). CD112 is positively correlated with neoantigens in LUAD, BLCA, READ, and COAD. To sum up, CD112 may be a viable therapeutic target for clinical application.
The therapeutic effectiveness of ICIs can be predicted by TMB (43), which reflects the mutations in tumor tissue (44). MSI is known as modifications to the length of the microsatellite sequence brought on by insertional or deletional mutations during DNA replication (45). DNA polymerase may result in the deletion or insertion of nucleotides at microsatellite sites during DNA replication (46). This process can be identified and repaired by the MMR system, which causes MSI when the MMR gene loses function due to hypermethylation of the promoter region or mutations in the coding region (47,48). This research revealed that CD112 markedly associates with MSI in 11 tumors, as well as five MMR genes in 11 malignancies. Additionally, TMB and four methyltransferases are both substantially correlated with CD112 expression in nine malignancies and six tumors, respectively. Moreover, the findings of the enrichment analysis showed CD112 relates to KRAS and EMT pathway. EMT is a biological process characterized by the transition of epithelial cells into mesenchymal stem cells, and this process is critical in numerous physiological and pathological settings, including embryogenesis, tissue remodeling, wound healing, and most notably, cancer progression (49). The hallmark of EMT in cancer progression is the loss of cell adhesion, increased cell mobility, and invasiveness, contributing to the metastatic potential of cancer cells (30). In our study, we observed that knockdown of CD112 led to an increase in E-cadherin protein expression, and a reduction in N-cadherin, MMP-9, and vimentin protein expression. E-cadherin is a key player in maintaining epithelial integrity and inhibiting EMT, while N-cadherin, MMP-9, and vimentin are recognized as mesenchymal markers, associated with increased cell motility and invasiveness. This inverse relationship between CD112 and E-cadherin, coupled with the downregulation of N-cadherin, MMP-9, and vimentin, could suggest that CD112 plays a role in promoting EMT in cancer progression. CD112 might facilitate the acquisition of mesenchymal traits by cancer cells, thus enhancing their motility, invasiveness, and metastatic potential. Collectively, aberrant CD112 expression in cancer is linked to EMT, TMB, MSI, MMR, and DNA methyltransferase, further fueling the metastasis and progression of malignancies.
By comparing the similarities and differences among diverse malignancies (50), we can get new cognition about cancer avoidance and unique tumor biomarkers by means of the pan-cancer study (32,51). Comprehensive evaluation of the CD112 expression in human malignancies revealed abnormally elevated levels in ACC, BLCA, BRCA, CESC, CHOL, COAD, ESCA, GBM, LGG, LIHC, LUAD, OV, PAAD, PRAD, SKCM, STAD, THCA, UCEC, and UCS compared with normal tissue. Furthermore, no matter OS or PFI, highly expressed CD112 indicates a poor prognosis in tumors. The IHC results of COAD, LIHC, BRCA, and STAD from HPA supported the elevated expression of CD112 in tumor cells. Moreover, RT-qPCR and western blotting experiments confirmed CD112 overexpresses in multiple CRC cell lines (HCT116, SW1116, DLD1, and SW480) compared with normal intestinal epithelial cells (NCM460). Additionally, clinical studies indicated that patients who are treated with anti-PD-1 and anti-CTLA-4 have lower OS rates when their CD112 expression is high. Together, our findings suggest that CD112 may be an invaluable cancer biomarker for promoting the effectiveness of immune therapy and early cancer identification.
Finally, we selected the intestinal cancer cell lines SW1116 and DLD1 and conducted CD112 knockdown using siRNA, verified with RT-qPCR, to assess the effect on cell functions. The CCK-8 assay, cell clone formation, and EdU assays all indicated that the proliferation of intestinal cancer cells is inhibited upon CD112 knockdown. Transwell and wound healing assays further suggested that the capacity of intestinal cancer cells to invade and migrate is markedly diminished with suppressed expression of CD112. This phenomenon might be associated with the role of CD112 as an adhesive molecule, which is considered to be involved in the proliferation of tumor cells.
Conclusions
In summary, our analysis has demonstrated that the expression of CD112 is strongly correlated with poor prognosis and is significantly higher in pan-cancer tissues compared to nearby tissues. Furthermore, through KEGG, Gene Ontology (GO), GSEA analyses, and western blot assays, it has been revealed that CD112 is involved in EMT pathway in cancer progression. Cell function experiments have also indicated that the knockdown of CD112 can effectively inhibit the growth and migration of intestinal cancer cells. Additionally, it has been discovered that the immune microenvironment and CD112 expression are adversely associated, and clinical data have demonstrated that patients with high CD112 expression had a lower OS rate when treated with anti-PD-1 and anti-CTLA-4 therapy. When considered collectively, our results strongly imply that CD112 may function as a promising independent predictor and therapeutic target for cancer patients.
Acknowledgments
We appreciate the clinical biobank platform provided by the Affiliated Hospital of Nantong University for its unwavering support of our investigation.
