
TP53 mutations as a biomarker for high-grade serous ovarian cancer: are we there yet?
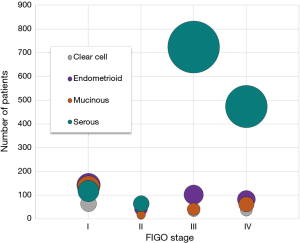
Ovarian cancer is in the top five causes of female cancer deaths in the developed world (1), and about 1 in 54 women will develop ovarian cancer based on lifetime risk (2). Symptoms can be vague, and most cases are diagnosed at an advanced stage. Ovarian cancer is staged surgically and pathologically according to the FIGO system (3), and the five-year survival rate is around 30% for patients diagnosed with advanced-stage ovarian cancer (4). The five-year survival increases to 92% when the cancer is confined to the ovary, supporting the theory that early detection via biomarkers has the potential to reduce mortality. However, accuracy in ovarian cancer detection methods can be problematic and also lead to mortality via unsuitable treatment. The US Preventative Services Task Force currently discourages ovarian cancer screening in asymptomatic women stating this can result in unnecessary interventions (5). In the absence of an ideal biomarker or screening method, an adequate adjuvant detection method may have the potential for ovarian cancer detection in the near future. Furthermore, there is a complete lack of biomarkers and screening methods for accurate early-stage detection of ovarian cancer. Screening may be particularly problematic for high-grade serous ovarian cancer (HGSOC). Detection of stage I serous carcinomas are rare, generally due to vague or absent symptoms or the lack of specific and sensitive biomarkers. Furthermore, the strong majority (up to 95%) of patients with ovarian carcinomas at an advanced stage have serous pathology (6). Similar statistics were obtained from the institutional tumor registry which indicated fairly common diagnosis early-stage for clear cell, mucinous, and endometrioid subtypes of epithelial ovarian cancer compared to a more frequent diagnosis of late-stage for high-grade serous ovarian carcinomas (Figure 1).
The tumor protein p53 (TP53) gene is mutated in over 96% of cases in HGSOC, the most common subtype of ovarian cancer. (7,8). Next generation sequencing methods to detect mutations can be limited by a low rate of sequencing errors. Moreover, amplification of limited amounts of DNA may introduce artificial mutations, and conventional next generation sequencing is not able to detect low-frequency mutations. Molecular “barcoding” to reduce intrinsic errors has been around for about a decade (9), and duplex sequencing further reduces false mutations that allows for ultra-deep sequencing (10). In a recent study, Krimmel et al. (11) aimed to determine whether duplex sequencing could detect TP53 mutations in cells present in the peritoneal fluid from HGSOC samples. They used ultra-deep sequencing to detect TP53 mutations in the peritoneal fluid from HGSOC patients with known TP53 mutations. This study built on their previous work (12,13) and demonstrated the proof-of-principal ability to detect mutations at low frequency with exceptional sequencing accuracy. This detection accuracy also uncovered low-frequency TP53 mutations in most peritoneal fluid samples (35/37) and in blood samples of chemotherapy-naïve patients (15/15), which included the non-cancer control samples (20 and 8 control samples, respectively). The study also evaluated the detection of TP53 mutations to distinguish HGSOC from controls.
Duplex sequencing, or “molecular tagging” both strands of DNA before amplification, was used to markedly reduce artifacts to a theoretical false positive rate of 4×10−10 for errors occurring at the same position for both strands of DNA (10). This approach is currently one of the most accurate sequencing technologies and has potential implications across the biomedical research spectrum and plausibly clinical relevance in the near future. Just as next generation sequencing changed the research landscape, this enhanced method allows investigators to address new biological questions by differentiating between genetic variation and detection errors. For example, the finding of TP53 mutations in noncancerous tissue (11) is intriguing and supports the concept proposed by Tomasetti et al. that many somatic mutations can be present and accrue with age prior to initiation of cancer (14). Although the study focused mainly on evaluating TP53 mutations in peritoneal wash samples for staging purposes, the finding of background TP53 mutations in control samples without cancer has serious implications for screening of early-stage HGSOC, which we discussed in later sections.
