
Significance of dysregulated M2 macrophage and ESR2 in the ovarian metastasis of gastric cancer
Highlight box
Key findings
• Dysregulated tumor immune microenvironment (TIME), especially the immunosuppressive and protumoral M2 macrophage, was investigated between primary gastric cancer (GC) and its ovarian metastasis (OM).
• Estrogen receptor 2 (ESR2), the protein-coding gene of estrogen receptor β, was potentially correlated with M2 accumulation, and more importantly, uniquely overexpressed in OM than in primary tumor.
• High expression of ESR2 was correlated with lymph node metastasis, more advanced pathological stage and a worse long-term survival of GC patients, and it promoted GC proliferation, migration and invasion in vitro.
What is known and what is new?
• It is already known that primary GC and its metastatic lesions exhibit distinctive TIME. Moreover, ESR2 is indicated to play a bi-faceted role of pro- and anti-tumorigenic, possibly dependent on the nature or mutation status of its downstream effectors in malignancies including GC.
• We revealed the unique enrichment of immunosuppressive and protumoral M2 macrophage in OM over primary GC. Moreover, we demonstrated that ESR2 potentially correlated with M2 accumulation and more importantly, contributed to GC severity and promoted GC cell metastasis in vitro.
What is the implication, and what should change now?
• Our study identified the dysregulated major immune cell subsets and its related key regulator in the OM of GC, which serves as potential targets for future treatment of this specific type of distant metastasis. More studies shall focus on investigating molecular mechanisms underlying not only ESR2-mediated M2 accumulation but also OM of GC.
Introduction
Distant metastasis poses a major challenge to the long-term survival of patients with gastric cancer (GC) (1,2). Notably, ovarian metastasis (OM), also known as Krukenberg tumor, is one of the most common patterns of metastasis, especially in young, premenopausal female population (3). OM synchronously occurs in 0.3–6.7% of GC patients undergoing radical surgery, and its incidence rate reaches up to 41% by autopsy (4). The median overall survival of these patients remains less than 19 months even though recent progressions in the comprehensive treatment modality have been made (5,6). Unfortunately, its underlying mechanism as well as the role of immune cell subsets in OM are still unclear. Hence, there is an urgent need to functionally characterize tumor immune microenvironment (TIME) and identify key regulators in this unique type of GC metastasis.
Recently, multiple landmark studies have demonstrated the efficacy of immune checkpoint inhibitors-based immunotherapy in the treatment of locally unresectable and metastatic GC (7,8), highlighting the significance of targeting GC from the perspectives of TIME. In fact, increasing evidence demonstrates that interactions between cancer cells and immune cell subsets within TIME play key roles in the progression and metastasis of malignancies including GC (9,10). The application of RNA-sequencing (RNA-seq) on paired primary and metastatic tumors facilitates the understanding of key components within TIME in cancer progression (11-13). Unfortunately, the comparative characterization of TIME between primary GC and matched OM has been very limited so far. We hereby conducted this comparative RNA-seq and follow-up analyses of paired primary and ovarian metastatic tumors to provide insights into the differential TIME as well as the underlying molecular pathogenesis in the OM of GC.
Our RNA-seq analyses for the first time unveiled the selective overexpression of estrogen receptor 2 (ESR2), the protein-coding gene of estrogen receptor β (ERβ), in OM over primary tumor. The role of estrogen and its receptor in GC progression has gained attention in recent years (14). For instance, it is reported that estrogen promotes the proliferation and invasion of GC cell through increasing the secretion of IL-6 by cancer-associated fibroblasts (15), indicating the oncogenic impact of estrogen in both tumor microenvironment and immunity. With respect to estrogen receptor, Zhou et al. reported that ESR2 plays pro-oncogenic role by suppressing GC cell apoptosis and autophagy through specific molecular mechanisms (16). Furthermore, retrospective studies indicated the prognostic role of ERβ in GC patients with OM (17) whereas others reported that abnormal expression of ERβ independently predicts the occurrence of OM of patients receiving radical gastrectomy (18). These findings suggest the significance of ESR2 in this unique type of GC metastasis although further mechanism investigations are highly demanded.
In this study, we conducted comparative transcriptome profiling of seven pairs of primary and ovarian metastatic lesions by performing high throughput RNA-seq. Follow-up analyses demonstrated the differential TIME between them, especially the significant up-regulation of M2 macrophage in the OM over primary tumors. Multiplex immunofluorescence (mIF) staining validated the overexpression of CD68+CD206+ M2 macrophage in OMs over primary tumors. Moreover, ESR2 was potentially correlated with M2 macrophage, and its expression was significantly up-regulated in the OMs. Cohort studies and functional assays validated the unique overexpression of ESR2 in the distant metastasis to ovary and further revealed its pro-oncogenic potential of promoting the migration and invasiveness of GC cells. These data indicated the significance of M2 macrophage and ESR2 in the OM of GC and suggested a novel therapeutic target for its potential treatment. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-124/rc).
Methods
Patient and sample collection
Patient samples were collected for either transcriptome sequencing or tissue microarray (TMA) in this study. Formalin-fixed paraffin-embedded (FFPE) and fresh tissues which were stored in liquid nitrogen from eligible GC patients including the paired primary tumors and metastatic ovarian lesions were submitted for transcriptome sequencing. The matched samples of individual patients were collected in the same extended radical gastrectomy between 2016 and 2020 at Fudan University Shanghai Cancer Center (Shanghai, China). Samples of malignancies were analyzed by two individual pathologists for histological and tumor cellularity determination (tumor cellularity ≥60%) before submission for sequencing. On the other hand, to evaluate clinical significance of candidate gene, we also collected surgically resected samples of primary gastric tumor and adjacent normal gastric mucosa of 90 GC patients (63 male and 27 female, average age 62.3 years old) without previous treatment between 2009 and 2012 from the same medical center. All samples were reviewed by two individual pathologists and then proceeded into FFPE tissue for TMA. In both cases, the pathological stage of studied patients was determined by the 8th edition of the Union for International Cancer Control (UICC) tumor-node-metastasis (TNM) classification, which records the primary tumor (T), its regional nodal extent (N) and whether distant metastasis is present or not (M). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the local ethics committee of the Fudan University Shanghai Cancer Center (No. 050432-4-1911D) and informed consent was taken from all patients.
