Overview of the interplay between m6A methylation modification and non-coding RNA and their impact on tumor cells
Introduction
Extensive research has demonstrated that non-coding RNAs (ncRNAs) play a crucial role in the regulation of phenotypic traits associated with tumor cell proliferation, apoptosis, drug resistance, and stem cell characteristics (1). For instance, the circular RNAs (circRNA) circGPR137B has been shown to inhibit the proliferation, invasion, and metastasis of tumor cells in liver cancer (2). The long non-coding RNA (lncRNA) CASC8 promotes chemoresistance in esophageal squamous cell carcinoma by upregulating heterogeneous nuclear ribonucleoprotein L (hnRNPL) (3). Additionally, upregulation of the lncRNA DDIT4-AS1 contributes to the maintenance of pancreatic cancer stemness (4).
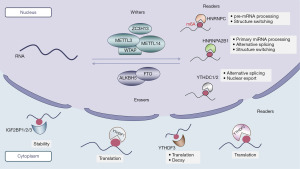
N6-methyladenosine (m6A), as a reversible epigenetic RNA modification, plays a vital role in the regulation of RNA translation, splicing, degradation, and other biological processes, closely associated with tumor development (5). Research has revealed that external factors such as radiation and hypoxia can disrupt the balance of m6A levels within cells, leading to cellular carcinogenesis. For instance, smoking can regulate the m6A modification of LINC0027 through low methylation by ALKBH5, promoting the progression of male esophageal squamous cell carcinoma (6). Cadmium exposure can inhibit m6A modification of pri-miR-374c, inducing breast cancer proliferation and metastasis (7). Furthermore, gene expression dysregulation following cellular carcinogenesis can further disrupt m6A levels within cells (8). Imbalanced m6A methylation levels in tumors have been identified as a critical factor contributing to the dysregulation of ncRNA expression. For example, increased m6A levels of lncRNA RHPN1-AS1 have been observed in ovarian cancer, resulting in upregulated RHPN1-AS1 expression (9). In hepatocellular carcinoma (HCC) cells, the m6A demethylase ALKBH5 decreases the m6A modification level of LINC02551, leading to increased LINC02551 expression (10). In esophageal cancer, METTL3 upregulates miR-20a-5p expression by enhancing its m6A modification level (11). Decreased m6A methylation levels in chronic myeloid leukemia (CML) accelerate the degradation of lncRNA NEAT1, which, through the NEAT1/miR-766-5p/CDKN1A axis, inhibits apoptosis and enhances the viability of CML cells (12). Furthermore, the enzymatic activity and stability of m6A regulatory factors are also influenced by ncRNAs. For example, miR-628-5p can suppress the expression of METTL14 in HCC (13). LncRNA LINRIS stabilizes IGF2BP2 in colorectal cancer (CRC). The m6A modification process is dependent on three important regulatory factors: writers, erasers, and readers, which respectively add, remove, or read an m6A site (Figure 1). The interplay between ncRNAs and m6A regulatory factors collectively influences tumor initiation and progression, providing insights into early cancer diagnosis, prognosis prediction, and therapeutic strategies.
M6A regulatory factors participate in the regulation of tumors through ncRNAs
M6A regulatory factors participate in the regulation of tumors through lncRNAs
M6A regulatory factors regulate tumor progression by influencing the stability of lncRNAs
The alteration of lncRNA stability is a primary regulatory method utilized by m6A regulatory factors. These factors modulate the proliferation, migration, and invasive capabilities of tumor cells by altering the stability of lncRNAs. For instance, METTL3 in various cancers increases the m6A methylation levels of target lncRNAs such as lncRNA LCAT3 (14), ZFAS1 (15), DANCR (16), LINC00958 (17), MEG3 (18), THAP7-AS1 (19), PCAT6 (20), lnc-CTHCC (21), lncRNA sil-as1 (22), lncRNA PVT1 (23) and LIFR-AS1 (24). This is done in synergy with corresponding m6A methylation readers like IGFBP1/IGFBP2 to enhance the stability of these lncRNAs. The increase in stability of lncRNAs mediated by METTL3 promotes the proliferation, migration, and invasion of the respective tumor cells both in vitro and in vivo. Another m6A-related methyltransferase, METTL14, is upregulated in gastric and liver cancers, enhancing the stability of LINC01320 (25) and ARHGAP5-AS1 (26) by increasing their methylation levels. In head and neck squamous carcinoma, METTL14 collaborates with METTL3 to mediate the methylation-dependent stability enhancement of LNCAROD (27), promoting tumor cell proliferation and invasive metastasis. WTAP, an m6A writer, ensures the positioning of the METTL3-METTL14 heterodimer at nuclear speckles (28). In nasopharyngeal carcinoma, WTAP-mediated m6A modification via IGF2BP2 enhances the stability of the target gene lncRNA DIAPH1-AS1 (29); in osteosarcoma, WTAP stabilizes lncRNA FOXD2-AS1 and enhances the stability of target gene FOXM1 mRNA (30), promoting tumor cell growth and metastasis. In HCC, METTL16, by upregulating the methylation level of lncRNA RAB11B-AS1, reduces its transcript stability, thereby suppressing RAB11B-AS1 expression and inhibiting the proliferation, migration, and invasion of HCC cells, and promoting apoptosis (31). The demethylase ALKBH5 in gastric cancer decreases the stability of lncRNA TP53TG1 (32), enhancing cancer cell proliferation. FTO catalyzes the demethylation of LINC00022, leading to a decrease in YTHDF2-mediated degradation of LINC00022 and promoting the proliferation of esophageal squamous cell carcinoma cells (32). The m6A reader IGF2BP3, with increased expression in glioblastoma and cervical cancer, stabilizes lncRNA WEE2-AS1 (33) and lncRNA KCNMB2-AS1 (34) in a methylation-dependent manner, facilitating tumor progression. The m6A writer YTHDF2 promotes the degradation of lncRNA THOR in the cytoplasm in a methylation-dependent manner, affecting the proliferation, migration, and invasion of various cancer cells, including melanoma (35), non-small cell lung cancer (36) and osteosarcoma (37). In breast and endometrial cancer, it enhances the degradation of lncRNA FGF14-AS2 (38) and lncRNA FENDRR (39), respectively, promoting tumor cell metastasis. YTHDF3 promotes the degradation of m6A-modified lncRNA GAS5, regulating the progression of CRC through a GAS5-YAP-YTHDF3 negative feedback loop (40).
M6A regulatory factors also influence the drug resistance of tumor cells by modifying the stability of lncRNAs. In HCC, the sorafenib resistance driver LINC01273 downregulates the expression of METTL3 through a ceRNA mechanism. The decreased expression of METTL3 leads to reduced m6A levels in LINC01273, inhibiting the YTHDF2-mediated degradation of LINC01273. This results in increased expression of LINC01273, contributing to drug resistance in tumor patients (41). In liver cancer, METTL3 can also enhance the stability of the sorafenib resistance driver lncRNA DUXAP8 in an m6A-dependent manner, promoting chemotherapy resistance (42). In lung adenocarcinoma, METTL3 increases the m6A levels of the target gene lncRNA SNHG17, enhancing its stability. This enhancement is achieved through the SNHG17/EZH2/LATS2 axis, increasing the resistance of lung adenocarcinoma cells to gefitinib (43). In breast cancer, WTAP binds to the m6A modification sites of DLGAP1-AS1 and enhances its stability. This process upregulates the expression of lncRNA DLGAP1-AS1, promoting doxorubicin resistance in breast cancer (44). In esophageal squamous cell carcinoma, ALKBH5 enhances the stability of lncRNA CASC8 in a methylation-dependent manner. This enhancement promotes the interaction between CASC8 and hnRNPL, activating the Bcl/caspase3 pathway and increasing the chemotherapy resistance of tumor cells (3).
A recent study has shown that m6A regulatory factors play a role in regulating the metabolism of tumor cells by modulating the stability of lncRNAs (45). Even when oxygen is abundant and mitochondrial function is intact, tumor cells tend to undergo “aerobic glycolysis”, a phenomenon known as the Warburg effect (46). The Warburg effect, a significant characteristic of tumor energy metabolism, is closely associated with the development and progression of cancer. In non-small cell lung cancer and prostate cancer, METTL3 enhances the stability of lncRNA ABHD11-AS1 (47) and lncRNA SNHG7 (48) in an m6A-dependent manner. This enhancement boosts the Warburg effect in tumor cells via the ABHD11-AS1/EZH2/KLF4 axis and the SNHG7/SRSF1/c-MYC axis, respectively. IGF2BP2 in glioblastoma and CRC interacts with m6A methylation sites on lncRNA CASC9 (49) and lncRNA ZFAS1 (50), increasing the stability of CASC9 and ZFAS1. This process promotes aerobic glycolysis in tumor cells through the IGF2BP2/CASC9/HK2 axis and the IGF2BP2/ZFAS1/OLA1 axis. In bladder cancer, METTL14 enhances the stability of lncRNA DBET in a methylation-dependent manner, activating the PPAR signaling pathway. This activation promotes lipid metabolism in tumor cells, accelerating cancer progression (51).