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK and MDAR reporting checklists. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2258/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2258/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2258/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2258/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Huang X, Qu P, Chen Y, et al. Low expression of CD112 is associated with poor overall survival in patients with hepatocellular carcinoma. Hum Pathol 2014;45:1944-50. [Crossref] [PubMed]
- Lin L, Pang W, Jiang X, et al. Light amplified oxidative stress in tumor microenvironment by carbonized hemin nanoparticles for boosting photodynamic anticancer therapy. Light Sci Appl 2022;11:47. [Crossref] [PubMed]
- Huang K, Lui WY. Nectins and nectin-like molecules (Necls): Recent findings and their role and regulation in spermatogenesis. Semin Cell Dev Biol 2016;59:54-61. [Crossref] [PubMed]
- Shibuya K, Shirakawa J, Kameyama T, et al. CD226 (DNAM-1) is involved in lymphocyte function-associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J Exp Med 2003;198:1829-39. [Crossref] [PubMed]
- Tahara-Hanaoka S, Shibuya K, Onoda Y, et al. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int Immunol 2004;16:533-8. [Crossref] [PubMed]
- Deuss FA, Gully BS, Rossjohn J, et al. Recognition of nectin-2 by the natural killer cell receptor T cell immunoglobulin and ITIM domain (TIGIT). J Biol Chem 2017;292:11413-22. [Crossref] [PubMed]
- Zhu Y, Paniccia A, Schulick AC, et al. Identification of CD112R as a novel checkpoint for human T cells. J Exp Med 2016;213:167-76. [Crossref] [PubMed]
- Whelan S, Ophir E, Kotturi MF, et al. PVRIG and PVRL2 Are Induced in Cancer and Inhibit CD8(+) T-cell Function. Cancer Immunol Res 2019;7:257-68. [Crossref] [PubMed]
- Oshima T, Sato S, Kato J, et al. Nectin-2 is a potential target for antibody therapy of breast and ovarian cancers. Mol Cancer 2013;12:60. [Crossref] [PubMed]
- Miao X, Yang ZL, Xiong L, et al. Nectin-2 and DDX3 are biomarkers for metastasis and poor prognosis of squamous cell/adenosquamous carcinomas and adenocarcinoma of gallbladder. Int J Clin Exp Pathol 2013;6:179-90. [PubMed]
- Stamm H, Klingler F, Grossjohann EM, et al. Immune checkpoints PVR and PVRL2 are prognostic markers in AML and their blockade represents a new therapeutic option. Oncogene 2018;37:5269-80. [Crossref] [PubMed]
- Murter B, Pan X, Ophir E, et al. Mouse PVRIG Has CD8(+) T Cell-Specific Coinhibitory Functions and Dampens Antitumor Immunity. Cancer Immunol Res 2019;7:244-56. [Crossref] [PubMed]
- Schoenfeld AJ, Hellmann MD. Acquired Resistance to Immune Checkpoint Inhibitors. Cancer Cell 2020;37:443-55. [Crossref] [PubMed]
- Scharping NE, Menk AV, Moreci RS, et al. The Tumor Microenvironment Represses T Cell Mitochondrial Biogenesis to Drive Intratumoral T Cell Metabolic Insufficiency and Dysfunction. Immunity 2016;45:374-88. [Crossref] [PubMed]
- Schietinger A, Philip M, Krisnawan VE, et al. Tumor-Specific T Cell Dysfunction Is a Dynamic Antigen-Driven Differentiation Program Initiated Early during Tumorigenesis. Immunity 2016;45:389-401. [Crossref] [PubMed]
- Chauvin JM, Zarour HM. TIGIT in cancer immunotherapy. J Immunother Cancer 2020;8:e000957. [Crossref] [PubMed]
- Loh CY, Chai JY, Tang TF, et al. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019;8:1118. [Crossref] [PubMed]
- Karabulut M, Gunaldi M, Alis H, et al. Serum nectin-2 levels are diagnostic and prognostic in patients with colorectal carcinoma. Clin Transl Oncol 2016;18:160-71. [Crossref] [PubMed]
- Fishel R. Mismatch repair. J Biol Chem 2015;290:26395-403. [Crossref] [PubMed]
- Spurr LF, Weichselbaum RR, Pitroda SP. Tumor aneuploidy predicts survival following immunotherapy across multiple cancers. Nat Genet 2022;54:1782-5. [Crossref] [PubMed]
- Hause RJ, Pritchard CC, Shendure J, et al. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016;22:1342-50. [Crossref] [PubMed]
- Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol 2010;7:153-62. [Crossref] [PubMed]
- Csiszar A, Balasubramanian P, Tarantini S, et al. Chemically induced carcinogenesis in rodent models of aging: assessing organismal resilience to genotoxic stressors in geroscience research. Geroscience 2019;41:209-27. [Crossref] [PubMed]