Transcriptome sequencing
RNAstormTM FFPE kit (CELLDATA, Fremont, USA) was applied for the isolation of total RNA from cancerous tissue. SMARTer Stranded Total RNA-Seq Kit—Pico Input Mammalian Library preparation kit (Clontech, Mountain View, USA) was applied for the preparation of strand-specific RNA-seq library. Qubit fluorometer (Thermo Fisher Scientific, Waltham, USA) and Qsep100 (BiOptic, New Taipei City) were applied for the check of library quality. Transcriptome sequencing was performed on an Illumina sequencing platform with 150 bp paired-end run metrics.
Transcriptome sequencing data analysis including identification and functional annotation of differentially expressed genes (DEGs)
FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to filter raw reads to remove low quality bases and adaptor sequences. HISAT2 was used to map reads to the GRCh38 human genome assembly. Gene expression levels were calculated with Fragments Per Kilobase of transcript per Million mapped reads (FPKM) by normalizing gene counts from feature counts. The Pearson correlation coefficient was calculated to compare the global gene expression in the samples. DEGs between primary and ovarian metastatic tumors were selected with log2 (fold change) ≥1 or log2 (fold change) ≤−1 and with statistical significance (P value <0.05). Further functional annotation of DEGs were conducted to understand their biological functions. Using the R package “org.Hs.eg.db (version 3.1.0)” we obtained the Gene Ontology (GO) annotations of genes. Moreover, the subset of “h.all.v7.4.symbols.gmt” was downloaded from Molecular Signatures Database (DOI: 10.1093/bioinformatics/btr260; http://www.gsea-msigdb.org/gsea/downloads.jsp). Genes were mapped to the GO and hallmark which were used as background set and the R package “package clusterProfiler (version 3.14.3)” was adopted to perform the gene set enrichment analysis. The minimum and maximum gene set were set to be 5 and 5,000, respectively. The CIBERSORT algorithm (https://cibersort.stanford.edu/) was adopted to calculate and compare the relative proportion of 22 immune cell subsets in each type of lesions to characterize the distinctive TIME between GC and OM, as previously described (19). P values <0.05 were considered statistically significant.
mIF
The mIF staining of CD68 (anti-CD68, 1:1,000, ab213363, Abcam, Cambridge, UK), CD206 (CD206 monoclonal antibody, 1:1,000, 60143-1-Ig, Proteintech, Wuhan, China), CK14 (cytokeratin 14 polyclonal antibody, 1:50, 10143-1-ap, Proteintech) and DAPI was conducted to visualize and quantify CD68+CD206+ M2 macrophage in the seven pairs of primary GCs and matching OMs. The similar methods as described by Chen et al. were adopted (20). Briefly, the dehydration, paraffin embedding, and antigen retrieval were routinely conducted. After serum blocking, the blocking solution was gently shaken off and the primary antibody prepared with phosphate buffered saline (PBS) was dropped on the slices, the slices was placed flat in a humidified box and incubated overnight at 4 ℃. Then, the slides were placed in PBS shaken on a destaining shaker for 3 times and for 5 minutes each time. After the slices were slightly dried, the horseradish peroxidase (HRP)-labeled secondary antibody corresponding to the primary antibody was added dropwise in the circle to cover the tissue and incubated at room temperature in the dark for 50 min. Next, the 532-TSA was added and treated with microwave to remove the primary and secondary antibodies that have been bound to the tissue. These steps were repeated for the second, third and fourth rounds of primary and secondary antibodies with 594-TSA (1:500, ab150080, Abcam), CY5-TSA (1:2,000, ab6564, Abcam) and 488-TSA (1:500, ab150113, Abcam) added at each time. The slides were placed in PBS, then shaken and washed 3 times on a decolorizing shaker for 5 min each time. After the slices were slightly dried, DAPI staining solution was added dropwise in the circle, and incubated at room temperature for 10 min in the dark. The process was repeated and then the slides were sealed with anti-fluorescence quenching mounting medium and stored in a light-proof section box at 4 ℃ after sealing. The nuclei stained by DAPI are blue under ultraviolet excitation, and positive expression is the corresponding fluorescein-labeled red light [594] or green light [fluorescein isothiocyanate] or orange light (CY5) or pink light [532].
Immunohistochemical (IHC) assay
TMA was applied to examine the clinical value of candidate by histological and IHC analysis, as previously described in detail (21). In general, the fixed and embedded tissue glass slides were deparaffinized and rehydrated by serial incubation in graded alcohol and water, followed by section blocking with 0.3% hydrogen peroxide for peroxidase activity and section boiling with 10 mM citrate buffer for antigen retrieval. Next, sections were incubated with primary antibody (anti-ERβ antibody, 1:500, 14007-1-AP, Proteintech), rinsed three times with phosphate-buffered saline (PBS) and then incubated with secondary antibody. Finally, the sections were incubated with avidin-alkaline phosphatase and then with red chromogen for visualization and further counterstained with Mayer hematoxylin method. ERβ (encoded by ESR2) was positively stained in both cytoplasm and nucleus of gastric cell. Brown cytoplasmic and/or nuclear staining were considered as positive. IHC scoring system was introduced to grade the staining intensity of the TMA by both the intensity and proportion of positive-staining cells, as previously reported (22): 0, no staining; 1, <10% positive, moderate or strong intensity; 2, 10–50% positive, moderate or strong intensity; 3, >50% positive, moderate intensity; and 4, >50% positive, strong intensity. IHC scoring was performed by two independent pathologists with no inform of clinical outcomes.
Quantitative real-time polymerase chain reaction (PCR)
TRIzol reagent (Invitrogen, Carlsbad, USA) and RNAstormTM FFPE kit (CELLDATA) were used to extract total RNA from GC cell lines and FFPE tissue, respectively. Superscript 3 Kit and oligo-dT primer (Thermo Fisher Scientific) were used to synthesize complementary DNA (cDNA). For quantitative real-time PCR, up to 500–1,000 ng total RNA was used for reverse transcription according to the protocol. Power SYBR Green PCR Master Mix kit (Thermo Fisher Scientific) was used for cDNA (dilution: 1:10) subjected to quantitative real-time PCR according to the protocol. The PCR primers used are listed as Table 1. The 2−ΔΔCt method was used to calculate relative expression level of candidate gene. GAPDH was used as endogenous reference for normalization.