M6A regulatory factors regulate tumor progression by influencing other biological processes of lncRNA
M6A regulatory factors can regulate the progression of tumor cells by influencing the splicing of lncRNA. METTL3 upregulates the m6A modification level of ANRIL, enhancing the splicing effect of SRSF3 on ANRIL, thereby promoting the DNA homologous recombination repair capacity of pancreatic cancer cells and facilitating resistance to gemcitabine in pancreatic cancer (52). Overexpression of YTHDF2 in HCC promotes the m6A-dependent splicing of PLXNB2 pre-mRNA into the iron death-related lncRNA lncFAL, upregulating the expression of lncFAL and reducing the susceptibility of liver cancer cells to iron death (53).
Furthermore, YTHDC1 has been found to selectively recognize the m6A methylation site A783 on lncRNA Hotair in breast cancer cell lines, promoting the binding of lncRNA Hotair to chromatin and driving the proliferation and invasion of triple-negative breast cancer (54). In CRC, increased m6A modification enhances the nuclear accumulation of lncRNA RP11, leading to increased expression of RP11 in CRC cells and positively regulating the migration, invasion, and epithelial-mesenchymal transition (EMT) of tumor cells both in vitro and in vivo (55). YTHDF1 also enhances the translation efficiency of lncRNA THOR by recognizing its m6A modification, promoting the proliferation, migration, and invasion of various cancer cells such as melanoma (35), non-small cell lung cancer (36), osteosarcoma (37), and renal cell carcinoma (56) in both in vitro and in vivo settings.
M6A regulatory factors participate in the regulation of tumors through microRNA (miRNA)
M6A regulatory factors regulate tumor progression by influencing the maturation of miRNA
M6A regulatory factors can modulate tumor cell proliferation, migration, and invasion by either promoting or delaying the maturation of miRNAs. Pri-miRNAs with m6A modifications are recognized by hnRNPA2B1 and interact with DGCR8, facilitating miRNA processing and thereby influencing the onset and development of cancer cells. For instance, METTL3 in various cancers promotes the maturation of pri-miRNAs, such as pri-miR-20a-5p (11), pri-miR-126-5p (57), and pri-miR-17-92 (58), through an m6A/DGCR8-dependent mechanism. These METTL3-mediated m6A-related miRNAs positively regulate tumor proliferation, migration, and invasion. This mechanism also explains how certain external factors disrupt m6A regulatory factors and affect tumor progression. For example, in pancreatic cancer, a smoke condensate induces the overexpression of METTL3, increasing m6A modification levels and promoting the maturation of pri-miR-25. miR-25-3p targets and activates the AKT-p70S6K signaling pathway, enhancing the proliferation and metastasis of pancreatic cancer cells, revealing a mechanism by which smoking contributes to the progression of pancreatic cancer (59). In CRC infected with Fusobacterium nucleatum, the bacterium elevates the m6A methylation level of pri-mir-4717 through METTL3, fostering its maturation. miR-4717 targets and suppresses the expression of the tumor suppressor MAP2K4, promoting cancer cell proliferation (60). Lin et al. discovered that deoxycholic acid, by binding with METTL3, disrupts the formation of the METTL3-METTL14-WTAP complex. This reduces the m6A methylation level of pri-mir-92b in gallbladder cancer cells, inhibiting the maturation of mir-92b-3p. Through its target gene PTEN, this leads to the deactivation of the oncogenic PI3K/AKT signaling pathway, ultimately exerting a tumor-suppressive effect (61). The hnRNPA2B1-DGCR8 complex binds with pri-miR-92a-2-5p or pri-miR-373-3p, transforming them into mature exosomal miRNAs. This activates osteoclastogenesis and inhibits osteoblastogenesis, driving osteolytic bone disease in multiple myeloma (62).
M6A regulatory factors modulate drug sensitivity in tumors by either promoting or delaying the maturation of miRNAs. In breast cancer cells, HNRNPA2/B1 enhances DROSHA processing of pre-miRNAs, leading to the downregulation of miR-29a-3p, miR-29b-3p, and miR-222, and the upregulation of miR-1266-5p, miR-1268a, and miR-671-3p. This results in decreased sensitivity of breast cancer cells to 4-hydroxytamoxifen and fulvestrant (63). In erlotinib-resistant lung adenocarcinoma cells, M6A acts through METTL3 to highly activate the miR-146a/Notch signaling pathway, while concurrently YTHDF2 suppresses TSUC7, countering the sponge-like regulatory effect of TUSC7 on miR-146a, which inhibits the Notch signaling function. These combined factors contribute to the drug resistance in lung adenocarcinoma (64). METTL14, in a methylation-dependent manner, promotes the maturation of miR-19a-5p (65) and miR-221-3p (66), respectively enhancing the resistance of non-small cell lung cancer to cisplatin and breast cancer to doxorubicin.
M6A regulatory factors control tumor cell metabolism, stemness, and radiotherapy sensitivity by promoting or delaying the maturation of miRNAs. Under hypoxic conditions, WTAP interacts with DGCR8 to facilitate the recognition of m6A-modified pri-miR-200 by DGCR8, leading to the processing of pri-miR-200 into mature miR-200. This significantly enhances the Warburg effect within cells (67). In esophageal squamous cell carcinoma cells, METTL14, through m6A modification, promotes the maturation of pri-miR-99a, increasing the stability of mir-99a-5p. This enhancement boosts the stem cell-like properties and radiation resistance of the cancer cells (68).
M6A regulatory factors regulate tumor progression by influencing other biological processes of miRNA
In addition to altering the processing and maturation of miRNA, m6A regulatory factors also regulate tumor progression by modulating pre-miRNA splicing and altering the local RNA structure in the terminal loop region of pri-miRNA. For instance, in lung cancer, METTL3 enhances the biogenesis of pre-miR-143-3p by increasing its splicing, thereby promoting lung cancer metastasis to the brain and angiogenesis through the miR-143-3p/VASH1 axis (69). In CRC, m6A, under the regulation of METTL3, modifies the local RNA structure in the terminal loop region of pri-miRNA, strengthening its interaction with RALY and facilitating the processing of specific miRNAs (such as miR-483, miR-676, and miR-877). These miRNAs systematically downregulate the expression of metabolism-related genes (ATP5I, ATP5G1, ATP5G3, and CYC1), reprogramming mitochondrial metabolism in cancer cells, inducing reactive oxygen species production, and promoting the growth and progression of CRC (70). In ovarian cancer, overexpression of miR-145 inhibits cell proliferation, migration, and apoptosis, while overexpression of YTHDF2 reduces the overall level of m6A mRNA in ovarian cancer cells, weakening the inhibitory effect of miR-145 on cell proliferation and migration. Furthermore, YTHDF2 is a direct target of miR-145, and a double-negative feedback loop exists between miR-145 and YTHDF2, collectively regulating the progression of ovarian cancer (71).
In certain circumstances, m6A regulatory factors also control the expression of miRNAs by regulating the levels of miRNA transcription factors, thereby impacting tumor progression. For example, in lung adenocarcinoma, METTL3 or YTHDF2 downregulates the transcription factor KLF4, which is a regulator of miR-1915, leading to the suppression of migration, invasion, and EMT in non-small cell lung cancer cells (72). FTO, in a methylation-dependent manner, enhances the stability and translation efficiency of the transcription factors MYC, which regulate mir-155/miR-23a, promoting the progression of glioma cells (73).
M6A regulatory factors participate in the regulation of tumors through circRNA
M6A regulatory factors regulate tumor progression by influencing the stability of circRNA
M6A regulatory factors can influence the stability of circRNA to regulate the proliferation, migration, and invasion abilities of tumor cells. In cervical cancer, ALKBH5-mediated m6A modification enhances the stability and expression of circCCDC134 in a YTHDF2-dependent manner. CircCCDC134 recruits p65 in the nucleus and acts as a miR-503-5P sponge to regulate MYB expression in the cytoplasm, ultimately stimulating HIF1A transcription and promoting cervical cancer cell proliferation and migration (74). In HCC, IGF2BP1 binds to the m6A site on circMDK, enhancing circMDK transcript stability. CircMDK, as a sponge for miR-346 and miR-874-3p, upregulates the expression of the target ATG16L1, activating the PI3K/AKT/mTOR signaling pathway and promoting liver cancer cell proliferation, migration, and invasion (75). In CRC, YTHDF2-mediated m6A modification leads to the degradation of circ_0003215. Through the circ-0003215/miR-663b/DLG4/G6PD axis, it inhibits the pentose phosphate pathway and the proliferation, migration, and invasion of CRC (76).
M6A regulatory factors can also modulate the therapeutic sensitivity of tumor cells by altering the stability of circRNA. In HCC, increased m6A modification levels enhance the stability of circRNA-SORE, which acts as a sponge for miR-103a-2-5p and miR-620-3p, competitively activating the Wnt/β-catenin pathway and promoting resistance to sorafenib in liver cancer cells (77). In hypopharyngeal squamous cell carcinoma, METTL3 enhances the stability of cirCUX1 in a methylation-dependent manner. Upregulated cirCUX1 binds to and inhibits Caspase1, reducing the release of inflammatory factors and inducing radioresistance in tumor cells (78).
