
Integrated bioinformatics investigation of adenylyl cyclase family co-expression network in bladder cancer followed by preliminary validation of member 2 (ADCY2) in tumorigenesis and prognosis
Highlight box
Key findings
• Adenylyl cyclase 2 (ADCY2), ADCY4, and ADCY5 exhibit a co-expression pattern, and jointly participate in the occurrence and prognosis of bladder cancer.
• ADCY2 can predict the efficacy of immunotherapy, and screen subgroups that may benefit from immune checkpoint blockades.
What is known and what is new?
• ADCYs are involved in various biological processes, involving tumour cell morphology, contact inhibition recovery, and decreased growth rate.
• ADCYs are first utilized in presenting pathogenesis and prognosis of bladder cancer.
What is the implication, and what should change now?
• Abundance of ADCY2 in bladder tissue indicates tendency of therapeutic benefits for patients with bladder cancer.
Introduction
Bladder urothelial carcinoma (BLCA) is a common urinary malignancy, with the ninth highest annual occurrence rate worldwide (1). Nearly 90% of all bladder malignant tumours are localised in the primary region (2); however, the relapse tendency has aroused a curve of long-term disease management (3). BLCA is clinically categorised into two subtypes: non-muscular invasive bladder cancer (NMIBC) and muscular invasive bladder cancer (MIBC). Most BLCA cases (>60%) are NMIBC, of which patients have a 5-year survival rate of approximately 90%. The unfavourable course of MIBC has a 5-year survival rate of less than 50%, owing to recurrence and early occult metastatic dissemination (4).
Traditional radical resection and medications have severe limitations in treating advanced bladder cancer. Owing to the high mutational burden, immune checkpoint blockades (ICBs) are used to obtain satisfactory curative effects in a subset of patients (5). New therapeutic strategies have been approved for several indications and have revolutionised the treatment landscape of advanced bladder cancer (6,7). Nevertheless, the main dilemma is the lack of predictors of immunotherapy efficacy and identification of a population with a durable immune response to immunotherapy (8). Therefore, identifying biomarkers for disease prognosis and adjusting therapeutic options for relapsed patients can benefit individual clinical treatment. Previous work had validated this viewpoint (9).
Adenylyl cyclases (ADCYs) are enzymes that generate second messengers and are involved in several biological processes, including cell cycle inhibition, apoptosis, and proliferation (10). Their activities are associated with tumour cell morphology, contact inhibition restoration, and decreased growth rates (11,12). The ADCY protein increases intracellular cyclic adenosine monophosphate (cAMP) levels, which regulate cellular transport by binding to protein kinase A (PKA). In addition, activated PKA-phosphatases regulate metabolic and transcription factors, such as the cAMP response element-binding protein (CREB), consequently activating the Wnt/β-catenin pathway, which is accompanied by cell cycle arrest, apoptosis, and survival. The ADCY-mediated calcium signalling pathway also participates in remodelling the tumour microenvironment (TME) and regulating immune checkpoints (13). The ADCY family plays an essential role in dyskinesia, diabetes, cardiovascular disease, and tumorigenesis (14-16).
Since first described by Earl W. Sutherland (17), a study on ADCY has explored its biological function, isoforms, and protein structures. Recently, the potential of ADCYs as an unelucidated therapeutic target has been reviewed (18). However, to our knowledge, limited studies have focused on the potential value of ADCYs as a biomarker for bladder cancer prognosis.
Therefore, this study aimed to characterize the prognostic value of ADCYs within BLCA and estimate ADCY-enriched patients’ response to ICBs. The ADCY2 would be used as a potential prognostic marker for bladder cancer and applied in the stratification for disease outcome. We present this article in accordance with the REMARK reporting checklist (available at https://trc.amegroups.com/article/view/10.21037/tcr-23-1796/rc).
Methods
Data source and pre-processing
Transcriptome profiling and clinical information were retrieved from The Cancer Genome Atlas (TCGA) database on the Genomic Data Commons (GDC) data portal, including 407 bladder tumour and 19 normal samples, respectively. To enrich the diversity of the sample sources, data from nine normal bladder samples were obtained from the Genotype-Tissue Expression (GTEx) database. The Toil workflow was used to normalize the RNA-sequencing (RNA-seq) data using the transcripts per million formats. The BLCA Gene Expression Omnibus (GEO) queue was retrieved from the GEO database (https://www.ncbi.nlm.nih.gov/geo/), which contains extensive survival data. GSE13507, including 256 bladder samples, was selected based on the GPL6102 (Illumina human-6 v2.0 expression beadchip) platform to build the validation group. All included patients received conventional treatment including resection and chemotherapy. Clinical information of these included patients involving outcome and prognosis were also available in the arrays.