This study focused on TP53 exons 4–10, where >95% of mutations of this gene cluster (11). Authors suggested that the peritoneal fluid samples may be suitable for early detection of TP53 mutations in HGSOC. HGSOC can disseminate through the peritoneal cavity, and aspects of FIGO staging are based on peritoneal washes or ascites fluid. Using ultra-deep sequencing in peritoneal fluid, this study revealed somatic TP53 mutations in nearly all ovarian cancer and control samples. The mutation burden, calculated as mutant TP53 molecules per nucleotides sequenced, was used to screen samples. Out of 17 peritoneal HGSOC samples with known TP53 mutations and 20 non-ovarian cancer control samples, TP53 tumor burden detections resulted in 82% sensitivity and 90% specificity without regard to disease stage. This study demonstrates a significant improvement in detecting mutations versus less accurate conventional next generation sequencing techniques, including detection of a TP53 mutation in eight samples that may have been equivocal with other methods. It should be noted that the study size is not large enough to provide reliable determination of the assay characteristics.
Even with highly accurate sequencing for the detection of mutations in TP53 as reported by Krimmel et al. (11), translating these assay characteristics to an ovarian cancer screening modality remains a challenge. Beyond the study size, current studies were designed to assess peritoneal cell mutation burden for the staging of HGSOC, and the use of peritoneal wash samples for screening is a non-starter. Earlier studies that utilized endometrial and vaginal swab samples may be more appropriate for the screening of early-stage HGSOC (15-17). However, the findings reported by Krimmel et al. that low-level TP53 mutations can be detected in control patients without ovarian cancer and that background mutations increase with age in control samples is concerning because these background mutations in normal cells may increase the background noise and lower the ability to detect cancer-specific mutation signal. A better understanding of background mutation patterns and rates relative to cancer-specific mutation rates is needed before mutation detection could be used to detect and screen for HGSOC.
Several large-scale trials have been undertaken to evaluate the clinical utility of traditional biomarkers such as cancer antigen-125 (CA-125) for the screening of early-stage epithelial ovarian cancer (4,18-20). In the latter trials, patients had initial screening and then were monitored over long periods of time with repeated measurements of CA-125 (18,20). With the improvement in risk algorithm that included the rate of rise for CA-125, screen-detected cancer rate doubled over a single-threshold for CA-125 (18). Nonetheless, there remains a substantial portion of false-negative screening results (22 patients out of 155 cases with invasive ovarian cancer were not detected by the screen). These CA-125 negative patients may benefit from TP53 mutation-based screening modalities.
Ovarian cancer screening is often initiated with CA-125 and transvaginal ultrasound screening (4). More recently, human epididymis secretory protein 4 has also been indicated as a biomarker, although further diagnostic clarification is needed to recommend this as a standard ovarian cancer biomarker (20). Furthermore, algorithms such as the risk of ovarian cancer (20) and risk of ovarian malignancy (21) are being used. Several large-scale trials have reached the common conclusion that the current ovarian cancer screen methods are not accurate enough to be recommended, at least in lower risk groups. These trials have exposed a need for improvement, and also a need for a higher standard of accuracy of any novel screening method.
Ovarian cancer screening trials
One of the major evaluations on the effect of screening on ovarian cancer mortality was the prostate, lung, colorectal, and ovarian (PLCO) cancer screening randomized control trial from 1993–2010 (4). This trial was designed to determine the effect of specific cancer screening tests in women aged 55 to 74 years on ovarian cancer-specific mortality. Over 78,000 participants were divided into a screening intervention or a usual care group. Screening consisted of serum CA-125 determination with the standard 35 U/mL threshold and transvaginal ultrasound. The intervention group had over 34,000 women of which 212 were diagnosed with ovarian cancer, and 37 of these were considered interval cases. Interval cancers were defined as cancers not detected by screening, which can be an indication of specificity. The majority of cancer cases in both screening and usual care groups were high-grade and at an advanced stage. The study reported 3,285 false-positive results, and 1,080 underwent surgery (32.9% for oophorectomy) as part of the diagnostic workup. Of these 1,080 women, 15% experienced distinct major complications. The PLCO trial concluded there was no statistically significant reduction in mortality from annual ovarian cancer screening of women at average risk. Also, there was a lack of an observed stage shift, suggesting that the screening modalities used were not effective in detecting ovarian cancers when the cancers were still in a non-advanced stage. The false-positive results in the PLCO trial were approximately 5% of those screened at each round (60% from transvaginal ultrasound). They further concluded that annual ovarian cancer screening does increase invasive medical procedures and associated harms.