M6A regulatory factors control tumor progression by influencing other biological processes of circRNA
M6A regulatory factors can also control tumor progression by influencing circRNA splicing and promoting the interaction between circRNA and miRNA. For instance, Rao et al. discovered that m6A modification also affects circRNA back-splicing and is involved in the process of HCC development caused by hepatitis B virus (HBV) infection. HBV protein upregulates METTL3 expression, increasing m6A modification of circ-ARL3. Subsequently, YTHDC1 binds to m6A-modified circ-ARL3, facilitating its back-splicing and biogenesis. Circ-ARL3 counteracts the inhibitory effect of miR-1305 on its target oncogenes, thereby promoting the proliferation and invasion of HBV-positive HCC (79). In CRC, METTL3-induced m6A modification enhances the binding ability of circALG1 to miR-342-5p, thereby enhancing its ceRNA function and upregulating the expression of placental growth factor (PGF), ultimately promoting the proliferation, migration, and invasion of CRC cells (80).
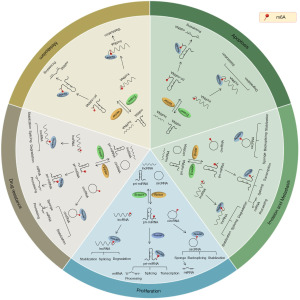
The process by which m6A regulators influence tumor cell biological behavior through ncRNA is summarized in Figure 2.
NcRNAs participate in the regulation of tumors through m6A regulatory factors
LncRNAs participate in the regulation of tumors through m6A regulatory factors
LncRNAs participate in the regulation of tumors by influencing the stability of m6A regulatory factors
LncRNAs can directly interact with m6A regulatory factors and impact their protein stability and other intrinsic properties, thereby regulating the proliferation, migration, and invasion capabilities of tumor cells.
For instance, research by Yang et al. demonstrated that in lung cancer cells, the lncRNA LCAT1 can directly bind to the IGF2BP2 protein, forming the LCAT1/IGF2BP2 complex. This complex stabilizes CDC6 mRNA in an m6A-dependent manner, leading to an increase in its expression level and promoting the growth and migration of lung cancer cells (81).
LncRNAs participate in the regulation of tumors by influencing the interaction between m6A regulatory factors and downstream target genes
In most cases, lncRNAs do not directly affect the expression levels and subcellular distribution of m6A regulatory factors in cancer cells. Instead, they modulate the binding capacity of m6A regulatory factors to target genes by directly interacting with them, thereby regulating tumor proliferation, migration, invasion, and other phenotypes. Specific lncRNAs, such as DMDRMR (82), LINC021 (83), lncRNA KB-1980E6.3 (84), LINC01605 (85), and LncRNA-PACERR (86), exhibit specific interactions with IGF2BP1/IGF2BP2/IGF2BP3, enhancing the binding capacity between IGF2BP1/IGF2BP2/IGF2BP3 and downstream target genes such as CDK4, MSX1, c-Myc, SPTBN2, and KLF12. This, in turn, increases the stability or accelerates the translation of target genes, promoting the proliferation, migration, invasion, and stem cell properties of CRC, breast cancer, and pancreatic cancer cells. LINC00460 (87) facilitates the interaction between the IGF3BP2/DHX9 complex and HMGA1, stabilizing HMGA1 mRNA in an m6A-dependent manner and promoting the proliferation, migration, and invasion of CRC cells. LncNRON (88) and lncRNA FOXM1-as (89) engage in RNA-protein interactions with ALKBH5, enhancing ALKBH5’s binding to target genes and increasing their expression levels, thereby promoting the pathological progression of gastric cancer and the stem cell characteristics of glioblastoma.
In HCC, lncRNA CASC11 enhances the binding ability of the demethylase ALKBH5 to UBE2T mRNA, leading to a decrease in UBE2T’s m6A modification level. Simultaneously, it weakens the binding ability of YTHDF2 to UBE2T mRNA, inhibiting YTHDF2-dependent mRNA degradation and ultimately increasing the stability of UBE2T, thus promoting the progression of HCC (90). In CRC cells, lncRNA STEAP3-as1 directly binds to YTHDF2, competitively inhibiting the interaction between YTHDF2 and downstream target gene STEAP3. This prevents m6A-mediated degradation of STEAP3 mRNA, thereby increasing STEAP3 expression levels and promoting the proliferation and metastasis of CRC cells (91). In renal cell carcinoma, lncRNA TRAF3IP2-AS1 directly interacts with the m6A methyltransferase complex composed of METTL3, METTL14, and WTAP, enhancing its binding to PARP1 mRNA. This interaction reduces the half-life of PARP1 mRNA, inhibiting the proliferation, migration, and invasion of renal cancer cells (92).
LncRNAs also regulate tumor treatment resistance by modulating the binding of m6A regulatory factors to downstream target genes. LncRNA ARHGAP5-as1 (93) and lncRNA DARS-AS1 (94) directly bind to METTL3/METTL14, regulating their binding capacity to target genes and increasing the m6A levels of target genes. This ultimately promotes chemoresistance in gastric cancer and hypoxia adaptation in cervical cancer. In CML, LINC00470 binds to METTL3, positively regulating the m6A modification level of PTEN mRNA and promoting its degradation in an m6A-dependent manner. This inhibition of cellular autophagy enhances chemoresistance in CML (95). In gastric cancer, lncRNA-CBSLR (96), and in CRC, lncRNA MIR100HG (97), interact with m6A writers such as YTHDF2/hnRNPA2B1 and target genes, promoting the binding between YTHDF2/hnRNPA2B1 and m6A modification sites of target genes. This m6A-dependent regulation affects the expression levels of target genes, leading to increased resistance to ferroptosis in gastric cancer and resistance to chemotherapy and EMT as well as resistance to cetuximab in CRC. Similarly, lncRNA JPX acts as a molecular scaffold, interacting with m6A eraser FTO and the target gene PDK1 mRNA, thereby enhancing FTO-mediated m6A demethylation of PDK1 mRNA and promoting temozolomide resistance in glioblastoma (98).
LncRNAs can also modulate tumor metabolism by regulating the binding of m6A regulatory factors to downstream target genes. For example, lncRNA NEAT1 interacts with hnRNPA2B1 and the target gene RPRD1B, facilitating the binding of hnRNPA2B1 to RPRD1B and increasing its stability in an m6A-dependent manner. This promotes fatty acid metabolism and lymph node metastasis in gastric cancer (99). LncRNA HOTAIR (100) and lncRNA JPX (98) enhance the interaction between m6A eraser FTO and downstream target genes, increasing FTO-mediated m6A demethylation and promoting aerobic glycolysis in lung cancer and glioblastoma, respectively.
MiRNAs participate in the regulation of tumors through m6A regulatory factors
MiRNAs participate in the regulation of tumors by directly targeting m6A regulatory factors
The majority of miRNAs regulate tumor proliferation, migration, and invasion by directly targeting m6A regulatory factors, leading to the downregulation of m6A regulatory factor expression. For instance, miR-186 (101), miR-320d (102), miR-338-5p (103,104), miR-302a (105), and miR-let-7g target and downregulate METTL3, thereby modulating the expression of downstream target genes in a methylation-dependent manner. This negative regulation ultimately affects the proliferation, migration, and invasion of HCC, prostate cancer, lung cancer, gastric cancer, and glioblastoma. In breast cancer, let-7g targets METTL3 and downregulates its expression. METTL3, in turn, promotes the expression of HBXIP through an m6A-dependent mechanism. HBXIP, which suppresses the expression of let-7g, forms a positive feedback loop involving HBXIP/let-7g/METTL3/HBXIP, thereby accelerating breast cancer cell proliferation (106). In liver cancer, exosomes derived from M1 macrophages transfer miR-628-5p to liver cancer cells. miR-628-5p targets and inhibits the expression of METTL14, suppressing the circFUT8/miR-552-3p/CHMP4B pathway mediated by METTL14 and thus inhibiting the progression of liver cancer (13). In acute myeloid leukemia, mir-550-1 targets and inhibits the expression of WTAP. This leads to tumor cell apoptosis and inhibition of tumor cell proliferation through the WTAP/WWTR1/BCL-2 and WTAP/WWTR1/CDK6/Rb/E2F1 pathways (107). miR-495, miR-6125, miR-362-3p, and miR-425-5p target and suppress the expression of YTHDF2 and ZC3H13, enhancing the stability of their target genes. This inhibition results in decreased proliferation, migration, and invasion of prostate cancer and colon cancer cells, as well as a reduction in tumor immune cell infiltration, immune cell biomarker expression, and immune checkpoint expression mediated by ZC3H13, ultimately leading to poor prognosis in liver cancer (108).
miRNAs can also regulate tumor drug resistance and metabolism by targeting and downregulating the expression of m6A regulatory factors. miR-4443 targets METTL3 and downregulates its expression, increasing the expression level of its target gene FSP1 in an m6A-dependent manner. This inhibits ferroptosis in non-small cell lung cancer cells and promotes the progression of non-small cell lung cancer and resistance to cisplatin (109). MiR-493-5p targets METTL3 and negatively regulates the METTL3/MYC axis, enhancing the sensitivity of acute myeloid leukemia cells to the chemotherapeutic agent cytarabine (AraC) (110). In breast cancer, miR-16-5p targets and downregulates the expression of YTHDF1, suppressing the protein translation of YTHDF1’s target gene PKM2 in an m6A-dependent manner. This, in turn, inhibits anaerobic glycolysis and the metastatic potential of breast cancer (111).
