
Deep sequencing reveals that low level TP53 mutations are ubiquitous in ovarian cancer patients and controls: successes and challenges for early detection
Introduction
Solid tumor cancers can have good prognosis if detected early, which has driven the search for screening techniques that are minimally invasive, inexpensive, and sensitive. Circulating tumor DNA (ctDNA) can be detected in the blood, even for localized tumors, as a result of tumor cell death. ctDNA can also be used to monitor response to therapy and relapse. Detection of low-level ctDNA in bodily fluids has been limited by artifactual mutations induced by the sequencing technologies or sample preparation protocols. Krimmel and colleagues (1) applied deep duplex sequencing (DS) to detect low levels of ctDNA in the peritoneal fluid (PF) of women with and without ovarian cancer. DS reduces many artifactual mutations by sequencing independently multiple copies of each strand of the same double-stranded molecule. This group not only found diagnostic mutations in nearly all ovarian cancer samples, but also low-level background TP53 mutations in all control samples. While the first finding may drive interest in duplex sequencing of liquid biopsies, the later findings shows that additional strategies will be needed to deal with the background mutations if deep-sequencing for mutations is to become an early detection biomarker.
ctDNA represents only a small fraction of the DNA circulating in bodily fluids and needs to be detected at levels far below the error levels of current high throughput sequencing technologies (1 in 104 for Illumina HiSeq). To overcome the sequencing errors of these technologies, many sample preparation protocols have been developed to sequence multiple copies of the same molecule and compute a consensus sequence that has greatly reduced sequencing artifacts. The methods are either Circle Sequencing (CS) (2), barcoding methods (3,4), or maximum depth sequencing (MDS) (5). Barcoding and MDS attaches a unique random barcode to each original DNA molecule prior to DNA manipulations and amplifications. Multiple copies of the same molecule are sequenced and clustered according to their barcodes to produce a consensus that has reduced sequencing artifacts. Barcoding methods can still misclassify PCR artifacts as mutations if they occur early in the PCR process. CS and MDS do not suffer from this problem, but they are still sensitive to artifacts introduced while the DNA is denatured as single-stranded templates or by oxidative reactions during DNA extraction or shearing (6), such as mutations to 8-oxoguianine (oxoG). These other artifacts only affect one strand of the double stranded DNA. Two newer double-stranded barcodes (1) and (7) methods eliminate those single-strand sequencing artifacts occurring before the DNA amplification steps. Figure 1 reviews the different methods. Notice that all those methods tremendously increase the cost of sequencing as multiple copies of each original molecule needs to be sequenced.
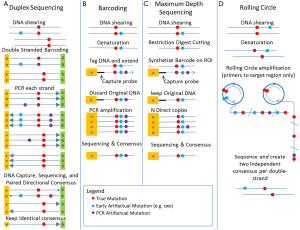
Krimmel and colleagues applied DS to the PF and blood of both patients with diagnosed High Grade Serous Ovarian Cancer (HGSOC) and normal controls selected from patients who underwent gynecological surgery and had no malignant pathology. While blood testing would be the ultimate non-invasive testing, PF is routinely collected during gynecological surgery and cytology of PF is routine to monitor disease progression. Clonal homozygous TP53 mutations is one of the hallmark of this cancer (8), even at an early stage (9) and should be present in all patients. They were able to detect the known TP53 clonal mutations in PF down to below 10−4 frequency in 16/17 patients (all with confirmed clonal TP53 mutation in primary tumor). Remarkably, 8 of the 16 had negative cytology, demonstrating the additional power of DS to detect low-level cancer activity when the TP53 frequency was below 0.01 (limit of detection for positive cytology). The application of DS to blood did not have the same level of success as the known clonal tumor TP53 mutation was only detected in the blood of one patient.
The authors made another seminal discovery with deep implication for the detection of low level mutation in bodily fluids. In addition to the known clonal mutation dominating the tumor they found several deleterious TP53 mutations in both the PF and even blood (16/17 patients and 19/20 controls). These mutations were consistent with the mutation signature of ovarian cancer (8) and unlikely to be sequencing artifacts. The number of mutation was related to the sequencing depth and the frequency of these mutations increased with age but was more abundant in cases, even after adjusting for BRCA status.