An earlier large-scale study of over 82,000 participants in Japan found that there is no evidence that screening results in a higher rate of detection of early-stage ovarian cancer and a reduction in mortality (22). In the screening intervention group, 27 cancers were detected, and this was not statistically different than the 32 cancers detected in the control group. Also, there was no statistical stage shift observed in this study. It was also statistically estimated with 33 surgeries one case of ovarian cancer would be detected in this study.
Recently, the final report of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) randomized control trial of over 200,000 women was published (20). They also found no significant mortality reduction in the primary screening analysis, although a significant mortality reduction of about 15% was noted with CA-125-based screening when prevalent cases from the initial screen were excluded. They concluded a longer duration of follow-up is needed to establish the magnitude of this death reduction. The study also excluded peritoneal cancers, which can be difficult to distinguish from ovarian cancer and the exclusion was needed for the mortality significance finding (23). It should be noted that the UKCTOCS study used a series of serum CA-125 measurements as part of the risk of ovarian cancer algorithm, and reported a sensitivity of 84% for ovarian cancer diagnosed within a year of screening.
The results of these trials have reinforced organizations to make recommendations against ovarian cancer screening (5). The European Group on Tumor Markers recently published an update on guidelines for serum markers in epithelial ovarian cancer (21). This group concluded that CA-125 is not recommended as a routine screening test in asymptomatic women based on accuracy concerns. These concerns included a low sensitivity for stage I disease and a low specificity, especially among premenopausal women. This group did note that serum CA-125 may be beneficial for monitoring patients. The limitations of current biomarkers and screening methods for ovarian cancer support an active area of research. However, the current detection methods and newer algorithms also set a standard for potential new screening methods and demonstrate a strong need for accuracy.
Conclusions
The technical accuracy of duplex sequencing is exceptional compared to conventional next generation sequencing. Accurate sequencing has the potential to uncover new biologically relevant mutations and precise assessment of mutation burden. Since HGSOC is primarily accompanied by TP53 mutations, TP53 mutation-based molecular test has the potential to overcome the challenges of early detection for HGSOC. Such a test might be limited in detecting early stage carcinomas of other subtypes, such as endometrioid, clear cell, and mucinous carcinomas. There are also other limitations to the viability of this technology for HGSOC detection. The cost of next generation sequencing, especially ultra-deep sequencing with duplex tagging, may be high for an asymptomatic patient considering ovarian cancer screening. As with many new technologies, the cost may gradually decrease, and the value may eventually have advantages over the cost and complication risks of surgery. However, statistical concerns related to specificity and sensitivity of these biomarkers need be considered in the context of prevalence for ovarian cancer. Considering the low prevalence of ovarian cancer with an incidence of approximately 50 per 100,000 (24), a screening test with 99% specificity and a sensitivity of 75% for the early disease is estimated to have a positive predictive value of 3.6%. This value translates into approximately 27 false-positives for each case of early-stage ovarian cancer. A high rate of false-positives can lead to unnecessary treatment and surgery, which can have major complications. Therefore, implementation of multi-modal screening strategies that improve both sensitivity and specificity for early-stage HGSOC is critically needed. Supplementation of current protein-based biomarkers with genome-based biomarkers may allow rapid realization of such a goal. Moreover, high-risk patients may be more appropriate for ovarian cancer screening although there is still a lack of statistical evidence to support the benefits outweighing the risks. Nonetheless, there is a general consensus that patients with early-stage epithelial ovarian cancer have superior overall survival compared to patients with advanced disease, and therefore the impact of detecting HGSOC before advanced stages is anticipated to improve overall survivorship from epithelial ovarian cancer. Regardless, an effective screening method for the detection of HGSOC at an early stage has not yet been established.