In addition to directly targeting mRNA of m6A regulatory factors to downregulate their expression, miRNAs can also indirectly regulate the expression of m6A regulatory factors by targeting their associated proteins, such as transcription factors. In breast cancer, ZNF217 upregulates the expression of METTL3, while miR-135 specifically targets and inhibits ZNF217 expression, indirectly reducing the expression levels of METTL3 and its mediated m6A modification. This ultimately promotes breast cancer cell migration, invasion, and EMT initiation (112). Similarly, miR-96 targets and downregulates the expression of AMPKα2 in CRC cells, inhibiting its ability to upregulate FTO expression. This AMPKα2/FTO/m6A/MYC axis promotes the occurrence and progression of CRC (113).
MiRNAs affect m6A regulatory factors through alternative pathways and participate in the regulation of tumors
In CRC, miR455-3p binds to the target gene HSF1 mRNA, competitively inhibiting the binding between METTL3 and HSF1, thereby suppressing HSF1 protein translation and impeding the progression of CRC (114). During glioblastoma stem cell differentiation, mir-145 forms a complex with FTO to recruit the FTO target gene CLIP3. This complex, consisting of FTO/AGO1/ILF3/miR-145, promotes m6A demethylation of CLIP3, induces CLIP3 translation, and facilitates glioblastoma progression (115).
CircRNAs participate in the regulation of tumors through m6A regulatory factors
CircRNAs participate in the regulation of tumors by modulating the expression of m6A regulatory factors through the ceRNA mechanism
CircRNAs commonly function as ceRNAs to influence the expression of m6A regulatory factors, thereby regulating tumor cell proliferation, migration, invasion, and drug resistance. For example, in breast cancer, CircBach2 acts as a sponge for has-miR-944, counteracting the inhibitory effect of has-mir-944 on hNRNPC and increasing hNRNPC expression, ultimately promoting breast cancer cell proliferation by upregulating the MAPK signaling pathway (116). In CRC, circEZH2 acts as a sponge for miR-133b and enhances the stability of CREB1 mRNA through the circEZH2/miR-133b/IGF2BP2/CREB1 axis, thereby promoting CRC proliferation and metastasis in both in vitro and in vivo settings (117). In non-small cell lung cancer, CircVMP1 serves as a sponge for miR-524-5p and upregulates METTL3, leading to increased stability of the target gene SOX2 mRNA in an m6A-dependent manner, thereby promoting pathological progression and cisplatin resistance in non-small cell lung cancer cells (118).
CircRNAs participate in the regulation of tumors by directly binding to m6A regulatory factors
CircRNAs can directly bind to m6A regulatory factors to enhance their protein stability and thereby regulate the pathological progression of tumors. In CRC, circEZH2 can directly interact with IGF2BP2, preventing ubiquitin-dependent degradation of IGF2BP2. Through the circEZH2/IGF2BP2/CREB1 axis, it enhances the stability of CREB1 mRNA and promotes the proliferation and metastasis of CRC in vitro and in vivo (117).
In HCC, CircRHBDD1 acts as a molecular scaffold, strengthening the interaction between YTHDF1 and its target gene PIK3R1 mRNA. It increases the translation of PIK3R1 mRNA in an m6A-dependent manner, activating the PI3K/AKT signaling pathway and promoting the proliferation and glycolysis of liver cancer cells (119). Another functional circRNA, rtcisE2F, in liver cancer, also acts as a molecular scaffold to facilitate the binding of IGF2BP2 to its target genes E2F6/E2F3 mRNA. It increases the stability of E2F6/E2F3 mRNA in an m6A-dependent manner, driving self-renewal of liver tumor-initiating cells (TICs), enhancing tumor heterogeneity, activating the Wnt/β-catenin pathway, and promoting the metastasis of liver cancer cells (120).
Other ncRNAs participate in the regulation of tumors through m6A regulatory factors
In addition to PIWI proteins, another class of non-coding small RNAs called piRNAs interact with Argonaute family proteins to regulate mRNA stability, protein synthesis, chromatin organization, and genome structure. Recent studies have discovered that certain piRNAs also participate in tumor progression through m6A regulatory factors. For instance, in cervical cancer cells, piRNA-14633 specifically binds to its target gene METTL14, increasing the stability of METTL14 mRNA, thereby enhancing the expression of the downstream target gene CYP1B1 in an m6A-dependent manner, promoting the proliferation, migration, and invasion of cervical cancer cells (121). In diffuse large B-cell lymphoma, piRNA-30473 directly targets WTAP and enhances its stability, leading to increased m6A levels of the HK2 mRNA, thereby promoting the proliferation and invasion of diffuse large B-cell lymphoma (122). Moreover, piRNA-17560 derived from senescent neutrophil-derived extracellular vesicles enhances the RNA stability of FTO by binding to the 3’UTR of FTO mRNA, resulting in the upregulation of the FTO target gene ZEB1 in an m6A methylation-dependent manner, thereby promoting chemotherapy resistance and EMT in breast cancer (123).
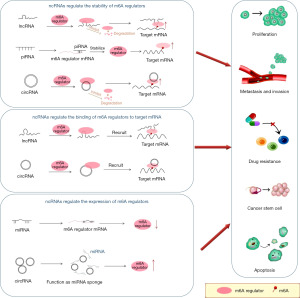
The process by which ncRNAs affect tumor phenotypes by regulating m6A regulators is summarized in Figure 3.
Clinical applications of M6A-associated ncRNAs
The role of m6A-associated ncRNAs as prognostic markers in cancer
Research has revealed a significant association between the dysregulation of many m6A-related ncRNAs and tumor grade and stage. The expression levels of these ncRNAs can serve as prognostic indicators for malignant tumors. For instance, a risk scoring system based on the differential expression of 11 m6A-related lncRNAs, including LINC01644, LINC01410, AL355574.1, AC091271.1, and LINC006303.2, has been identified as an independent prognostic factor for oral squamous cell carcinoma (124). Upregulation of nine m6A-related lncRNAs (e.g., linc01117, linc00592, tmpo.as1) in lung adenocarcinoma is associated with lower overall survival. The risk score generated from these lncRNAs can serve as an independent prognostic marker for lung adenocarcinoma, with patients exhibiting higher scores demonstrating increased sensitivity to PD-1/L1 immunotherapy and targeted therapy (125). Current research suggests that risk scoring models based on m6A-related lncRNAs can serve as prognostic indicators for malignant tumors in various cancers, including gastric cancer, bladder cancer, and breast cancer. This review article provides an overview of these findings (Figure 4) (124-147).
The role of m6A-related ncRNAs as therapeutic targets in cancer
Current research suggests that m6A-related ncRNAs can serve as therapeutic targets in cancer either independently or in combination with chemotherapy drugs, other targeted anti-cancer agents, and immunotherapy, thereby enhancing anti-tumor efficacy. This article provides a comprehensive review of these approaches (Figure 5).
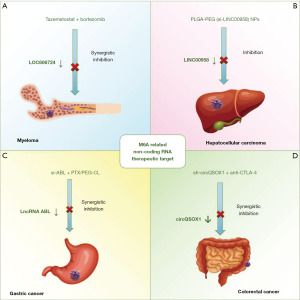
In multiple myeloma cells, EZH2 promotes the enrichment of LOC606724 in adipocyte-derived exosomes through METTL7A-mediated m6A methylation. The exosomal LOC606724 enhances c-MYC mRNA translation via eIF4E, protecting myeloma cells from chemotherapy-induced apoptosis. In vitro experiments demonstrated that the EZH2 inhibitor tazemetostat significantly reduces the expression of LOC606724 in adipocyte-derived exosomes, and when combined with bortezomib, it synergistically inhibits tumor growth in a mouse model of multiple myeloma (Figure 5A) (148). In HCC, the m6A-related lncRNA LINC00958 promotes lipid accumulation, proliferation, migration, and invasion of liver cancer cells. A polyethylene glycolyzed poly (lactic acid/glycolic acid) nanoplatform (PLGA-PEG NP) loaded with LINC00958 siRNA (si-LINC00958) PLGA-PEG (si-LINC00958) NPs effectively suppress tumor growth in a mouse model of liver cancer and significantly prolong overall survival, suggesting the potential of PLGA-PEG (si-LINC00958) NPs as nanotherapeutic candidates for HCC (Figure 5B) (17). In gastric cancer cells, m6A modification enhances the stability of LncRNA ABL through m6A methylation reader IGF2BP1, inhibiting apoptosis and promoting multidrug resistance. In vitro experiments demonstrated the development of polyethylene glycol-modified cationic liposomes (PEG-CLs) loaded with ABL-specific siRNA. Combined with paclitaxel, targeting ABL significantly enhances chemosensitivity of gastric cancer cells and improves anti-tumor efficacy (Figure 5C) (149). In CRC cells, circQSOX1 is upregulated in an m6A methylation-dependent manner and regulates glycolysis through miR-326/miR-330-5p, promoting immune evasion of CRC cells and suppressing anti-CTLA-4 therapy response in CRC patients, leading to immune therapy resistance. In in vivo experiments, the combination of sh-circQSOX1 and anti-CTLA-4 significantly inhibits tumor volume and weight in a mouse model of CRC, enhancing the efficacy of immunotherapy (Figure 5D) (150).