Table 1
| Gene | Forward | Reverse |
|---|---|---|
| CLDN11 | 5'-CGCGATTGGTCGGCGCGTTTC-3' | 5'-GACGAAAACAACAACGCTACT-3' |
| HOXD8 | 5'-CCTGACTGTAAATCGTCCAGTGGTA-3' | 5'-AGTTTGGAAGCGACTGTAGGTTTG-3' |
| CXCL14 | 5'-CGCTACAGCGACGTGAAGAA-3' | 5'-GTTCCAGGCGTTGTACCAC-3' |
| SPINK1 | 5'-TGTCTGTGGGACTGATGGAA-3' | 5'-TCAACAATAAGGCCAGTCAGG-3' |
| ESR2 | 5'-TGGGCACCTTTCTCCTTTAG-3' | 5'-TGAGCATCCCTCTTTGAACC-3' |
| GAPDH | 5'-GGACCTGACCTGCCGTCTAG-3' | 5'-GTAGCCCAGGATGCCCTTGA-3' |
PCR, polymerase chain reaction.
Western blot analysis
Details of Western blot analysis were described previously (21). Briefly, RIPA buffer (Beyotime, Haimen, China) with protease and phosphatase inhibitor (ratio: 10:1) were used to extract protein from both tissues and cell lines. BCA protein assay kit (Thermo Fisher Scientific) was used to determine the protein concentration. After the pre-treatment of cell lysates by mixing with Laemmli sample buffer, boiling and instant cooling, they were subjected to 10% sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a polyvinylidene difluoride (PVDF) membrane by using the Trans-Blot® TurboTM device (Bio-Rad, Hercules, USA). Next, the PVDF membrane was blocked with 5% milk solution, rinsed with 1× PBS Tween-20 (PBST) buffer and then incubated with the primary antibody against ERβ (1:1,000, 14007-1-AP, Proteintech). After rinsing with 1× PBST buffer, the membrane was incubated with HRP-conjugated secondary antibody. ChemiDoc XRS (Bio-Rad) with Image Lab software were used for the visualization and record of blots.
Small-interfering RNAs (siRNAs)-induced ESR2 knockdown
siRNAs targeting ESR2 were synthesized by Thermo Fisher Scientific. Two sets of stealth siRNAs were HSS103378 (targeting CCC UGC UGU GAU GAA UUA CAG CAU U) and HSS103380 (targeting CCU UUA GUG GUC CAU CGC CAG UUA U). Lipofectamine 3000 (Thermo Fisher Scientific) was used to perform transient transfection of siRNAs according to the instruction of manufacturer. Quantitative reverse transcriptase-PCR (qRT-PCR) and Western blot analysis were performed to examine knockdown efficiency of siRNA targeting ESR2.
In vitro functional assay
A series of in vitro functional assays including cell proliferation, cell migration and cell invasion assay were conducted according to previous description (21). Two human GC cell lines, AGS and HGC27, were purchased from Shanghai Institutes for Biological Sciences. For cell proliferation assay, Cell Counting Kit-8 (CCK-8) solution (Dojindo Molecular Technologies, Kumamoto, Japan) was added at certain interval to individual well seeded with 1×103 to 2×103 cells. After 2 hours of cell incubation, a multi-well spectrophotometer (microtiter plate reader) was used to measure the optical density which correlates with the activity of cell proliferation. On the other hand, Transwell assays were used to evaluate both cell migration and cell invasion. Chambers of 8 µm Transwell inserts (BD FalconTM, BD, Franklin Lakes, USA) with and without Matrigel coating were used for cell migration (insert without the Matrigel coating) and cell invasion (insert with the Matrigel coating). Cells (1×104 to 4×104) suspended in serum-free medium were seeded in the upper chamber while serum-containing medium was in the lower chamber of the 24-well plate for attracting cell. After 48–72 hours of incubation, cells remained in the upper chamber were removed while penetrated cells were fixed with 4% polyformaldehyde and then stained with 0.1% crystal violet. IX71 inverted microscope (Olympus Corp, Tokyo, Japan) and Image J software were used to calculate and analyze the square of stained crystal violet according to previous description (23).
Statistical analysis
Transcriptome sequencing data was analyzed as described above (section “Transcriptome sequencing data analysis including identification and functional annotation of differentially expressed genes (DEGs)”). The potential association between expression level (high/moderate/low or negative) of ESR2 and clinicopathological parameters of GC patients was examined by Chi-square test (χ2 test) or Fisher’s exact test. Kaplan-Meier method with a log-rank test was used to analyze the survival data of GC patients. The results of in vitro functional assays (including cell proliferation, cell migration and cell invasion assay) were demonstrated as the mean ± standard deviation (SD) from three independent experiments for technical replication. Significance of difference between experimental group (siESR2) and control group (siNC) was analyzed by the Student’s t-test. Differences of IHC score as well as ESR2 expression between paired ovarian metastases and primary tumors were analyzed by Wilcoxon signed-rank test. A two-tailed P value less than 0.05 was recognized as statistical significance. SPSS software (version 23.0, IBM SPSS Statistics lnc., Armonk, USA) was used to conduct these statistical analyses.
Results
Transcriptome characterization of GC OM
Surgically resected cancerous tissues including both primary GCs and matched OMs from GC patients (n=7) were collected from Fudan University Shanghai Cancer Center. Paired samples were submitted for high-throughput paired-end transcriptome sequencing. The normalized expression level of each gene was measured by FPKM. The threshold of average fold change >2.0 and P<0.05 was used to detect DEGs between primary and ovarian metastatic tumors (Figure 1A,1B). Several selected significantly up-regulated genes [claudin 11 (CLDN11), homeobox D8 (HOXD8) and ESR2] and down-regulated [C-X-C motif chemokine ligand 14 (CXCL14), serine peptidase inhibitor kazal type 1 (SPINK1)] in OMs over primary tumors were further validated by qRT-PCR (Figure 1C).
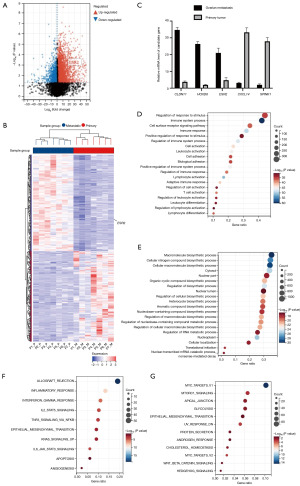
Next, we aimed to characterize the biological functions of the DEGs between primary tumors and OMs. It was identified in the GO enrichment analysis that the DEGs were classified into multiple functional categories. Notably, multiple immune-related pathways including regulation of response to stimulus, immune system process, immune response and regulation of immune system process were significantly down-regulated in OMs when compared to primary tumors. On the contrary, several biosynthetic process-related pathways were up-regulated in OMs (Figure 1D,1E). Similarly, we conducted hallmark enrichment analysis and found that a variety of immune-related pathways including allograft rejection, inflammatory response, interferon gamma response, etc. were remarkably down-regulated in the OMs (Figure 1F,1G), strongly indicating the dysregulation of certain immune cell subsets within TIME in the specific type of GC metastasis.