Moreover, a cohort of IMvigor210 queues linked with immunotherapy obtained from http://research-pub.Gene.com/imvigor210corebiologies/ was compiled. The patients in IMvigor210 cohort received treatment of immune checkpoint inhibitors.
Tissue microarrays (TMAs)
Formalin-fixed paraffin-embedded (FFPE) samples were obtained from patients with BLCA receiving conventional first-line therapeutic resection. The TMAs were constructed using FFPE samples by Shanghai Outdo Biotech Co., Ltd. (https://www.superchip.com.cn/). Details of the TMA cohort are provided in Figure S1. The hole-making instrument was applied to the recipient array blocks and used to acquire tissue cores from the donor blocks. A stereotactic microscope was used to identify the areas of interest, with an additional bright light source under each block. After block construction, 8 µm sections of the resulting tumour TMA block were cut using a microtome knife, cutting underneath a tape piece placed over the block surface. The thin tissue section was rolled upon an adhesive-coated microscope slide. The malignant tumour tissues retained their histological pattern throughout the entire 1.5-mm-deep block. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of Second Military Medical University (No. CHEC2019-134). Individual consent for this retrospective analysis was waived due to the retrospective nature.
Immunohistochemical (IHC) staining evaluation
IHC staining was graded by two skilled pathologists for blinded rechecking. The staining index (SI) was used to integrate the intensity and percentage of positive cells for semiquantitative assessment of protein marker expression.
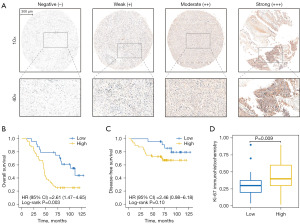
The staining intensity was graded as follows: 0, no staining; 1, pale yellow; 2, yellow; and 3, light brown. The percentage of cells stained was graded as follows: 0, <5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; and 4, >75% in tumour cells. The SI was calculated by multiplying the two scores and was classified into the following grades: 0, negative (−); 1–4, weak (+); 5–8, moderate (++); and 9–12, strong (+++). Specimens with SIs ≥5 and ≤4 were defined as high and low ADCY2 expression, respectively. A third pathologist was consulted in case of a disagreement between the two observers. Low-magnification images and examples of high- and low-expression strains are shown in Figures S1,S2.
Enrichment analysis and annotation of differentially expressed genes (DEGs)
DEGs were analysed using the limma package (19). Gene Ontology (GO) enrichment and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway analyses were performed with hub genes using the R package “clusterprofiler”. GO terms and KEGG pathways were visualised using the package “ggplot2”. The “clusterprofiler” package was used to conduct gene set enrichment analysis (GSEA) for signature genes, and the “gene set variation analysis (GSVA)” package was used to identify significantly correlated pathways. The gene sets “c2.cp.kegg.v6.2.symbols.gmt” and “h.all.v7.2.symbols.gmt” were selected as reference gene sets.
Genome alteration and DNA methylation
CBioPortal was used to retrieve the genomic data to query the genome alterations in ADCY gene family. The CBioPortal Cancer Genomics is an open-access resource (https://www.cbioportal.org) for interactive exploration of multiple cancer genomics data sets. Cytosine-phosphate-guanine (CpG) site information in the promoter region of ADCY2, ADCY4, and ADCY5 was extracted using Illumina Human Methylation 450 data, examining the Pearson correlation between the methylation level (beta value) and gene expression.
Immune cell infiltration analysis
Single-sample GSEA (ssGSEA) (20) was proposed for a single sample that could not be utilised for GSEA. The R package GSVA can be used to implement it. The extent of tumour immune cell infiltration was calculated using GSVA. We retrieved the expression properties of each immune cell using a deconvolution technique. The method of CIBERPORT (21) was used to determine immune cell infiltration. To estimate the abundance of immune cells, it uses linear support vector regression to deconvolute the expression matrix of immune cell subtypes.
TME analysis
We used ESTIMATE (22) to evaluate the tumour immune microenvironment scores of samples, and comparisons of their differential distribution in different subtypes was performed. Based on expression profiles, ESTIMATE provided us with tumour purity scores and level of stromal cells and immune cell infiltration in tumour tissues. Linear regression analysis was used to determine the link between the components and the expression of each immune cell and the final combination.