- Georgakopoulos-Soares I, Koh G, Momen SE, et al. Transcription-coupled repair and mismatch repair contribute towards preserving genome integrity at mononucleotide repeat tracts. Nat Commun 2020;11:1980. [Crossref] [PubMed]
- Kim S, Wyckoff J, Morris AT, et al. DNA methylation associated with healthy aging of elderly twins. Geroscience 2018;40:469-84. [Crossref] [PubMed]
- Butler M, Pongor L, Su YT, et al. MGMT Status as a Clinical Biomarker in Glioblastoma. Trends Cancer 2020;6:380-91. [Crossref] [PubMed]
- Feng M, Jiang W, Kim BYS, et al. Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat Rev Cancer 2019;19:568-86. [Crossref] [PubMed]
- Li L, Goedegebuure SP, Gillanders WE. Preclinical and clinical development of neoantigen vaccines. Ann Oncol 2017;28:xii11-7. [Crossref] [PubMed]
- Germano G, Lamba S, Rospo G, et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 2017;552:116-20. [Crossref] [PubMed]
- Tang Q, Chen J, Di Z, et al. TM4SF1 promotes EMT and cancer stemness via the Wnt/β-catenin/SOX2 pathway in colorectal cancer. J Exp Clin Cancer Res 2020;39:232. [Crossref] [PubMed]
- Yang C, Dou R, Wei C, et al. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol Ther 2021;29:2088-107. [Crossref] [PubMed]
- Wang J, Cai H, Liu Q, et al. Cinobufacini Inhibits Colon Cancer Invasion and Metastasis via Suppressing Wnt/β-Catenin Signaling Pathway and EMT. Am J Chin Med 2020;48:703-18. [Crossref] [PubMed]
- Hegde S, Leader AM, Merad M. MDSC: Markers, development, states, and unaddressed complexity. Immunity 2021;54:875-84. [Crossref] [PubMed]
- Weber R, Groth C, Lasser S, et al. IL-6 as a major regulator of MDSC activity and possible target for cancer immunotherapy. Cell Immunol 2021;359:104254. [Crossref] [PubMed]
- Bruger AM, Vanhaver C, Bruderek K, et al. Protocol to assess the suppression of T-cell proliferation by human MDSC. Methods Enzymol 2020;632:155-92. [Crossref] [PubMed]
- Li B, Chan HL, Chen P. Immune Checkpoint Inhibitors: Basics and Challenges. Curr Med Chem 2019;26:3009-25. [Crossref] [PubMed]
- Postow MA, Sidlow R, Hellmann MD. Immune-Related Adverse Events Associated with Immune Checkpoint Blockade. N Engl J Med 2018;378:158-68. [Crossref] [PubMed]
- Zhang Y, Zheng J. Functions of Immune Checkpoint Molecules Beyond Immune Evasion. Adv Exp Med Biol 2020;1248:201-26. [Crossref] [PubMed]
- Topalian SL, Taube JM, Anders RA, et al. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016;16:275-87. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science 2011;331:1553-8. [Crossref] [PubMed]
- McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. [Crossref] [PubMed]
- Friedman CF, Hainsworth JD, Kurzrock R, et al. Atezolizumab Treatment of Tumors with High Tumor Mutational Burden from MyPathway, a Multicenter, Open-Label, Phase IIa Multiple Basket Study. Cancer Discov 2022;12:654-69. [Crossref] [PubMed]
- Bai Y, Xie T, Wang Z, et al. Efficacy and predictive biomarkers of immunotherapy in Epstein-Barr virus-associated gastric cancer. J Immunother Cancer 2022;10:e004080. [Crossref] [PubMed]
- O'Malley DM, Bariani GM, Cassier PA, et al. Pembrolizumab in Patients With Microsatellite Instability-High Advanced Endometrial Cancer: Results From the KEYNOTE-158 Study. J Clin Oncol 2022;40:752-61. [Crossref] [PubMed]
- Palluk S, Arlow DH, de Rond T, et al. De novo DNA synthesis using polymerase-nucleotide conjugates. Nat Biotechnol 2018;36:645-50. [Crossref] [PubMed]
- Kawazu M, Ueno T, Saeki K, et al. HLA Class I Analysis Provides Insight Into the Genetic and Epigenetic Background of Immune Evasion in Colorectal Cancer With High Microsatellite Instability. Gastroenterology 2022;162:799-812. [Crossref] [PubMed]
- Bennett JA, Fleming GF, Kurnit KC, et al. Tumor board presentation of a woman with metastatic, hormone receptor-positive, mismatch repair-deficient endometrial cancer. CA Cancer J Clin 2022;72:102-11. [Crossref] [PubMed]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014;15:178-96. [Crossref] [PubMed]
- Schaub FX, Dhankani V, Berger AC, et al. Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas. Cell Syst 2018;6:282-300.e2. [Crossref] [PubMed]
- Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet 2013;45:1134-40. [Crossref] [PubMed]