While there was unmitigated success in detecting the clonal TP53 mutations in PF that are important in monitoring recurrence, the presence of so many deleterious mutations in the PF and blood of controls raises some challenges for early detection and monitoring in PF or blood. Because the controls were selected from patients that had suspected ovarian malignancies, it could be that some of those controls have low level cancer, but this is unlikely to be the case for all controls. Even for the confirmed HGSC patients, only one had in blood the same clonal TP53 mutation known from the tumor, the other patients having other TP53 mutation in blood than the clonal mutation in their tumor. To use DS results in the as an early detection biomarker, we have to ask at what level are TP53 mutations indicative of normal background processes? Are those ctDNA TP53 mutations produced from the death of cells targeted by apoptosis or the immune system? What strategies can we employ to deal with the background of mutations? The strategy to use will likely be cancer dependent.
From studies of early HGSOC (9) and even in the precursors to HGSOC (10), we know that TP53 mutations must become homozygous to inactivate the tumor suppressor function (unless there is silencing of the active copy). So for diagnosis purposes, cells could be isolated to assess whether TP53 mutations are homozygous. For early detection from blood samples, the Krimmel paper clearly indicates that deeper sequencing should reveal more TP53 mutations. But at which level is it actionable? In ovarian cancer, it may be necessary to extend methods developed for prenatal diagnostic (11) to detect arm-level Loss of Heterozygocity (LOH) at the TP53 loci or isolate circulating tumor cells (CTC) from blood in order to assess if detected mutations are homozygous. However, waiting until the disease is at the stage where CTC’s are detectable in blood may lower prognosis. For HGSOC, another possible non-invasive early detection strategy would be to use intravaginal tampons or Tao brushings (12) to detect mutations in precursor cells.
Conclusions
Krimmel and colleagues have described the application of DS to ovarian cancer patients and controls. Their study showed the power of DS to detect the presence of cancer from PF but also highlighted that low-level TP53 mutations present in both blood and PF can present a real challenge to the use of DS for early detection. The presence of ubiquitous background mutations will likely require the adoption of disease-specific strategies to further elucidate which patients have early stage disease versus non-malignant background processes.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Da Li (Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.08.11). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Krimmel JD, Schmitt MW, Harrell MI, et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc Natl Acad Sci U S A 2016;113:6005-10. [Crossref] [PubMed]
- Lou DI, Hussmann JA, McBee RM, et al. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc Natl Acad Sci U S A 2013;110:19872-7. [Crossref] [PubMed]
- Jabara CB, Jones CD, Roach J, et al. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc Natl Acad Sci U S A 2011;108:20166-71. [Crossref] [PubMed]
- Kinde I, Wu J, Papadopoulos N, et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:9530-5. [Crossref] [PubMed]
- Jee J, Rasouly A, Shamovsky I, et al. Rates and mechanisms of bacterial mutagenesis from maximum-depth sequencing. Nature 2016;534:693-6. [Crossref] [PubMed]
- Costello M, Pugh TJ, Fennell TJ, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res 2013;41:e67 [Crossref] [PubMed]
- Gregory MT, Bertout JA, Ericson NG, et al. Targeted single molecule mutation detection with massively parallel sequencing. Nucleic Acids Res 2016;44:e22 [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609-15. [Crossref] [PubMed]
- Chien J, Sicotte H, Fan JB, et al. TP53 mutations, tetraploidy and homologous recombination repair defects in early stage high-grade serous ovarian cancer. Nucleic Acids Res 2015;43:6945-58. [Crossref] [PubMed]
- Ahmed AA, Etemadmoghadam D, Temple J, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol 2010;221:49-56. [Crossref] [PubMed]
- Webb A, Madgett T, Miran T, et al. Non invasive prenatal diagnosis of aneuploidy: next generation sequencing or fetal DNA enrichment? Balkan J Med Genet 2012;15:17-26. [PubMed]
- Bakkum-Gamez JN, Wentzensen N, Maurer MJ, et al. Detection of endometrial cancer via molecular analysis of DNA collected with vaginal tampons. Gynecol Oncol 2015;137:14-22. [Crossref] [PubMed]