Statistical analysis
Differences in clinicopathological features between the two ADCY groups were investigated using the chi-square test. Differences between the high- and low-ADCY2 groups were investigated using a Student’s t-test. Correlation analysis was performed using the Pearson correlation coefficient. Statistical analysis was conducted in R (version 4.0.2). Significance was set as P<0.05. The cut-off for DEGs was set as |log2(fold change)| >1 and false discovery rate (FDR) <0.05.
Results
Expression of ADCY gene family in bladder specimens
The BLCA datasets from TCGA and GTEx were integrated to analyse the messenger RNA (mRNA) levels of ADCYs in bladder tissues. The expression of ADCY2, ADCY4, ADCY5, and ADCY9 was significantly downregulated in tumour tissues (P<0.001); however, no statistically significant difference was observed in the other ADCY genes (Figure 1A). To further elucidate the potential function of ADCY2, ADCY4, ADCY5, and ADCY9 in tumour malignancy, the relationship between ADCY expression levels and clinicopathological characteristics, including pathological stage, T stage, histologic grade, lymphovascular invasions, and pathological subtypes, were analysed (Figure 1B, Figure S3A). In contrast to the differential expression levels between normal and tumour tissues, ADCY expression levels, especially ADCY2, ADCY4, and ADCY5, were positively associated with clinical features indicating tumour progression. ADCY2 levels were significantly higher in patients with metastatic risk than in those with focal lesions. The non-papillary subtypes showed significantly higher ADCY2 and ADCY9 expression than the papillary subtypes. ADCY expression levels were also associated with earlier T stage and histological grade. Moreover, the mRNA expression levels in 21 bladder cancer cell lines were compared with those in telomerase reverse transcriptase-normal human urothelial cells (TERT-NHUCs) bladder epithelial cells, and the results confirmed the above conclusion (Figure 1C).
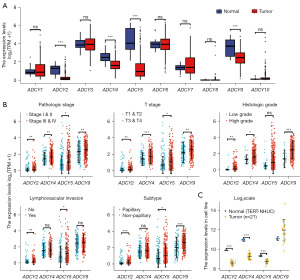
Elevated ADCYs expression is associated with poor clinical outcomes in BLCA patients
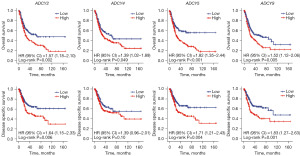
To further elucidate the relationship between survival promotion and transcriptional levels, a prognostic analysis was performed by retrieving TCGA clinical data (Figure 2, Figure S3B). Kaplan-Meier (KM) curves showed that multiple ADCY genes were correlated with improved prognosis. Patients with higher ADCY2, ADCY4, ADCY5, and ADCY9 expressions had significantly prolonged overall survival (OS) and disease-specific survival (DSS) time.
TMA cohort validated the relation between ADCY2 expression level and clinical outcomes
To validate the hypothesis that ADCY2, ADCY4, ADCY5, and ADCY9 are prognostic indicators, pathological tissues of 80 in-patients in urological department of Changhai Hospital were included into TMA. ADCY2 was selected as a candidate for IHC staining, and IHC scores were quantitatively evaluated (Figure S1A). The staining standard was classified into three levels (Figure 3A). The clinical outcome of the TMA cohort was illustrated using the KM curve, showing a significant difference between the high- and low-ADCY2 expression groups (Figure 3B). The KM curve of disease-free survival (DFS) further demonstrated that ADCY2 expression had no statistical relationship with tumour recurrence (Figure 3C), consistent with the KM curve of disease-free interval (DFI) in TCGA cohort (Figure S1B). In the high-ADCY2 expression group, Ki67, a proliferative indicator of cancer cells, was significantly increased (Figure 3D).
To verify ADCY2 expression in other datasets, the GSE13507 dataset (23) was downloaded from the GEO database. In addition to observing significant differences in ADCY2 expression between tumour and normal tissues, we counted and combined the expression in tumour occurrence sites in GSE13507; the difference between the recurrent and primary sites was not statistically significant (P>0.05, Figure S1C,S1D).
ADCYs co-expression analysis
To explore the interconnection among ADCY isoforms in BLCA regulation, Spearman’s rank correlation analysis was used to calculate the correlation scores among the ADCY gene family members at the transcriptional level. The heatmap (Figure 4A) displayed two clustered gene groups with significant correlations (ADCY2, ADCY4, and ADCY5; and ADCY3, ADCY7, and ADCY9), indicating a potential biological connection among these clustered genes. Further analysis of this phenomenon by overlapping the 500 most correlated genes of each ADCY family revealed a probable co-expression relationship among ADCY2, ADCY4, and ADCY5, containing 165 candidate genes at the intersection, ranking the highest in the clustered groups (Figure 4B).