Differential TIME between primary and ovarian metastatic tumors
Recently, CIBERSORT (https://cibersort.stanford.edu/), a computational algorithm for quantifying cell fractions from bulk tissue gene expression profiles, has been developed and widely applied to profile the relative expression level of tumor infiltrating immune cell subsets (19). Considering the remarkably dysregulated immune-related signaling pathways between primary gastric and ovarian metastatic tumor samples, we hereby compared the relative proportion of 22 subsets of immune cells to characterize the distinctive TIME between them. It was shown that various key subsets were differentially expressed, such as plasma cells, mast cells, CD4+ naïve and memory T cells. In particular, M1 macrophages were significantly up-regulated in the primary tumors whereas M2 macrophages were significantly up-regulated in the OMs (Figure 2A). In fact, M2 macrophage was the highest expressed subset in the metastatic lesions and its differential expression between primary GC and OM ranked the top among all the dysregulated subsets of immune cells. To further validate the finding, we conducted the mIF staining of CD68 and CD206, two key markers of M2 macrophage, on the paired primary and ovarian metastatic lesions. In line with the CIBERSORT analysis, we quantified the CD68+CD206+ M2 macrophage on both tumors (Figure 2B,2C) and found that M2 macrophage was significantly enriched in the OMs than primary tumors (Figure 2D), reinforcing the observation that M2 macrophage was overexpressed in the OMs according to our RNA-seq analysis.
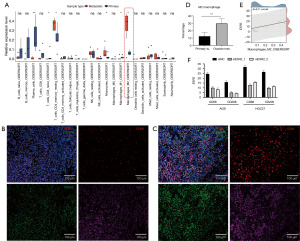
With respect to the DEGs between primary tumors and OMs, we noted that ESR2, the protein-coding gene of ERβ, was significantly up-regulated in the OMs. Considering the reported oncogenic role of estrogen receptors in the progression of multiple malignancies (24), especially in such unique site of distant metastasis occurred merely in female GC patients (18), as well as the differential expression of M2 macrophage shown above, we then focused on the role of ESR2 in the OM of GC. We evaluated their potential correlation and found that ESR2 expression was potentially correlated with the proportion of M2 macrophage (Pearson correlation coefficient r=0.44, P=0.11, Figure 2E). To further validate their correlation, we suppressed ESR2 expression in AGS and HGC27 GC cell lines by siRNA (Figure S1A,S1B) and compared the mRNA level of CD68 and CD206 between GC cell lines with and without the down-regulation of ESR2. In both AGS and HGC27 cell lines, we found that suppressing ESR2 correspondingly decreased the expression of CD68 and CD206 (Figure 2F), indicating the potential correlation between ESR2 expression and M2 macrophage in GC. In addition, we also investigated their correlation in The Cancer Genome Atlas Stomach Adenocarcinoma (TCGA-STAD) dataset but conversely found that ESR2 expression was negatively correlated with M2 macrophage (Figure S1C). As this study was based on our own cohort of paired primary and ovarian metastatic GCs, which was remarkably different from the TCGA study that excluded any metastatic patients, such contrast rightly reflected the significance of ESR2 and its correlation with M2 macrophage in the circumstance of this unique type of GC metastasis. Therefore, we focused on ESR2 to further examine its potential role in GC progression and metastasis.
ESR2 was selectively overexpressed in OM and correlated with the severity of GC
To confirm the overexpression of ESR2 in OMs over primary tumors, we examined the expression of ESR2 in a cohort of GC patients with primary tumors and matching OMs (N=21) as well as a cohort of GC patients with primary tumors and matching hepatic and peritoneal metastases [hepatic metastasis (HM) and peritoneal metastasis (PM) in short, respectively] (N=15). Remarkably, it was noted in the IHC staining-based cohort validation that ESR2 expression were significantly increased in the OM in comparison with primary tumor (P<0.05) (Figure 3A,3B). On the contrary, ESR2 expression was comparable between paired primary tumor and HM/PM (P=0.84) (Figure 3C,3D). We observed similar results of dysregulated ESR2 expression between primary GC and OM rather than PM/HM based on Western blot analysis (Figure S1D-S1G). These results suggested the selective overexpression of ESR2 in the distant metastases to ovary rather than other sites, indicating the specific correlation of ESR2 with ovarian metastatic GC. To further elucidate the role of ESR2 in GC progression, we examined its expression in 90 pairs of surgically resected primary GC tissues and normal gastric mucosae. Expression of ESR2 on mRNA was significantly higher in tumors when compared with non-tumor tissues (Figure 3E). Similarly, IHC staining of ESR2 demonstrated its higher expression in cancerous tissue than normal mucosae as high expression of ESR2 was 68.9% (62/90) in GC tissue and 54.4% (49/90) in adjacent normal gastric mucosa (Figure 3F,3G). Clinicopathological analysis revealed the significant association between high expression of ESR2 and younger age (P=0.03), lymph node metastasis (P=0.02) as well as advanced pathological stage (P=0.03) (Table 2). Kaplan-Meier survival analysis demonstrated that high expression of ESR2 correlated with a worse long-term survival of 283 GC patients from the Asian Cancer Research Group (ACRG) cohort (GSE62254) (Figure 3H). Taken together, these results indicated the strong association between high expression of ESR2 and severity of GC.