Conclusions
M6A methylation modification is a reversible epigenetic RNA modification regulated by m6A-related regulatory factors, and it plays a crucial role in regulating RNA processes such as translation, splicing, and degradation. NcRNAs are important components of the transcriptome and play critical regulatory roles in tumor cell proliferation, apoptosis, drug resistance, and stemness maintenance. Dysregulation of ncRNAs is closely associated with cancer progression. In tumor cells, ncRNAs interact with m6A regulatory factors, collectively influencing the pathological processes of cancer. M6A regulatory factors can regulate the expression of ncRNAs through various mechanisms, such as stabilizing lncRNAs, promoting the maturation of pri-miRNAs, etc. Similarly, ncRNAs can modulate the expression and function of m6A regulatory factors through various pathways, including enhancing the binding capacity of m6A regulatory factors to downstream target genes, regulating the stability of m6A regulatory factors, and targeting the degradation of m6A regulatory factor mRNAs.
This review summarizes the interaction between ncRNAs and m6A regulatory factors in tumor cells and highlights m6A-related ncRNAs that can serve as prognostic markers, providing insights into exploring novel therapeutic targets and prognostic indicators in cancer associated with m6A modifications. Currently, there are therapeutic strategies targeting m6A-related ncRNAs as tumor therapy targets, but these studies are limited to the animal level, and the clinical treatment efficacy remains unknown, requiring further validation. Furthermore, numerous studies have elucidated the role of ncRNAs in cancer prognosis through m6A-seq and risk models. For instance, a prognostic risk model based on nine m6A-related lncRNAs in glioblastoma achieved an AUC (areas under the receiver operating characteristic curve) of 0.928, indicating the tremendous potential of m6A-related ncRNAs as tumor prognostic markers. However, the current sample size for m6A-related ncRNAs as tumor prognostic markers is limited, and further investigations with larger sample sizes are needed to assess the accuracy of m6A-related ncRNAs as tumor prognostic markers.
Acknowledgments
Funding: This work is supported by
Footnote
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2401/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2401/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem 2021;65:625-39. [Crossref] [PubMed]
- Liu L, Gu M, Ma J, et al. CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol Cancer 2022;21:149. [Crossref] [PubMed]
- Wu Q, Zhang H, Yang D, et al. The m6A-induced lncRNA CASC8 promotes proliferation and chemoresistance via upregulation of hnRNPL in esophageal squamous cell carcinoma. Int J Biol Sci 2022;18:4824-36. [Crossref] [PubMed]
- Zhang Y, Liu X, Wang Y, et al. The m(6)A demethylase ALKBH5-mediated upregulation of DDIT4-AS1 maintains pancreatic cancer stemness and suppresses chemosensitivity by activating the mTOR pathway. Mol Cancer 2022;21:174. [Crossref] [PubMed]
- Fazi F, Fatica A. Interplay Between N6-Methyladenosine (m6A) and Non-coding RNAs in Cell Development and Cancer. Front Cell Dev Biol 2019;7:116. [Crossref] [PubMed]
- Wu S, Zhang L, Deng J, et al. A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell Carcinoma. Cancer Res 2020;80:2790-803. [Crossref] [PubMed]
- Yue Y, Deng P, Xiao H, et al. N6-methyladenosine-mediated downregulation of miR-374c-5p promotes cadmium-induced cell proliferation and metastasis by targeting GRM3 in breast cancer cells. Ecotoxicol Environ Saf 2022;229:113085. [Crossref] [PubMed]
- Hao CC, Xu CY, Zhao XY, et al. Up-regulation of VANGL1 by IGF2BPs and miR-29b-3p attenuates the detrimental effect of irradiation on lung adenocarcinoma. J Exp Clin Cancer Res 2020;39:256. [Crossref] [PubMed]
- Wang J, Ding W, Xu Y, et al. Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging (Albany NY) 2020;12:4558-72. [Crossref] [PubMed]
- Zhang H, Liu Y, Wang W, et al. ALKBH5-mediated m(6)A modification of lincRNA LINC02551 enhances the stability of DDX24 to promote hepatocellular carcinoma growth and metastasis. Cell Death Dis 2022;13:926. [Crossref] [PubMed]
- Liang X, Zhang Z, Wang L, et al. Mechanism of methyltransferase like 3 in epithelial-mesenchymal transition process, invasion, and metastasis in esophageal cancer. Bioengineered 2021;12:10023-36. [Crossref] [PubMed]
- Yao FY, Zhao C, Zhong FM, et al. m(6)A Modification of lncRNA NEAT1 Regulates Chronic Myelocytic Leukemia Progression via miR-766-5p/CDKN1A Axis. Front Oncol 2021;11:679634. [Crossref] [PubMed]
- Wang L, Yi X, Xiao X, et al. Exosomal miR-628-5p from M1 polarized macrophages hinders m6A modification of circFUT8 to suppress hepatocellular carcinoma progression. Cell Mol Biol Lett 2022;27:106. [Crossref] [PubMed]
- Qian X, Yang J, Qiu Q, et al. LCAT3, a novel m6A-regulated long non-coding RNA, plays an oncogenic role in lung cancer via binding with FUBP1 to activate c-MYC. J Hematol Oncol 2021;14:112. [Crossref] [PubMed]
- Peng J, Zheng H, Liu F, et al. The m6A methyltransferase METTL3 affects autophagy and progression of nasopharyngeal carcinoma by regulating the stability of lncRNA ZFAS1. Infect Agent Cancer 2022;17:1. [Crossref] [PubMed]
- Zhou X, Yang Y, Li Y, et al. METTL3 Contributes to Osteosarcoma Progression by Increasing DANCR mRNA Stability via m6A Modification. Front Cell Dev Biol 2022;9:784719. [Crossref] [PubMed]
- Zuo X, Chen Z, Gao W, et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol 2020;13:5. [Crossref] [PubMed]
- Wu J, Pang R, Li M, et al. m6A-Induced LncRNA MEG3 Suppresses the Proliferation, Migration and Invasion of Hepatocellular Carcinoma Cell Through miR-544b/BTG2 Signaling. Onco Targets Ther 2021;14:3745-55. [Crossref] [PubMed]
- Liu HT, Zou YX, Zhu WJ, et al. lncRNA THAP7-AS1, transcriptionally activated by SP1 and post-transcriptionally stabilized by METTL3-mediated m6A modification, exerts oncogenic properties by improving CUL4B entry into the nucleus. Cell Death Differ 2022;29:627-41. [Crossref] [PubMed]
- Lang C, Yin C, Lin K, et al. m(6) A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin Transl Med 2021;11:e426. [Crossref] [PubMed]
- Xia A, Yuan W, Wang Q, et al. The cancer-testis lncRNA lnc-CTHCC promotes hepatocellular carcinogenesis by binding hnRNP K and activating YAP1 transcription. Nat Cancer 2022;3:203-18. [Crossref] [PubMed]
- Hu Z, Zhu L, Zhang Y, et al. N6-methyladenosine-induced SVIL antisense RNA 1 restrains lung adenocarcinoma cell proliferation by destabilizing E2F1. Bioengineered 2022;13:3093-107. [Crossref] [PubMed]
- Chen B, Liu C, Long H, et al. N(6)-methyladenosine-induced long non-coding RNA PVT1 regulates the miR-27b-3p/BLM axis to promote prostate cancer progression. Int J Oncol 2023;62:16. [Crossref] [PubMed]
- Chen JQ, Tao YP, Hong YG, et al. M(6)A-mediated up-regulation of LncRNA LIFR-AS1 enhances the progression of pancreatic cancer via miRNA-150-5p/ VEGFA/Akt signaling. Cell Cycle 2021;20:2507-18. [Crossref] [PubMed]
- Hu N, Ji H. N6-methyladenosine (m6A)-mediated up-regulation of long noncoding RNA LINC01320 promotes the proliferation, migration, and invasion of gastric cancer via miR495-5p/RAB19 axis. Bioengineered 2021;12:4081-91. [Crossref] [PubMed]
- Liu J, Zhang N, Zeng J, et al. N(6) -methyladenosine-modified lncRNA ARHGAP5-AS1 stabilises CSDE1 and coordinates oncogenic RNA regulons in hepatocellular carcinoma. Clin Transl Med 2022;12:e1107. [Crossref] [PubMed]
- Ban Y, Tan P, Cai J, et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol Oncol 2020;14:1282-96. [Crossref] [PubMed]
- Chang YZ, Chai RC, Pang B, et al. METTL3 enhances the stability of MALAT1 with the assistance of HuR via m6A modification and activates NF-κB to promote the malignant progression of IDH-wildtype glioma. Cancer Lett 2021;511:36-46. Erratum in: Cancer Lett 2023;569:216309. [Crossref] [PubMed]