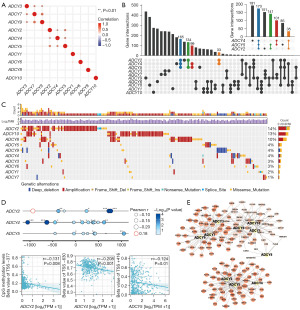
The mechanism of co-expression of ADCY2, ADCY4, and ADCY5 was preliminarily explored from genetics and epigenetics perspectives, respectively; no apparent coherence in copy number variation (CNV) was observed among the genes (Figure 4C), and each gene had a unique chromosomal distribution (Figure S2A). The methylation levels of CpG sites in the ADCY2, ADCY4, and ADCY5 promoter regions were assessed using bladder cancer samples (n=440) from TCGA database. A significant inverse correlation was observed between methylation level and mRNA expression in almost all CpG sites, indicating that ADCY2, ADCY4, and ADCY5 are regulated by DNA methylation (Figure 4D). However, direct evidence that this co-expression phenomenon is regulated by epigenetics requires further exploration.
To better identify the transcriptional co-expression patterns in ADCY genes, co-expression network was constructed using correlated genes (Figure 4E); the network containing ADCY2, ADCY4, and ADCY5 were clearly separated from the other ADCYs. Overall, the transcriptional regulatory network validated the close correlation among ADCYs in BLCA, and co-expression was potentially regulated by epigenetics.
Differential expression gene enrichment analysis
To evaluate the clinical values and understand the biological functions of ADCY2, ADCY4, and ADCY5 in BLCA, DEGs were extracted based on the median expression values, and the pathways enriched using DEGs were investigated. The receiver operating characteristic (ROC) curve revealed that ADCY2, ADCY4, and ADCY5 were favourable diagnostic biomarkers for bladder cancer, and the area under the curve (AUC) were 0.96, 0.92, and 0.88 for ADCY5, ADCY2, and ADCY4, respectively (Figure 5A). Moreover, 2,017, 2,904, and 2,496 DEGs of ADCY2, ADCY4, and ADCY5, respectively, displayed by volcano plots to separate the up- and down-regulated groups, were identified (Figure S2B). DEGs were intersected using a Venn diagram to observe the overlap between these three groups (Figure 5B). The common genes were used for subsequent enrichment analysis. The significantly correlated genes and DEGs of the three groups were intersected, and five common genes ADRA2A, SNED1, RASL12, LIMS2, and JAM2 were obtained (Figure 5C).
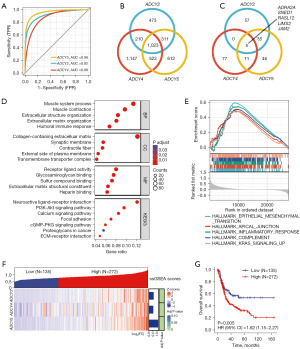
KEGG analysis revealed that the ADCY2, ADCY4, and ADCY5 DEGs were enriched in the calcium and PI3K-Akt signalling pathways (Figure 5D). GO analysis revealed phenotypic features, including collagen-containing extracellular matrix, extracellular structure organization, and a series of components focusing on epithelial-mesenchymal transition (EMT) and tumour metastasis phenotypes (Figure 5D). Considering the difference between the absolute expression trends in specific pathways and general expression alterations, GSEA was performed. The GSEA results further validated that the ADCY2, ADCY4, and ADCY5-related gene sets were associated with metastasis and tumour immunosuppressive-related process, such as hallmarks of EMT and the interleukin 2-signal transducer and activator of transcription 5 (IL2-STAT5) pathway (Figure 5E). ssGSEA clustered samples with relatively high or low ADCY2, ADCY4, and ADCY5 expressions (Figure 5F). KM plots revealed a significant survival difference between the upregulated and downregulated groups (Figure 5G).