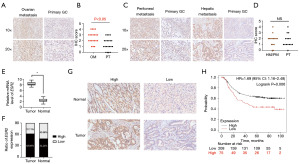
Table 2
| Variable | Case number | Immunostaining of ERβ | P value | |
|---|---|---|---|---|
| High | Low | |||
| Surgically resected samples | 0.046* | |||
| Non-tumor tissue | 90 | 62 | 28 | |
| GC tissue | 90 | 49 | 41 | |
| Age | 0.03* | |||
| <50 years old | 35 | 27 | 8 | |
| ≥50 years old | 55 | 30 | 25 | |
| Gender | 0.62 | |||
| Male | 63 | 43 | 20 | |
| Female | 27 | 17 | 10 | |
| Tumor size | 0.39 | |||
| <5 cm | 52 | 32 | 20 | |
| ≥5 cm | 38 | 20 | 18 | |
| Differentiation | 0.46 | |||
| Low | 66 | 37 | 14 | |
| Medium/high | 24 | 9 | 5 | |
| Lauren’s classification | 0.21 | |||
| Diffuse | 39 | 30 | 9 | |
| Intestinal | 51 | 33 | 18 | |
| T stage | 0.20 | |||
| T1 + T2 | 19 | 13 | 6 | |
| T3 + T4 | 71 | 37 | 34 | |
| Lymph node metastasis | 0.02* | |||
| Negative | 24 | 12 | 12 | |
| Positive | 66 | 49 | 17 | |
| Pathological stage | 0.03* | |||
| I + II | 35 | 20 | 15 | |
| III + IV | 55 | 43 | 12 | |
*, P<0.05, indicating that difference between two groups is statistically significant. ERβ, estrogen receptor β; GC, gastric cancer.
siRNA-induced ESR2 knockdown inhibited GC cell migration and invasion in vitro
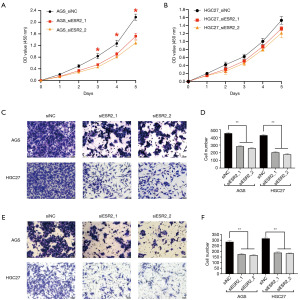
Considering the association of increased ESR2 expression with GC severity, especially in the OM of GC, we speculated that ESR2 played pro-oncogenic roles in GC progression and metastasis. To explore this, we suppressed ESR2 expression in AGS and HGC27 GC cell lines by siRNA (Figure S1A,S1B). It was shown in the CCK-8 cell proliferation assays that ESR2 knockdown significantly hindered the growth of AGS but not HGC27 cell lines 72, 96 and 108 hours after cell seeding (Figure 4A,4B), implying that the impact of ESR2 on cancer cell proliferation varied between different cell lines. With respect to cell migration and invasiveness, it was observed in the Transwell migration assays that siRNA-induced knockdown of ESR2 expression exerted significant inhibitory effect on the migration of both AGS and HGC27 cells (Figure 4C,4D). Similarly, these observations were resembled in the Transwell invasion assay that siRNA-induced knockdown of ESR2 in both AGS and HGC27 cells significantly decreased number of cells invading through the chamber (Figure 4E,4F). Collectively, these results demonstrated that enforced down-regulation of ESR2 expression hindered GC cell migration and invasion in vitro, indicating that ESR2 played pro-oncogenic and pro-metastatic roles in GC.
Discussion
Ovarian metastatic tumors originated from non-genital malignancies such as gastrointestinal, bile duct, breast cancer are not rarely seen, with stomach remains the most frequent site of primary tumors (25). Although OM confers an extremely unsatisfying prognosis of GC patient, the molecular pathogenesis governing its occurrence remains poorly understood, let alone the distinctive characteristics of immune microenvironment between primary GC and its ovarian metastatic lesion. In this study, we identified distinctive sets of DEGs in between them by conducting transcriptome sequencing of paired primary tumors and OMs of seven eligible GC patients. Follow-up analyses revealed multiple immune- and cancer-related pathways which were activated and/or inhibited in OMs when compared with primary tumors. Among several dysregulated subsets of immune cells between them, pro-oncogenic and immunosuppressive M2 macrophage was significantly up-regulated in OMs, which was validated by mIF-based quantification of CD68+CD206+ M2 macrophage between primary tumors and OMs. Furthermore, we identified that ESR2, which encodes ERβ, was not only overexpressed in OMs but also potentially correlated with M2 macrophage in our analyses. Investigation of ESR2 by clinical cohort validation and functional assays indicated its unique overexpression in OM and demonstrated its potential of promoting metastasis of GC cells. To our knowledge, this is the first integrative TIME-related study of OM in GC by combining comparative RNA-seq study of paired tumors with mIF and IHC-based clinical cohort studies as well as in vitro functional assays of candidate gene in this specific pattern of deadly metastasis.
It is widely recognized that TIME plays crucial roles in cancer progression and metastasis (26). In GC, a large body of studies have uncovered that various types of infiltrating subsets of immune cells, such as CD4+ regulatory T cell and tumor-associated macrophage (TAM), are shown to promote cancer progression and metastasis, whereas other immune effector cells including CD8+ cytotoxic T cells, natural killer cells and etc. play the opposite role in this process (27,28). Among the dysregulated immune cell subsets, the immunosuppressive and protumoral M2 macrophage is particularly known to actively promote GC metastasis through various mechanisms (29). For instance, Yamaguchi et al. compared the ascites and peritoneal lavage samples of GC patients with and without peritoneal metastasis (PM) and found that CD68+CD163+ or CD68+CD206+ M2 macrophages were significantly enriched in patients with PM than without PM (30). They also found that co-culture of GC cell lines with M2 macrophages promoted the progression of GC both in vitro and in vivo (30). Moreover, M2 macrophage-secreted CHI3L1 interacted with interleukin-13 receptor α2 chain (IL-13Rα2) located on the membranes of cancer cells, which then activated the MAPK signaling pathway and up-regulated matrix metalloproteinase genes, leading to the enhanced GC metastasis (31). Li et al. found that GC cell-secreted mesenchymal stromal cells promoted the M2 polarization of macrophages by secreting IL-6 and IL-8 via the JAK2/STAT3 pathway. The polarized M2, in turn, promoted GC cell metastasis through advancing the epithelial-mesenchymal transition (EMT) process (32). However, little is known when it comes to the relationship between TIME, especially M2 macrophage, and OM, even though a few studies explored its underlying mechanism (33,34). As comparative transcriptome profiling of primary and ovarian metastatic tumor was rarely introduced in previous studies, key immune cell subsets and molecules underlying OM are yet to be unveiled. Consequently, we profiled gene expression of paired primary GC and OM and revealed that plasma cells, mast cells, CD4+ naïve, memory T cells, etc. were dysregulated between them. It is worth noting that the pro-inflammatory and tumor-inhibiting M1 macrophages were significantly up-regulated in the primary tumors whereas the anti-inflammatory and tumor-promoting M2 macrophages were significantly up-regulated in the OMs. We also validated these findings by conducting mIF staining to quantify the differential amounts of CD68+CD206+ M2 macrophages between paired tumors.