- Li ZX, Zheng ZQ, Yang PY, et al. WTAP-mediated m(6)A modification of lncRNA DIAPH1-AS1 enhances its stability to facilitate nasopharyngeal carcinoma growth and metastasis. Cell Death Differ 2022;29:1137-51. [Crossref] [PubMed]
- Ren Z, Hu Y, Sun J, et al. N(6)-methyladenosine methyltransferase WTAP-stabilized FOXD2-AS1 promotes the osteosarcoma progression through m(6)A/FOXM1 axis. Bioengineered 2022;13:7963-73. [Crossref] [PubMed]
- Dai YZ, Liu YD, Li J, et al. METTL16 promotes hepatocellular carcinoma progression through downregulating RAB11B-AS1 in an m(6)A-dependent manner. Cell Mol Biol Lett 2022;27:41. [Crossref] [PubMed]
- Fang D, Ou X, Sun K, et al. m6A modification-mediated lncRNA TP53TG1 inhibits gastric cancer progression by regulating CIP2A stability. Cancer Sci 2022;113:4135-50. [Crossref] [PubMed]
- Li B, Zhao R, Qiu W, et al. The N(6)-methyladenosine-mediated lncRNA WEE2-AS1 promotes glioblastoma progression by stabilizing RPN2. Theranostics 2022;12:6363-79. [Crossref] [PubMed]
- Zhang Y, Wang D, Wu D, et al. Long Noncoding RNA KCNMB2-AS1 Stabilized by N(6)-Methyladenosine Modification Promotes Cervical Cancer Growth Through Acting as a Competing Endogenous RNA. Cell Transplant 2020;29:963689720964382. [Crossref] [PubMed]
- Liu H, Xu Y, Yao B, et al. A novel N6-methyladenosine (m6A)-dependent fate decision for the lncRNA THOR. Cell Death Dis 2020;11:613. [Crossref] [PubMed]
- Hosono Y, Niknafs YS, Prensner JR, et al. Oncogenic Role of THOR, a Conserved Cancer/Testis Long Non-coding RNA. Cell 2017;171:1559-1572.e20. [Crossref] [PubMed]
- Chen W, Chen M, Xu Y, et al. Long non-coding RNA THOR promotes human osteosarcoma cell growth in vitro and in vivo. Biochem Biophys Res Commun 2018;499:913-9. [Crossref] [PubMed]
- Zhang M, Wang J, Jin Y, et al. YTHDF2-mediated FGF14-AS2 decay promotes osteolytic metastasis of breast cancer by enhancing RUNX2 mRNA translation. Br J Cancer 2022;127:2141-53. [Crossref] [PubMed]
- Shen J, Feng XP, Hu RB, et al. N-methyladenosine reader YTHDF2-mediated long noncoding RNA FENDRR degradation promotes cell proliferation in endometrioid endometrial carcinoma. Lab Invest 2021;101:775-84. [Crossref] [PubMed]
- Ni W, Yao S, Zhou Y, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer 2019;18:143. [Crossref] [PubMed]
- Kong H, Sun J, Zhang W, et al. Long intergenic non-protein coding RNA 1273 confers sorafenib resistance in hepatocellular carcinoma via regulation of methyltransferase 3. Bioengineered 2022;13:3108-21. [Crossref] [PubMed]
- Liu Z, Lu J, Fang H, et al. m6A Modification-Mediated DUXAP8 Regulation of Malignant Phenotype and Chemotherapy Resistance of Hepatocellular Carcinoma Through miR-584-5p/MAPK1/ERK Pathway Axis. Front Cell Dev Biol 2021;9:783385. [Crossref] [PubMed]
- Zhang H, Wang SQ, Wang L, et al. m6A methyltransferase METTL3-induced lncRNA SNHG17 promotes lung adenocarcinoma gefitinib resistance by epigenetically repressing LATS2 expression. Cell Death Dis 2022;13:657. [Crossref] [PubMed]
- Huang T, Cao L, Feng N, et al. N(6)-methyladenosine (m(6)A)-mediated lncRNA DLGAP1-AS1enhances breast canceradriamycin resistance through miR-299-3p/WTAP feedback loop. Bioengineered 2021;12:10935-44. [Crossref] [PubMed]
- Cusenza VY, Tameni A, Neri A, et al. The lncRNA epigenetics: The significance of m6A and m5C lncRNA modifications in cancer. Front Oncol 2023;13:1063636. [Crossref] [PubMed]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029-33. [Crossref] [PubMed]
- Xue L, Li J, Lin Y, et al. m(6) A transferase METTL3-induced lncRNA ABHD11-AS1 promotes the Warburg effect of non-small-cell lung cancer. J Cell Physiol 2021;236:2649-58. [Crossref] [PubMed]
- Liu J, Yuan JF, Wang YZ. METTL3-stabilized lncRNA SNHG7 accelerates glycolysis in prostate cancer via SRSF1/c-Myc axis. Exp Cell Res 2022;416:113149. [Crossref] [PubMed]
- Liu H, Qin S, Liu C, et al. m(6)A reader IGF2BP2-stabilized CASC9 accelerates glioblastoma aerobic glycolysis by enhancing HK2 mRNA stability. Cell Death Discov 2021;7:292. [Crossref] [PubMed]
- Lu S, Han L, Hu X, et al. N6-methyladenosine reader IMP2 stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect: implication in colorectal cancer. J Hematol Oncol 2021;14:188. [Crossref] [PubMed]
- Liu P, Fan B, Othmane B, et al. m(6)A-induced lncDBET promotes the malignant progression of bladder cancer through FABP5-mediated lipid metabolism. Theranostics 2022;12:6291-307. [Crossref] [PubMed]
- Wang ZW, Pan JJ, Hu JF, et al. SRSF3-mediated regulation of N6-methyladenosine modification-related lncRNA ANRIL splicing promotes resistance of pancreatic cancer to gemcitabine. Cell Rep 2022;39:110813. [Crossref] [PubMed]
- Yuan J, Lv T, Yang J, et al. HDLBP-stabilized lncFAL inhibits ferroptosis vulnerability by diminishing Trim69-dependent FSP1 degradation in hepatocellular carcinoma. Redox Biol 2022;58:102546. [Crossref] [PubMed]
- Porman AM, Roberts JT, Duncan ED, et al. A single N6-methyladenosine site regulates lncRNA HOTAIR function in breast cancer cells. PLoS Biol 2022;20:e3001885. [Crossref] [PubMed]
- Wu Y, Yang X, Chen Z, et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer 2019;18:87. [Crossref] [PubMed]
- Ye XT, Huang H, Huang WP, et al. LncRNA THOR promotes human renal cell carcinoma cell growth. Biochem Biophys Res Commun 2018;501:661-7. [Crossref] [PubMed]
- Bi X, Lv X, Liu D, et al. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther 2021;28:335-49. [Crossref] [PubMed]
- Sun Y, Li S, Yu W, et al. N(6)-methyladenosine-dependent pri-miR-17-92 maturation suppresses PTEN/TMEM127 and promotes sensitivity to everolimus in gastric cancer. Cell Death Dis 2020;11:836. [Crossref] [PubMed]
- Zhang J, Bai R, Li M, et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun 2019;10:1858. [Crossref] [PubMed]
- Xu Q, Lu X, Li J, et al. Fusobacterium nucleatum induces excess methyltransferase-like 3-mediated microRNA-4717-3p maturation to promote colorectal cancer cell proliferation. Cancer Sci 2022;113:3787-800. [Crossref] [PubMed]
- Lin R, Zhan M, Yang L, et al. Deoxycholic acid modulates the progression of gallbladder cancer through N(6)-methyladenosine-dependent microRNA maturation. Oncogene 2020;39:4983-5000. [Crossref] [PubMed]
- Liu R, Zhong Y, Chen R, et al. m(6)A reader hnRNPA2B1 drives multiple myeloma osteolytic bone disease. Theranostics 2022;12:7760-74. [Crossref] [PubMed]
- Klinge CM, Piell KM, Tooley CS, et al. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci Rep 2019;9:9430. [Crossref] [PubMed]
- Li K, Peng ZY, Gao S, et al. M6A associated TSUC7 inhibition contributed to Erlotinib resistance in lung adenocarcinoma through a notch signaling activation dependent way. J Exp Clin Cancer Res 2021;40:325. [Crossref] [PubMed]
- Gong S, Wang S, Shao M. Mechanism of METTL14-mediated m(6)A modification in non-small cell lung cancer cell resistance to cisplatin. J Mol Med (Berl) 2022;100:1771-85. [Crossref] [PubMed]
- Pan X, Hong X, Li S, et al. METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. Exp Mol Med 2021;53:91-102. [Crossref] [PubMed]
- Lyu Y, Zhang Y, Wang Y, et al. HIF-1α Regulated WTAP Overexpression Promoting the Warburg Effect of Ovarian Cancer by m6A-Dependent Manner. J Immunol Res 2022;2022:6130806. [Crossref] [PubMed]
- Liu Z, Wu K, Gu S, et al. A methyltransferase-like 14/miR-99a-5p/tribble 2 positive feedback circuit promotes cancer stem cell persistence and radioresistance via histone deacetylase 2-mediated epigenetic modulation in esophageal squamous cell carcinoma. Clin Transl Med 2021;11:e545. [Crossref] [PubMed]
- Wang H, Deng Q, Lv Z, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer 2019;18:181. [Crossref] [PubMed]
- Sun L, Wan A, Zhou Z, et al. RNA-binding protein RALY reprogrammes mitochondrial metabolism via mediating miRNA processing in colorectal cancer. Gut 2021;70:1698-712. [Crossref] [PubMed]