Boosting immune infiltration status and TME formation
Investigating the immunological role of ADCY2, ADCY4, and ADCY5 could elucidate their potentially critical role in treating BLCA, especially in immunotherapy. Thus, the difference in immune cell enrichment between the two groups (Figure 5F) was evaluated; the upregulated group was characterised by increased immune infiltration by memory T cells, natural killer (NK) cells, macrophages, and other innate immune cells (Figure 6A). GSEA illustrated the pathways involved in the recruitment of innate immune cells, focusing on effector CD8+ T, memory CD4+ T cells, and macrophages (Figure 6B).
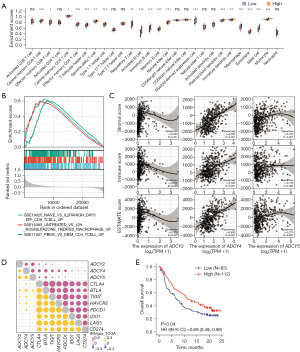
Next, ADCY2, ADCY4, and ADCY5 enhancements in the formation of the TME were explored. Regression analysis showed that the mRNA levels of ADCY2, ADCY4, and ADCY5 were positively correlated with the number of stromal cells present and infiltration of immune cells (Figure 6C). Markers of immune cells were also significantly correlated with ADCY2, ADCY4, and ADCY5 expression levels, especially in CD4+/CD8+ T, memory B, and mast cells (Figure S4A). Survival analyses were performed; patients with higher infiltration of CD4+/CD8+ T, memory B, and mast cells had a relatively short OS, suggesting that the upregulation of ADCY2, ADCY4, and ADCY5 co-expression affected the immune-mediated process and influenced survival time (Figure S4B). To further elucidate the role of ADCY in immunotherapy (particularly ICBs) (24), an immunotherapy cohort of bladder cancer patients was used to further explore the associations between ADCYs and several typical immune checkpoint genes. In TCGA-BLCA cohort, ADCY expression was positively correlated with programmed death ligand 1 (PD-L1), cytotoxic T lymphocyte-associated antigen-4 (CTLA-4), indoleamine 2,3-dioxygenase-1 (IDO1), B and T lymphocyte attenuator (BTLA), hepatitis A virus cellular receptor 2 (HAVCR2), and T cell immunoreceptor with Ig and ITIM domains (TIGIT) expression, and strong agreement was observed in the IMvigor210 cohort (Figure 6D). The population with higher ADCY expression in this cohort had a better prognosis and higher responsiveness to immunotherapy (Figure 6E, Figure S4C). It is noteworthy that the correlation between ADCYs and immunomodulators, chemokines, and chemokine receptors in both the TCGA and IMvigor datasets is consistent (Figure S5).
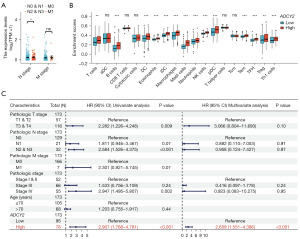
In order to explore whether ADCY2 has the ability to independently predict the prognosis of BLCA and its role in the immune microenvironment, we first analysed the correlation between the clinical indicators of ADCY2 and BLCA, and found that its expression levels were significantly different in lymph node metastasis of bladder cancer at different levels (Figure 7A). Subsequently, we conducted an in-depth analysis of the patterns and prognostic roles of tumour mutation burden (TMB) in bladder cancer (Figure S6A-S6C). The plot illustrated that TMB was significantly leading to a bad outcome, especially in bladder cancer. Our findings revealed a significant correlation between ADCY2 and tumour purity (R=0.37, P<0.001, as illustrated in Figure 7B). Then, we assessed the infiltration levels of diverse immune cells in both high and low ADCY2 expression groups. In particular, the infiltration levels of T cells, B cells, dendritic cell (DC), Eosinophils, immature DC (iDC), macrophages, mast cells, neutrophils, NK cells, plasmacytoid DC (pDC), effector memory T cell (Tem), T follicular helper (TFH), T helper type 1 (Th1) cells, and regulatory T cell (Treg) were all found to be correlated with the expression of ADCY2, and this phenomenon is universal in bladder cancer compared to the normal tissues (Figure 7C, Figure S6D,S6E). Figure 7C summarized the results, incorporating tumour-node-metastasis (TNM) stage, pathological grade, age, and categorization based on high and low ADCY2 expression levels. Remarkably, ADCY2 stands as an independent risk factor for BLCA. These findings collectively underscore the pivotal role of ADCY2 in the immune microenvironment and its potential as a prognostic indicator for BLCA.