According to our RNA-seq analyses, ESR2 was significantly overexpressed in OMs over primary tumors. In fact, emerging studies have indicated that GC is a hormone-associated malignancy (16,35,36) whereas the broad impact of estrogen on multiple aspects of malignancies is mainly mediated by its interaction with estrogen receptors, which act as transcription factors that bind to the regulatory domains of target oncogenic or tumor-suppressing genes and influence their activity of transcription (37). Considering the ovary as a major target of elevated activity of sex hormone in young female GC patients (38), our finding aroused our interest to further investigate its role in the occurrence of OM. In fact, the correlation between high expression of ESR2 and younger age was observed in our clinicopathological analysis, which was in line with previous report that ERβ is more frequently seen in younger GC patients (39). Additionally, the correlation analyses uncovered that up-regulated expression of ESR2 potentially correlated with higher proportion of M2 macrophage. Although their correlation was not statistically significant, which was possibly due to the limited number of samples, the tendency of correlation was clear (r=0.44) and previous studies have also shown that estrogen and its receptors are involved in cancer progression and metastasis through mediating certain immune cell subsets and potentially act as an innate regulator of polarization of TAM (40-42). More importantly, we have demonstrated that siRNA-induced down-regulation of ESR2 correspondingly decreased the expression level of CD68 and CD206, both of which were key markers of M2 macrophage, further implying that increased expression of ESR2 could induce M2 macrophage in GC. In fact, previous studies have shown the close ties between estrogen and its receptors and the polarization and function of TAMs (43). For instance, Jing et al. found that ERα in TAMs played an oncogenic role by expediting the mTOR/KIF5B-induced EMT as well as promoting cancer immune evasion in endometrial cancer cells (44). Additionally, to respond to the TLR4 signaling in macrophages, estrogen is reported to induce the release of inflammatory cytokines (IL-1β, IL-6, and TNF-α) and facilitate both tumor-associated inflammation and cancer immune evasion (45). In summary, these findings suggest that estrogen and its receptors contribute to immunosuppression by promoting M2 polarization of TAMs via multiple molecular mechanisms. Consequently, ESR2 was selected as a key candidate regulator of M2 macrophage in GC and its OM for further clinical and functional investigation.
So far, the exact role of ESR2 in GC remains in controversy. For instance, Ryu et al. profiled expression status of ERβ in 148 GC patients and revealed the association between its positive expression with lower tumor stage, negative perineural invasion and more importantly, a better 3-year survival (46). This is in line with conclusions from a meta-analysis that high expression of ERβ was negatively associated with lymph node metastasis (47). On the contrary, Takano et al. demonstrated the preferential expression of ERβ in GC as well as the association of elevated ERβ expression with increased metastatic potential (48). Moreover, Zhou et al. observed that positive expression of ERβ was more frequently observed in younger patients with advanced pathological TNM stages in early-onset GC (39). Polymorphism analyses of both east Asian and north American cohorts by Sunakawa et al. uncovered the association between genetic variation in ESR2 and higher survival rate of patients with locally advanced GC (49). With respect to the significance of ERβ in the OM of GC, there has only been a few retrospective studies based on IHC staining of ERβ on resected GC samples. Yu et al. reviewed the expression status of ERβ in 152 GC patients with synchronous and metachronous OM and identified positive expression of ERβ as a favorable prognostic factor for overall survival (17). The controversy of ERβ in cancer research is partially attributed to methodological limitations such as unspecific commercial antibodies, the structural complexity of this gene, starting material for the study, etc. (50). Mechanistically, it is increasingly recognized that ESR2 plays a bi-faceted role of pro- and anti-tumorigenic, possibly dependent on the nature or mutation status of its downstream effectors (51). Reportedly, ERβ plays an oncogenic role in GC cells via specific key regulators and pathway (16). Enforced knockdown of ESR2 not only leads to apoptosis via the activation of GADD45α, a canonical p53 target gene that can induce cell cycle arrest in a p53-independent manner, but also induces autophagy in GC cells, which is partially mediated through MAPK pathway. Moreover, GC cell growth is hormone-dependent as low-dose estrogen (10 nM) induces the proliferative effect on GC cells in addition to activation of Erk1/2 (16), highlighting the role of estrogen and its interaction with ESR2 in GC progression.
A major limitation of our study is the lack of faithful in vivo ovarian metastatic model to validate the potential of ESR2 to promote distant metastasis of primary GC cells to the ovary. In fact, the establishment of in vivo model regarding hepatic, peritoneal and abdominal metastasis of GC was reportedly successful by orthotopic transplantation of GC cell lines or tissues from patient-derived xenograft (52-54). Unfortunately, modeling metastasis to ovary is yet to be accomplished and should be urgently addressed. Secondly, we did not add estrogen to GC cells in the functional assays. Therefore, it remains unclear whether the interaction between estrogen and its receptor ERβ exerts additional effect on the proliferation, migration and invasion of GC cells. In addition, as the fast-developing single-cell sequencing enables a more sensitive and deeper transcriptomic characterization of primary GC as well as its distant metastasis (55,56), this technique should be applied to specifically decipher the molecular pathogenesis of OM in the next step. Lastly, although ESR2 expression was potentially correlated with M2 macrophage (Pearson correlation coefficient r=0.44), such correlation did not reach the statistical significance. Limited number of samples (n=7) possibly underlies the insignificance so that further collection of more samples is required to confirm the correlation between them.
In summary, the present study unveiled distinctive gene expression, immune- and cancer-related pathways as well as TIME in the OM of GC by comparative transcriptomic profiling of paired primary and ovarian metastatic lesions. M2 macrophages were significantly enriched in OMs than primary tumors. ESR2, the protein-coding gene of ERβ, was significantly up-regulated in the OMs and potentially correlated with M2 macrophage accumulation in the metastatic lesions. Following clinical cohort study verified the unique overexpression of ESR2 in the OMs while in vitro functional assays demonstrated its potential to promote GC cell migration and invasion. Future research shall not only address the above-mentioned limitations but more importantly, focus on not only investigating the molecular mechanism underlying ESR2-mediated dysregulation of TIME, especially M2 macrophage, but also identifying key regulators and pathways of OM for potential targeted therapy in the future.