- Li J, Wu L, Pei M, et al. YTHDF2, a protein repressed by miR-145, regulates proliferation, apoptosis, and migration in ovarian cancer cells. J Ovarian Res 2020;13:111. [Crossref] [PubMed]
- Pan H, Pan Z, Guo F, et al. MicroRNA-1915-3p inhibits cell migration and invasion by targeting SET in non-small-cell lung cancer. BMC Cancer 2021;21:1218. [Crossref] [PubMed]
- Xiao L, Li X, Mu Z, et al. FTO Inhibition Enhances the Antitumor Effect of Temozolomide by Targeting MYC-miR-155/23a Cluster-MXI1 Feedback Circuit in Glioma. Cancer Res 2020;80:3945-58. [Crossref] [PubMed]
- Liang L, Zhu Y, Li J, et al. ALKBH5-mediated m6A modification of circCCDC134 facilitates cervical cancer metastasis by enhancing HIF1A transcription. J Exp Clin Cancer Res 2022;41:261. [Crossref] [PubMed]
- Meng X, Deng Y, He S, et al. m(6)A-Mediated Upregulation of LINC00857 Promotes Pancreatic Cancer Tumorigenesis by Regulating the miR-150-5p/E2F3 Axis. Front Oncol 2021;11:629947. [Crossref] [PubMed]
- Chen B, Hong Y, Gui R, et al. N6-methyladenosine modification of circ_0003215 suppresses the pentose phosphate pathway and malignancy of colorectal cancer through the miR-663b/DLG4/G6PD axis. Cell Death Dis 2022;13:804. [Crossref] [PubMed]
- Xu J, Wan Z, Tang M, et al. N(6)-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer 2020;19:163. [Crossref] [PubMed]
- Wu P, Fang X, Liu Y, et al. N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis 2021;12:298. [Crossref] [PubMed]
- Rao X, Lai L, Li X, et al. N(6) -methyladenosine modification of circular RNA circ-ARL3 facilitates Hepatitis B virus-associated hepatocellular carcinoma via sponging miR-1305. IUBMB Life 2021;73:408-17. [Crossref] [PubMed]
- Lin C, Ma M, Zhang Y, et al. Correction to: The N6-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Mol Cancer 2022;21:101. [Crossref] [PubMed]
- Yang J, Qian X, Qiu Q, et al. LCAT1 is an oncogenic LncRNA by stabilizing the IGF2BP2-CDC6 axis. Cell Death Dis 2022;13:877. [Crossref] [PubMed]
- Gu Y, Niu S, Wang Y, et al. DMDRMR-Mediated Regulation of m(6)A-Modified CDK4 by m(6)A Reader IGF2BP3 Drives ccRCC Progression. Cancer Res 2021;81:923-34. [Crossref] [PubMed]
- Wu H, Ding X, Hu X, et al. LINC01021 maintains tumorigenicity by enhancing N6-methyladenosine reader IMP2 dependent stabilization of MSX1 and JARID2: implication in colorectal cancer. Oncogene 2022;41:1959-73. [Crossref] [PubMed]
- Zhu P, He F, Hou Y, et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene 2021;40:1609-27. [Crossref] [PubMed]
- Yue M, Liu T, Yan G, et al. LINC01605, regulated by the EP300-SMYD2 complex, potentiates the binding between METTL3 and SPTBN2 in colorectal cancer. Cancer Cell Int 2021;21:504. [Crossref] [PubMed]
- Liu Y, Shi M, He X, et al. LncRNA-PACERR induces pro-tumour macrophages via interacting with miR-671-3p and m6A-reader IGF2BP2 in pancreatic ductal adenocarcinoma. J Hematol Oncol 2022;15:52. [Crossref] [PubMed]
- Hou P, Meng S, Li M, et al. LINC00460/DHX9/IGF2BP2 complex promotes colorectal cancer proliferation and metastasis by mediating HMGA1 mRNA stability depending on m6A modification. J Exp Clin Cancer Res 2021;40:52. [Crossref] [PubMed]
- Wang S, Wang Y, Zhang Z, et al. Long Non-Coding RNA NRON promotes Tumor Proliferation by regulating ALKBH5 and Nanog in Gastric Cancer. J Cancer 2021;12:6861-72. [Crossref] [PubMed]
- Zhang S, Zhao BS, Zhou A, et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017;31:591-606.e6. [Crossref] [PubMed]
- Chen F, Li M, Wang L. LncRNA CASC11 Promotes Hepatocellular Carcinoma Progression via Upregulation of UBE2T in a m(6)A-Dependent Manner. Front Oncol 2021;11:772671. [Crossref] [PubMed]
- Zhou L, Jiang J, Huang Z, et al. Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m(6)A-mediated degradation of STEAP3 mRNA. Mol Cancer 2022;21:168. [Crossref] [PubMed]
- Yang L, Chen Y, Liu N, et al. Low expression of TRAF3IP2-AS1 promotes progression of NONO-TFE3 translocation renal cell carcinoma by stimulating N(6)-methyladenosine of PARP1 mRNA and downregulating PTEN. J Hematol Oncol 2021;14:46. [Crossref] [PubMed]
- Zhu L, Zhu Y, Han S, et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis 2019;10:383. [Crossref] [PubMed]
- Shen W, Zhu M, Wang Q, et al. DARS-AS1 recruits METTL3/METTL14 to bind and enhance DARS mRNA m(6)A modification and translation for cytoprotective autophagy in cervical cancer. RNA Biol 2022;19:751-63. [Crossref] [PubMed]
- Lai X, Wei J, Gu XZ, et al. Dysregulation of LINC00470 and METTL3 promotes chemoresistance and suppresses autophagy of chronic myelocytic leukaemia cells. J Cell Mol Med 2021;25:4248-59. [Crossref] [PubMed]
- Yang H, Hu Y, Weng M, et al. Hypoxia inducible lncRNA-CBSLR modulates ferroptosis through m6A-YTHDF2-dependent modulation of CBS in gastric cancer. J Adv Res 2021;37:91-106. [Crossref] [PubMed]
- Liu H, Li D, Sun L, et al. Interaction of lncRNA MIR100HG with hnRNPA2B1 facilitates m(6)A-dependent stabilization of TCF7L2 mRNA and colorectal cancer progression. Mol Cancer 2022;21:74. [Crossref] [PubMed]
- Li XD, Wang MJ, Zheng JL, et al. Long noncoding RNA just proximal to X-inactive specific transcript facilitates aerobic glycolysis and temozolomide chemoresistance by promoting stability of PDK1 mRNA in an m6A-dependent manner in glioblastoma multiforme cells. Cancer Sci 2021;112:4543-52. [Crossref] [PubMed]
- Jia Y, Yan Q, Zheng Y, et al. Long non-coding RNA NEAT1 mediated RPRD1B stability facilitates fatty acid metabolism and lymph node metastasis via c-Jun/c-Fos/SREBP1 axis in gastric cancer. J Exp Clin Cancer Res 2022;41:287. [Crossref] [PubMed]
- Sun X, Li Q, Yang L. Sevoflurane Inhibits lncRNA HOTAIR-Modulated Stability of HK2 mRNA in a m6A-Dependent Manner to Dampen Aerobic Glycolysis and Proliferation in Lung Cancer. Biomed Res Int 2022;2022:4668774. [Crossref] [PubMed]
- Cui X, Wang Z, Li J, et al. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif 2020;53:e12768. [Crossref] [PubMed]
- Ma H, Zhang F, Zhong Q, et al. METTL3-mediated m6A modification of KIF3C-mRNA promotes prostate cancer progression and is negatively regulated by miR-320d. Aging (Albany NY) 2021;13:22332-44. [Crossref] [PubMed]
- Wu H, Li F, Zhu R. miR-338-5p inhibits cell growth and migration via inhibition of the METTL3/m6A/c-Myc pathway in lung cancer. Acta Biochim Biophys Sin (Shanghai) 2021;53:304-16. [Crossref] [PubMed]
- Zhang F, Yan Y, Cao X, et al. Methylation of microRNA-338-5p by EED promotes METTL3-mediated translation of oncogene CDCP1 in gastric cancer. Aging (Albany NY) 2021;13:12224-38. [Crossref] [PubMed]
- Zhong C, Tao B, Yang F, et al. Histone demethylase JMJD1C promotes the polarization of M1 macrophages to prevent glioma by upregulating miR-302a. Clin Transl Med 2021;11:e424. [Crossref] [PubMed]
- Cai X, Wang X, Cao C, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett 2018;415:11-9. [Crossref] [PubMed]
- Hu C, Yu M, Li C, et al. miR-550-1 functions as a tumor suppressor in acute myeloid leukemia via the hippo signaling pathway. Int J Biol Sci 2020;16:2853-67. [Crossref] [PubMed]
- Wu S, Liu S, Cao Y, et al. Downregulation of ZC3H13 by miR-362-3p/miR-425-5p is associated with a poor prognosis and adverse outcomes in hepatocellular carcinoma. Aging (Albany NY) 2022;14:2304-19. [Crossref] [PubMed]
- Song Z, Jia G, Ma P, et al. Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis. Life Sci 2021;276:119399. [Crossref] [PubMed]
- Wang A, Chen Y, Shi L, et al. Tumor-suppressive MEG3 induces microRNA-493-5p expression to reduce arabinocytosine chemoresistance of acute myeloid leukemia cells by downregulating the METTL3/MYC axis. J Transl Med 2022;20:288. [Crossref] [PubMed]
- Yao X, Li W, Li L, et al. YTHDF1 upregulation mediates hypoxia-dependent breast cancer growth and metastasis through regulating PKM2 to affect glycolysis. Cell Death Dis 2022;13:258. [Crossref] [PubMed]