Discussion
This study aimed to evaluate the potential value of ADCYs as diagnostic biomarkers or therapeutic targets for bladder cancer. Four of the 10 members of the ADCY gene family exhibited transcriptional differences between tumour and normal specimens. Among them, ADCY2, ADCY4, and ADCY5 exhibited closer interactions than other members in the corresponding expression network, indicating a joint interaction mechanism or similar functions. Thus, these members can be used as prognostic biomarkers to indicate OS/DSS time in BLCA patients; the clinical data of the TMA cohort validated ADCY2 efficacy in disease prognosis. Moreover, ADCYs can predict the efficacy of immunotherapy and screen for the subpopulation that may benefit from ICBs.
Abnormal methylation of ADCY2 can affect clinical outcomes in colorectal cancer (25). SNP(rs4702484) in ADCY2 region was identified significantly associated with capecitabine/5-fluorouracil (5-FU) susceptibility, which is directly influencing the chemotherapy outcome of bladder cancer (26). ADCY4 mediated the activation of intracellular Ca2+, which might influence carcinogenesis and adverse invasion of lung adenocarcinoma cells (27). Some studies have reported that ADCY5 plays a role in the genesis and development of neurobiological diseases, including dyskinesia, early onset autosomal dominant chorea, and dystonia (28,29).
This study explored the correlation between features of ADCY2, ADCY4, and ADCY5 gene and BLCA prognosis. The effect of ADCY2 on survival was validated using real-world clinical data and verified using TMA experiments. In the TMA cohort, the correlation between included clinical characteristics and ADCY2 expression was investigated; ADCY2 levels were not associated with distant metastasis but were positively correlated with lymph node invasion.
A previous study has demonstrated that ADCYs are not homologous regarding degrees of amino acid sequence and can be divided into distinct isoform subgroups with similar amino acid sequences and biochemical properties (30). Therefore, we explored co-expression from a genetic and epigenetic perspective. Amplification and missense mutation were the major DNA alteration in ADCY2, while deep deletion was the main genomic alteration in ADCY4. Although the mutation distribution could account for the downregulation in BLCA samples compared to normal tissues, the DNA alteration types in ADCY2, ADCY4, and ADCY5 were not coincident. Given that the transcription of these different ADCY genes is regulated by distinct promoter/enhancer elements (30,31), we sought clues from a DNA methylation perspective. We found that methylation of the transcriptional start site was negatively correlated with ADCY gene expression, similarly to the results of previous studies (32,33). Owing to the consistent trend in ADCY2, ADCY4, and ADCY5, methylated modifications might function in constructing the co-expression network and work as a synchronous system. These results are consistent with those of previous studies (31,34), suggesting that the expansion of the ADCY gene family is not due to gene duplication. Subsequent studies should explore the regulatory mechanism of epigenetic modifiers on the ADCY co-expression network to evaluate the potential therapeutic value of targeting ADCY genes.
Pathway enrichment in neuroactive ligand-receptor interaction, EMT, PI3K-AKT signalling, inflammation response, and IL2-STAT5 signalling might account for the worsened prognosis in the ADCY-upregulated groups. These pathways modulate tumour cells to transfer and escape immune surveillance (35,36). The intersection of DEGs also revealed five correlated genes (ADRA2A, SNED1, RASL12, LIMS2, and JAM2). These screened genes have been reported as intermediate elements in tumour biological processes and are involved in tumour metastasis and drug resistance (37-39). Potential regulatory mechanisms exist among these five correlated genes in the interaction networks, in which ADCY2 might also affect tumour malignancies.
The activities of anticancer immunity comprehensively reflect the ability of tumours to escape immune surveillance and develop drug resistance, which is closely related to the complicated immunomodulatory interactions in the TME. ADCY2 was associated with several steps in tumour evolution from innate immunity. Consequently, the infiltration levels of effector immune cells, such as CD8+ T cells, NK cells, mast cells, and macrophages, were also significantly increased in the high-ADCY2 enrichment population. These patients were classified as better responders to the immune-killing effect. Thus, this subset of patients with advanced bladder cancer may benefit from immunotherapy and targeted therapy (40). The effect of ADCY on the regulation of immune checkpoint activity was tentatively analysed; mRNA expression of ADCY2, ADCY4, and ADCY5 indicated a significant correlation with well-known immune checkpoint markers, and the survival data conformed to the immune status evaluated. Thus, our results improve the selection of populations that may achieve better clinical outcomes and durable responses to immunotherapy.
TME was demonstrated to largely participate into the process of tumour recurrence. The relationship between ADCY2 level and TME formation has been statistically disclosed in the research. We expected that the succedent studies could further validated the role of ADCY2 in formation of TME and promoting long-term recurrence. ADCY2 could be identified as an indirect bridge to measure long-term tumour recurrence from the perspective of TME formation.