Conclusions
Comparative RNA sequencing analysis unveiled dysregulated TIME, especially the immunosuppressive and protumoral M2 macrophage subset between primary GC and its ovarian metastatic lesions. Notably, ESR2 was potentially correlated with M2 macrophage and was indicated to promote oncogenic processes during the progression and metastasis of GC.
Acknowledgments
We thank the tissue bank of the Pathology Department of Fudan University Shanghai Cancer Center for their generous help.
Funding: This work was supported by the grant from
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-124/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-124/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-124/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-124/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the local ethics committee of the Fudan University Shanghai Cancer Center (No. 050432-4-1911D) and informed consent was taken from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet 2016;388:2654-64. [Crossref] [PubMed]
- Yasufuku I, Tsuchiya H, Fujibayashi S, et al. Oligometastasis of Gastric Cancer: A Review. Cancers (Basel) 2024;16:673. [Crossref] [PubMed]
- Kubeček O, Laco J, Špaček J, et al. The pathogenesis, diagnosis, and management of metastatic tumors to the ovary: a comprehensive review. Clin Exp Metastasis 2017;34:295-307. [Crossref] [PubMed]
- Zhang C, Hou W, Huang J, et al. Effects of metastasectomy and other factors on survival of patients with ovarian metastases from gastric cancer: a systematic review and meta-analysis. J Cell Biochem 2019;120:14486-98. [Crossref] [PubMed]
- Yan D, Du Y, Dai G, et al. Management Of Synchronous Krukenberg Tumors From Gastric Cancer: a Single-center Experience. J Cancer 2018;9:4197-203. [Crossref] [PubMed]
- Ma F, Li Y, Li W, et al. Metastasectomy Improves the Survival of Gastric Cancer Patients with Krukenberg Tumors: A Retrospective Analysis of 182 patients. Cancer Manag Res 2019;11:10573-80. [Crossref] [PubMed]
- Moehler M, Shitara K, Garrido M, et al. LBA6_PR Nivolumab (nivo) plus chemotherapy (chemo) versus chemo as first-line (1L) treatment for advanced gastric cancer/gastroesophageal junction cancer (GC/GEJC)/esophageal adenocarcinoma (EAC): First results of the CheckMate 649 study. Ann Oncol 2020;31:S1191. [Crossref]
- Xu J, Jiang H, Pan Y, et al. LBA53 Sintilimab plus chemotherapy (chemo) versus chemo as first-line treatment for advanced gastric or gastroesophageal junction (G/GEJ) adenocarcinoma (ORIENT-16): First results of a randomized, double-blind, phase III study. Ann Oncol 2021;32:S1331. [Crossref]
- Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res 2019;79:4557-66. [Crossref] [PubMed]
- Lei X, Lei Y, Li JK, et al. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett 2020;470:126-33. [Crossref] [PubMed]
- Varešlija D, Priedigkeit N, Fagan A, et al. Transcriptome Characterization of Matched Primary Breast and Brain Metastatic Tumors to Detect Novel Actionable Targets. J Natl Cancer Inst 2019;111:388-98. [Crossref] [PubMed]
- Zhang Q, Abdo R, Iosef C, et al. The spatial transcriptomic landscape of non-small cell lung cancer brain metastasis. Nat Commun 2022;13:5983. [Crossref] [PubMed]
- Gambaro K, Marques M, McNamara S, et al. Copy number and transcriptome alterations associated with metastatic lesion response to treatment in colorectal cancer. Clin Transl Med 2021;11:e401. [Crossref] [PubMed]
- Ur Rahman MS, Cao J. Estrogen receptors in gastric cancer: Advances and perspectives. World J Gastroenterol 2016;22:2475-82. [Crossref] [PubMed]
- Zhang Y, Cong X, Li Z, et al. Estrogen facilitates gastric cancer cell proliferation and invasion through promoting the secretion of interleukin-6 by cancer-associated fibroblasts. Int Immunopharmacol 2020;78:105937. [Crossref] [PubMed]
- Zhou F, Jin J, Zhou L, et al. Suppression of estrogen receptor-beta promotes gastric cancer cell apoptosis with induction of autophagy. Am J Transl Res 2020;12:4397-409. [PubMed]
- Yu P, Huang L, Cheng G, et al. Treatment strategy and prognostic factors for Krukenberg tumors of gastric origin: report of a 10-year single-center experience from China. Oncotarget 2017;8:82558-70. [Crossref] [PubMed]
- Gao J, Weng W, Qu X, et al. Risk factors predicting the occurrence of metachronous ovarian metastasis of gastric cancer. Ann Transl Med 2021;9:1049. [Crossref] [PubMed]
- Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59. [Crossref] [PubMed]
- Chen Y, Jia K, Sun Y, et al. Predicting response to immunotherapy in gastric cancer via multi-dimensional analyses of the tumour immune microenvironment. Nat Commun 2022;13:4851. [Crossref] [PubMed]
- Gao J, Pan H, Zhu Z, et al. Guanine nucleotide-binding protein subunit beta-4 promotes gastric cancer progression via activating Erk1/2. Acta Biochim Biophys Sin (Shanghai) 2020;52:975-87. [Crossref] [PubMed]
- Kang X, Li W, Liu W, et al. LIMK1 promotes peritoneal metastasis of gastric cancer and is a therapeutic target. Oncogene 2021;40:3422-33. [Crossref] [PubMed]
- Pijuan J, Barceló C, Moreno DF, et al. In vitro Cell Migration, Invasion, and Adhesion Assays: From Cell Imaging to Data Analysis. Front Cell Dev Biol 2019;7:107. [Crossref] [PubMed]
- Zhang D, Ku J, Yi Y, et al. The prognostic values of estrogen receptor alpha and beta in patients with gastroesophageal cancer: A meta-analysis. Medicine (Baltimore) 2019;98:e17954. [Crossref] [PubMed]
- Xie H, Erickson BJ, Sheedy SP, et al. The diagnosis and outcome of Krukenberg tumors. J Gastrointest Oncol 2021;12:226-36. [Crossref] [PubMed]