- Xu LM, Zhang J, Ma Y, et al. MicroRNA-135 inhibits initiation of epithelial-mesenchymal transition in breast cancer by targeting ZNF217 and promoting m6A modification of NANOG. Oncogene 2022;41:1742-51. [Crossref] [PubMed]
- Yue C, Chen J, Li Z, et al. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J Exp Clin Cancer Res 2020;39:240. [Crossref] [PubMed]
- Song P, Feng L, Li J, et al. β-catenin represses miR455-3p to stimulate m6A modification of HSF1 mRNA and promote its translation in colorectal cancer. Mol Cancer 2020;19:129. [Crossref] [PubMed]
- Zepecki JP, Karambizi D, Fajardo JE, et al. miRNA-mediated loss of m6A increases nascent translation in glioblastoma. PLoS Genet 2021;17:e1009086. [Crossref] [PubMed]
- Lv W, Tan Y, Xiong M, et al. Analysis and validation of m6A regulatory network: a novel circBACH2/has-miR-944/HNRNPC axis in breast cancer progression. J Transl Med 2021;19:527. [Crossref] [PubMed]
- Yao B, Zhang Q, Yang Z, et al. CircEZH2/miR-133b/IGF2BP2 aggravates colorectal cancer progression via enhancing the stability of m(6)A-modified CREB1 mRNA. Mol Cancer 2022;21:140. [Crossref] [PubMed]
- Xie H, Yao J, Wang Y, et al. Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Deliv 2022;29:1257-71. [Crossref] [PubMed]
- Cai J, Chen Z, Zhang Y, et al. CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m(6)A modification in hepatocellular carcinoma. Mol Ther Oncolytics 2022;24:755-71. [Crossref] [PubMed]
- Chen Z, Huang L, Wang K, et al. rtcisE2F promotes the self-renewal and metastasis of liver tumor-initiating cells via N(6)-methyladenosine-dependent E2F3/E2F6 mRNA stability. Sci China Life Sci 2022;65:1840-54. [Crossref] [PubMed]
- Xie Q, Li Z, Luo X, et al. piRNA-14633 promotes cervical cancer cell malignancy in a METTL14-dependent m6A RNA methylation manner. J Transl Med 2022;20:51. [Crossref] [PubMed]
- Han H, Fan G, Song S, et al. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood 2021;137:1603-14. [Crossref] [PubMed]
- Ou B, Liu Y, Gao Z, et al. Senescent neutrophils-derived exosomal piRNA-17560 promotes chemoresistance and EMT of breast cancer via FTO-mediated m6A demethylation. Cell Death Dis 2022;13:905. [Crossref] [PubMed]
- Yang Q, Cheng C, Zhu R, et al. A N6-methyladenosine-related long noncoding RNAs model for predicting prognosis in oral squamous cell carcinoma: Association with immune cell infiltration and tumor metastasis. Oral Oncol 2022;127:105771. [Crossref] [PubMed]
- Shen Y, Wang S, Wu Y. A Novel m6A-Related LncRNA Signature for Predicting Prognosis, Chemotherapy and Immunotherapy Response in Patients with Lung Adenocarcinoma. Cells 2022;11:2399. [Crossref] [PubMed]
- Shan L, Lu Y, Song Y, et al. Identification of Nine M6A-Related Long Noncoding RNAs as Prognostic Signatures Associated with Oxidative Stress in Oral Cancer Based on Data from The Cancer Genome Atlas. Oxid Med Cell Longev 2022;2022:9529814. [Crossref] [PubMed]
- Zhu Y, Yu T, Huang J, et al. Development and validation of prognostic m6A-related lncRNA and mRNA model in thyroid cancer. Am J Cancer Res 2022;12:3259-79. [PubMed]
- Zhang W, Zhang Q, Xie Z, et al. N (6) -Methyladenosine-Related Long Non-Coding RNAs Are Identified as a Potential Prognostic Biomarker for Lung Squamous Cell Carcinoma and Validated by Real-Time PCR. Front Genet 2022;13:839957. [Crossref] [PubMed]
- Xie H, Shi M, Liu Y, et al. Identification of m6A- and ferroptosis-related lncRNA signature for predicting immune efficacy in hepatocellular carcinoma. Front Immunol 2022;13:914977. [Crossref] [PubMed]
- Zhang C, Zhang W, Shui Y, et al. Implications of m6A-associated snRNAs in the prognosis and immunotherapeutic responses of hepatocellular carcinoma. Front Immunol 2022;13:1001506. [Crossref] [PubMed]
- Chen B, Yang Z, Lang Z, et al. M6A-related lncRNAs predict clinical outcome and regulate the tumor immune microenvironment in hepatocellular carcinoma. BMC Cancer 2022;22:867. [Crossref] [PubMed]
- Ma T, Wang X, Wang J, et al. N6-Methyladenosine-Related Long Non-coding RNA Signature Associated With Prognosis and Immunotherapeutic Efficacy of Clear-Cell Renal Cell Carcinoma. Front Genet 2021;12:726369. [Crossref] [PubMed]
- Song Y, Qu H. Identification and validation of a seven m6A-related lncRNAs signature predicting prognosis of ovarian cancer. BMC Cancer 2022;22:633. [Crossref] [PubMed]
- Li M, Zhang L, Feng M, et al. m6A-Related lncRNA Signature Is Involved in Immunosuppression and Predicts the Patient Prognosis of the Age-Associated Ovarian Cancer. J Immunol Res 2022;2022:3258400. [Crossref] [PubMed]
- Shan L, Lu Y, Xiang CC, et al. Identification of Five m6A-Related lncRNA Genes as Prognostic Markers for Endometrial Cancer Based on TCGA Database. J Immunol Res 2022;2022:2547029. [Crossref] [PubMed]
- Jia H, Cao M, Hao S, et al. Prediction of prognosis, immune infiltration and immunotherapy response with N6-methyladenosine-related lncRNA clustering patterns in cervical cancer. Sci Rep 2022;12:17256. [Crossref] [PubMed]
- Chen Y, Guo Y, Li S, et al. Identification of N6-Methyladenosine-Related lncRNAs as a Prognostic Signature in Glioma. Front Oncol 2022;12:789283. [Crossref] [PubMed]
- Zhou LQ, Shen JX, Zhou JY, et al. The prognostic value of m6A-related LncRNAs in patients with HNSCC: bioinformatics analysis of TCGA database. Sci Rep 2022;12:579. [Crossref] [PubMed]
- Wang W, Dong D, Yu P, et al. Prognostic model based on m6A-associated lncRNAs in esophageal cancer. Front Endocrinol (Lausanne) 2022;13:947708. [Crossref] [PubMed]
- Lei L, Li N, Yuan P, et al. A new risk model based on a 11-m(6)A-related lncRNA signature for predicting prognosis and monitoring immunotherapy for gastric cancer. BMC Cancer 2022;22:365. [Crossref] [PubMed]
- Zhang X, Wang C, Liu Z, et al. Identification of m(6)A-related long non-coding RNAs for predicting prognosis and immune characterizations in gastric cancer. Front Genet 2022;13:1011716. [Crossref] [PubMed]
- Xu S, Chen W, Wang Y, et al. N6-methyladenosine-related lncRNAs identified as potential biomarkers for predicting the overall survival of Asian gastric cancer patients. BMC Cancer 2022;22:721. [Crossref] [PubMed]
- Liu Y, Wang T, Fang Z, et al. Analysis of N6-methyladenosine-related lncRNAs in the tumor immune microenvironment and their prognostic role in pancreatic cancer. J Cancer Res Clin Oncol 2022;148:1613-26. [Crossref] [PubMed]
- Cao PW, Liu L, Li ZH, et al. Prognostic Value of Drug Targets Predicted Using Deep Bioinformatic Analysis of m6A-Associated lncRNA-Based Pancreatic Cancer Model Characteristics and Its Tumour Microenvironment. Front Genet 2022;13:853471. [Crossref] [PubMed]
- Song W, Ren J, Xiang R, et al. Cross-Talk Between m(6)A- and m(5)C-Related lncRNAs to Construct a Novel Signature and Predict the Immune Landscape of Colorectal Cancer Patients. Front Immunol 2022;13:740960. [Crossref] [PubMed]
- Zhang L, Ke W, Hu P, et al. N6-Methyladenosine-Related lncRNAs Are Novel Prognostic Markers and Predict the Immune Landscape in Acute Myeloid Leukemia. Front Genet 2022;13:804614. [Crossref] [PubMed]
- Feng ZH, Liang YP, Cen JJ, et al. m6A-immune-related lncRNA prognostic signature for predicting immune landscape and prognosis of bladder cancer. J Transl Med 2022;20:492. [Crossref] [PubMed]
- Wang Z, He J, Bach DH, et al. Induction of m(6)A methylation in adipocyte exosomal LncRNAs mediates myeloma drug resistance. J Exp Clin Cancer Res 2022;41:4. [Crossref] [PubMed]
- Wang Q, Chen C, Xu X, et al. APAF1-Binding Long Noncoding RNA Promotes Tumor Growth and Multidrug Resistance in Gastric Cancer by Blocking Apoptosome Assembly. Adv Sci (Weinh) 2022;9:e2201889. [Crossref] [PubMed]
- Liu Z, Zheng N, Li J, et al. N6-methyladenosine-modified circular RNA QSOX1 promotes colorectal cancer resistance to anti-CTLA-4 therapy through induction of intratumoral regulatory T cells. Drug Resist Updat 2022;65:100886. [Crossref] [PubMed]