Nevertheless, there are still some limitations in our study. Firstly, owing to the difficulty in retrieving datasets containing both transcriptomic and clinical survival data from public databases, the sample size of TCGA raw data was small and thus unpersuasive. Some differences were observed between the cohorts. Then, the clinical cohort of TMA mainly comprised patients with invasive advanced bladder cancer, whereas TCGA cohort had a higher proportion of patients at earlier stages. Finally, the majority of our research findings were based on bioinformatics analysis, without experimental validation of the associated conclusions. We will conduct laboratory studies in the future to further investigate and validate the phenomenon of co-expression and molecular mechanisms of ADCYs in EMT.
Conclusions
In this research, the role of ADCY2 in predicting prognosis and immunotherapy responsiveness in bladder cancer was investigated. ADCY2 could be used as a prognostic marker for BLCA. We will further explore the potential molecular mechanism of ADCY2 in modulating and alternating the biologic features of bladder cancer cells.
Acknowledgments
We would like to thank the Editage (https://www.editage.com/) for English language editing and Professor Xu Gao for his guidance on conceiving the study and funding support.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://trc.amegroups.com/article/view/10.21037/tcr-23-1796/rc
Data Sharing Statement: Available at https://trc.amegroups.com/article/view/10.21037/tcr-23-1796/dss
Peer Review File: Available at https://trc.amegroups.com/article/view/10.21037/tcr-23-1796/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://trc.amegroups.com/article/view/10.21037/tcr-23-1796/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of Second Military Medical University (No. CHEC2019-134). Individual consent for this retrospective analysis was waived due to the retrospective nature.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fernández MI, Brausi M, Clark PE, et al. Epidemiology, prevention, screening, diagnosis, and evaluation: update of the ICUD-SIU joint consultation on bladder cancer. World J Urol 2019;37:3-13. [Crossref] [PubMed]
- Grayson M. Bladder cancer. Nature 2017;551:S33. [Crossref] [PubMed]
- Guo CC, Lee S, Lee JG, et al. Molecular profile of bladder cancer progression to clinically aggressive subtypes. Nat Rev Urol 2024; Epub ahead of print. [Crossref] [PubMed]
- Hurst CD, Knowles MA. Mutational landscape of non-muscle-invasive bladder cancer. Urol Oncol 2022;40:295-303. [Crossref] [PubMed]
- Huang G, Xiong H, Li S, et al. The efficacy of immune checkpoint inhibitors therapy versus chemotherapy in the treatment of advanced and metastatic urothelial carcinoma: a meta-analysis. J Cancer Res Clin Oncol 2024;150:5. [Crossref] [PubMed]
- Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J Clin 2020;70:404-23. [Crossref] [PubMed]
- Coelho JQ, Ramos MJ, Ranchor R, et al. What's new about the tumor microenvironment of urothelial carcinoma? Clin Transl Oncol 2024; Epub ahead of print. [Crossref] [PubMed]
- Witjes JA, Bruins HM, Cathomas R, et al. European Association of Urology Guidelines on Muscle-invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. Eur Urol 2021;79:82-104. [Crossref] [PubMed]
- Wu Z, Wang D, Zhang Y, et al. SPP1 mRNA determination based on molecular beacon for the recurrence prognosis of bladder cancer. Transl Androl Urol 2023;12:1834-44. [Crossref] [PubMed]
- Tang G, Li S, Zhang C, et al. Clinical efficacies, underlying mechanisms and molecular targets of Chinese medicines for diabetic nephropathy treatment and management. Acta Pharm Sin B 2021;11:2749-67. [Crossref] [PubMed]
- Aldehni F, Tang T, Madsen K, et al. Stimulation of renin secretion by catecholamines is dependent on adenylyl cyclases 5 and 6. Hypertension 2011;57:460-8. [Crossref] [PubMed]
- Pluznick JL, Zou DJ, Zhang X, et al. Functional expression of the olfactory signaling system in the kidney. Proc Natl Acad Sci U S A 2009;106:2059-64. [Crossref] [PubMed]