Acknowledgments
Funding: The work is funded by the University of Kansas Endowment Association, the University of Kansas Cancer Center Support Grant (P30-CA168524), the American Cancer Society Research Scholar (125618-RSG-14-067-01-TBE), and the Department of Defense Ovarian Cancer Research Program under award number (W81XWH-10-1-0386). Views and opinions of, and endorsements by the author(s) do not reflect those of the US Army or the Department of Defense.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Da Li (Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.07.45). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jayson GC, Kohn EC, Kitchener HC, et al. Ovarian cancer. Lancet 2014;384:1376-88. [Crossref] [PubMed]
- Ledermann JA, Raja FA, Fotopoulou C, et al. Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2013;24:vi24-32. [Crossref] [PubMed]
- Prat JFIGO Committee on Gynecologic Oncology. Staging classification for cancer of the ovary, fallopian tube, and peritoneum. Int J Gynaecol Obstet 2014;124:1-5. [Crossref] [PubMed]
- Buys SS, Partridge E, Black A, et al. Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA 2011;305:2295-303. [Crossref] [PubMed]
- Moyer VAU.S. Preventive Services Task Force. Screening for ovarian cancer: U.S. Preventive Services Task Force reaffirmation recommendation statement. Ann Intern Med 2012;157:900-4. [Crossref] [PubMed]
- Colombo N, Peiretti M, Parma G, et al. Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2010;21:v23-30. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609-15. [Crossref] [PubMed]
- Ahmed AA, Etemadmoghadam D, Temple J, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol 2010;221:49-56. [Crossref] [PubMed]
- McCloskey ML, Stöger R, Hansen RS, et al. Encoding PCR products with batch-stamps and barcodes. Biochem Genet 2007;45:761-7. [Crossref] [PubMed]
- Schmitt MW, Kennedy SR, Salk JJ, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A 2012;109:14508-13. [Crossref] [PubMed]
- Krimmel JD, Schmitt MW, Harrell MI, et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc Natl Acad Sci U S A 2016;113:6005-10. [Crossref] [PubMed]
- Ahn EH, Hirohata K, Kohrn BF, et al. Detection of Ultra-Rare Mitochondrial Mutations in Breast Stem Cells by Duplex Sequencing. PLoS One 2015;10:e0136216 [Crossref] [PubMed]
- Schmitt MW, Fox EJ, Prindle MJ, et al. Sequencing small genomic targets with high efficiency and extreme accuracy. Nat Methods 2015;12:423-5. [Crossref] [PubMed]
- Tomasetti C, Vogelstein B, Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci U S A 2013;110:1999-2004. [Crossref] [PubMed]
- Kinde I, Bettegowda C, Wang Y, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med 2013;5:167ra4 [Crossref] [PubMed]
- Kinde I, Wu J, Papadopoulos N, et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:9530-5. [Crossref] [PubMed]
- Erickson BK, Kinde I, Dobbin ZC, et al. Detection of somatic TP53 mutations in tampons of patients with high-grade serous ovarian cancer. Obstet Gynecol 2014;124:881-5. [Crossref] [PubMed]
- Menon U, Ryan A, Kalsi J, et al. Risk Algorithm Using Serial Biomarker Measurements Doubles the Number of Screen-Detected Cancers Compared With a Single-Threshold Rule in the United Kingdom Collaborative Trial of Ovarian Cancer Screening. J Clin Oncol 2015;33:2062-71. [Crossref] [PubMed]
- Menon U, Gentry-Maharaj A, Hallett R, et al. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS). Lancet Oncol 2009;10:327-40. [Crossref] [PubMed]
- Jacobs IJ, Menon U, Ryan A, et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet 2016;387:945-56. [Crossref] [PubMed]
- Sölétormos G, Duffy MJ, Othman Abu Hassan S, et al. Clinical Use of Cancer Biomarkers in Epithelial Ovarian Cancer: Updated Guidelines From the European Group on Tumor Markers. Int J Gynecol Cancer 2016;26:43-51. [Crossref] [PubMed]
- Kobayashi H, Yamada Y, Sado T, et al. A randomized study of screening for ovarian cancer: a multicenter study in Japan. Int J Gynecol Cancer 2008;18:414-20. [Crossref] [PubMed]
- Narod SA, Sopik V, Giannakeas V. Should we screen for ovarian cancer? A commentary on the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) randomized trial. Gynecol Oncol 2016;141:191-4. [Crossref] [PubMed]
- Nossov V, Amneus M, Su F, et al. The early detection of ovarian cancer: from traditional methods to proteomics. Can we really do better than serum CA-125? Am J Obstet Gynecol 2008;199:215-23. [Crossref] [PubMed]