- El-Kenawi A, Hänggi K, Ruffell B. The Immune Microenvironment and Cancer Metastasis. Cold Spring Harb Perspect Med 2020;10:a037424. [Crossref] [PubMed]
- Ma M, Sun J, Liu Z, et al. The Immune Microenvironment in Gastric Cancer: Prognostic Prediction. Front Oncol 2022;12:836389. [Crossref] [PubMed]
- Wang W, Ye LF, Bao H, et al. Heterogeneity and evolution of tumour immune microenvironment in metastatic gastroesophageal adenocarcinoma. Gastric Cancer 2022;25:1017-30. [Crossref] [PubMed]
- Gambardella V, Castillo J, Tarazona N, et al. The role of tumor-associated macrophages in gastric cancer development and their potential as a therapeutic target. Cancer Treat Rev 2020;86:102015. [Crossref] [PubMed]
- Yamaguchi T, Fushida S, Yamamoto Y, et al. Tumor-associated macrophages of the M2 phenotype contribute to progression in gastric cancer with peritoneal dissemination. Gastric Cancer 2016;19:1052-65. [Crossref] [PubMed]
- Chen Y, Zhang S, Wang Q, et al. Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein. J Hematol Oncol 2017;10:36. [Crossref] [PubMed]
- Li W, Zhang X, Wu F, et al. Gastric cancer-derived mesenchymal stromal cells trigger M2 macrophage polarization that promotes metastasis and EMT in gastric cancer. Cell Death Dis 2019;10:918. [Crossref] [PubMed]
- Nadauld LD, Garcia S, Natsoulis G, et al. Metastatic tumor evolution and organoid modeling implicate TGFBR2 as a cancer driver in diffuse gastric cancer. Genome Biol 2014;15:428. [Crossref] [PubMed]
- Lee EK, Song KA, Chae JH, et al. GAGE12 mediates human gastric carcinoma growth and metastasis. Int J Cancer 2015;136:2284-92. [Crossref] [PubMed]
- Jang YC, Leung CY, Huang HL. Association of hormone replacement therapy with risk of gastric cancer: a systematic review and meta-analysis. Sci Rep 2022;12:12997. [Crossref] [PubMed]
- Jang Y, Mok Y. The estrogenic hormone effect in gastric cancer. J Clin Oncol 2018;36:85. [Crossref]
- Hua H, Zhang H, Kong Q, et al. Mechanisms for estrogen receptor expression in human cancer. Exp Hematol Oncol 2018;7:24. [Crossref] [PubMed]
- Folkerd EJ, Dowsett M. Influence of sex hormones on cancer progression. J Clin Oncol 2010;28:4038-44. [Crossref] [PubMed]
- Zhou F, Xu Y, Shi J, et al. Expression profile of E-cadherin, estrogen receptors, and P53 in early-onset gastric cancers. Cancer Med 2016;5:3403-11. [Crossref] [PubMed]
- Orzołek I, Sobieraj J, Domagała-Kulawik J. Estrogens, Cancer and Immunity. Cancers (Basel) 2022;14:2265. [Crossref] [PubMed]
- Milette S, Hashimoto M, Perrino S, et al. Sexual dimorphism and the role of estrogen in the immune microenvironment of liver metastases. Nat Commun 2019;10:5745. [Crossref] [PubMed]
- Chakraborty B, Byemerwa J, Shepherd J, et al. Inhibition of estrogen signaling in myeloid cells increases tumor immunity in melanoma. J Clin Invest 2021;131:e151347. [Crossref] [PubMed]
- Wang T, Jin J, Qian C, et al. Estrogen/ER in anti-tumor immunity regulation to tumor cell and tumor microenvironment. Cancer Cell Int 2021;21:295. [Crossref] [PubMed]
- Jing X, Peng J, Dou Y, et al. Macrophage ERα promoted invasion of endometrial cancer cell by mTOR/KIF5B-mediated epithelial to mesenchymal transition. Immunol Cell Biol 2019;97:563-76. [Crossref] [PubMed]
- Calippe B, Douin-Echinard V, Delpy L, et al. 17Beta-estradiol promotes TLR4-triggered proinflammatory mediator production through direct estrogen receptor alpha signaling in macrophages in vivo. J Immunol 2010;185:1169-76. [Crossref] [PubMed]
- Ryu WS, Kim JH, Jang YJ, et al. Expression of estrogen receptors in gastric cancer and their clinical significance. J Surg Oncol 2012;106:456-61. [Crossref] [PubMed]
- Ge H, Yan Y, Tian F, et al. Prognostic value of estrogen receptor α and estrogen receptor β in gastric cancer based on a meta-analysis and The Cancer Genome Atlas (TCGA) datasets. Int J Surg 2018;53:24-31. [Crossref] [PubMed]
- Takano N, Iizuka N, Hazama S, et al. Expression of estrogen receptor-alpha and -beta mRNAs in human gastric cancer. Cancer Lett 2002;176:129-35. [Crossref] [PubMed]
- Sunakawa Y, Cao S, Berger MD, et al. Estrogen receptor-beta genetic variations and overall survival in patients with locally advanced gastric cancer. Pharmacogenomics J 2017;17:36-41. [Crossref] [PubMed]
- Božović A, Mandušić V, Todorović L, et al. Estrogen Receptor Beta: The Promising Biomarker and Potential Target in Metastases. Int J Mol Sci 2021;22:1656. [Crossref] [PubMed]
- Mukhopadhyay UK, Oturkar CC, Adams C, et al. TP53 Status as a Determinant of Pro- vs Anti-Tumorigenic Effects of Estrogen Receptor-Beta in Breast Cancer. J Natl Cancer Inst 2019;111:1202-15. [Crossref] [PubMed]
- Kang W, Maher L, Michaud M, et al. Development of a Novel Orthotopic Gastric Cancer Mouse Model. Biol Proced Online 2021;23:1. [Crossref] [PubMed]
- Miwa T, Kanda M, Umeda S, et al. Establishment of Peritoneal and Hepatic Metastasis Mouse Xenograft Models Using Gastric Cancer Cell Lines. In Vivo 2019;33:1785-92. [Crossref] [PubMed]
- Zhu Y, Hu Y, Cheng M, et al. Establishment and Characterization of a Nude Mouse Model of Subcutaneously Implanted Tumors and Abdominal Metastasis in Gastric Cancer. Gastroenterol Res Pract 2017;2017:6856107. [Crossref] [PubMed]
- Zhang M, Hu S, Min M, et al. Dissecting transcriptional heterogeneity in primary gastric adenocarcinoma by single cell RNA sequencing. Gut 2021;70:464-75. [Crossref] [PubMed]
- Wang B, Zhang Y, Qing T, et al. Comprehensive analysis of metastatic gastric cancer tumour cells using single-cell RNA-seq. Sci Rep 2021;11:1141. [Crossref] [PubMed]