- Sheng QJ, Tian WY, Dou XG, et al. Programmed death 1, ligand 1 and 2 correlated genes and their association with mutation, immune infiltration and clinical outcomes of hepatocellular carcinoma. World J Gastrointest Oncol 2020;12:1255-71. [Crossref] [PubMed]
- Abdel-Halim SM, Al Madhoun A, Nizam R, et al. Increased Plasma Levels of Adenylate Cyclase 8 and cAMP Are Associated with Obesity and Type 2 Diabetes: Results from a Cross-Sectional Study. Biology (Basel) 2020;9:244. [Crossref] [PubMed]
- Baizabal-Carvallo JF, Cardoso F. Chorea in children: etiology, diagnostic approach and management. J Neural Transm (Vienna) 2020;127:1323-42. [Crossref] [PubMed]
- Zhang X, Yu K, Ma L, et al. Endogenous glutamate determines ferroptosis sensitivity via ADCY10-dependent YAP suppression in lung adenocarcinoma. Theranostics 2021;11:5650-74. [Crossref] [PubMed]
- Rall TW, Sutherland EW. Formation of a cyclic adenine ribonucleotide by tissue particles. J Biol Chem 1958;232:1065-76. [Crossref] [PubMed]
- Guo R, Liu T, Shasaltaneh MD, et al. Targeting Adenylate Cyclase Family: New Concept of Targeted Cancer Therapy. Front Oncol 2022;12:829212. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108-12. [Crossref] [PubMed]
- Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Kim WJ, Kim EJ, Kim SK, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer 2010;9:3. [Crossref] [PubMed]
- Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544-8. [Crossref] [PubMed]
- Yu SJ, Yu JK, Ge WT, et al. SPARCL1, Shp2, MSH2, E-cadherin, p53, ADCY-2 and MAPK are prognosis-related in colorectal cancer. World J Gastroenterol 2011;17:2028-36. [Crossref] [PubMed]
- O'Donnell PH, Stark AL, Gamazon ER, et al. Identification of novel germline polymorphisms governing capecitabine sensitivity. Cancer 2012;118:4063-73. [Crossref] [PubMed]
- Liu J, Hao Y, Wang Y, et al. Candidate methylated genes in osteoarthritis explored by bioinformatics analysis. Knee 2016;23:1035-43. [Crossref] [PubMed]
- Ferrini A, Steel D, Barwick K, et al. An Update on the Phenotype, Genotype and Neurobiology of ADCY5-Related Disease. Mov Disord 2021;36:1104-14. [Crossref] [PubMed]
- Vijiaratnam N, Bhatia KP, Lang AE, et al. ADCY5-Related Dyskinesia: Improving Clinical Detection of an Evolving Disorder. Mov Disord Clin Pract 2019;6:512-20. [Crossref] [PubMed]
- Ishikawa Y, Homcy CJ. The adenylyl cyclases as integrators of transmembrane signal transduction. Circ Res 1997;80:297-304. [Crossref] [PubMed]
- Hellevuo K, Berry R, Sikela JM, et al. Localization of the gene for a novel human adenylyl cyclase (ADCY7) to chromosome 16. Hum Genet 1995;95:197-200. [Crossref] [PubMed]
- Van Tongelen A, Loriot A, De Smet C. Oncogenic roles of DNA hypomethylation through the activation of cancer-germline genes. Cancer Lett 2017;396:130-7. [Crossref] [PubMed]
- Moore LE, Pfeiffer RM, Poscablo C, et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol 2008;9:359-66. [Crossref] [PubMed]
- Gaudin C, Homcy CJ, Ishikawa Y. Mammalian adenylyl cyclase family members are randomly located on different chromosomes. Hum Genet 1994;94:527-9. [Crossref] [PubMed]
- Bakir B, Chiarella AM, Pitarresi JR, et al. EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol 2020;30:764-76. [Crossref] [PubMed]
- Mak MP, Tong P, Diao L, et al. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin Cancer Res 2016;22:609-20. [Crossref] [PubMed]
- Haam K, Kim HJ, Lee KT, et al. Epigenetic silencing of BTB and CNC homology 2 and concerted promoter CpG methylation in gastric cancer. Cancer Lett 2014;351:206-14. [Crossref] [PubMed]
- Naba A, Clauser KR, Lamar JM, et al. Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. Elife 2014;3:e01308. [Crossref] [PubMed]
- Zhao H, Yu H, Martin TA, et al. The role of JAM-B in cancer and cancer metastasis Oncol Rep 2016;36:3-9. (Review). [Crossref] [PubMed]
- Tran L, Xiao JF, Agarwal N, et al. Advances in bladder cancer biology and therapy. Nat Rev Cancer 2021;21:104-21. [Crossref] [PubMed]


