
LncRNA CBR3-AS1 is associated with the BCR::ABL1 kinase independent mechanism of tyrosine kinase inhibitor (TKI) resistance in chronic myeloid leukemia patients
Highlight box
Key findings
• Long non-coding CBR3-AS1 might be involved in BCR::ABL1 independent tyrosine kinase inhibitor (TKI) resistance of chronic myeloid leukemia (CML) patients through targeting KCNA6, and CBR3-AS1 may be a novel target for intervention treatment of CML patients with BCR::ABL1 independent TKI resistance.
What is known and what is new?
• Long non-coding RNA (lncRNA) showed function in BCR::ABL1 dependent TKI resistance and pathogenesis of CML patients.
• CBR3-AS1 overexpression is associated with BCR::ABL1 independent TKI resistance of CML patients by targeting KCNA6.
What is the implication, and what should change now?
• Further in vitro and in vivo are needed to validate the function of CBR3-AS1-KCNA6 in BCR::ABL1 independent TKI resistance of CML patients.
Introduction
Chronic myeloid leukemia (CML) is a group of malignant clonal blood system disease derived from hematopoietic stem cells and is the most prevalent type of chronic leukemia, accounting for approximately 15% of adult leukemia (1). CML occurs due to t(9; 22)(q34; q11) chromosome translocation producing the BCR::ABL1 fusion gene, detected in around 90% of CML patients (2). The fusion protein encoded by BCR::ABL1 fusion gene has robust tyrosine kinase activity, which can activate cell proliferation-related signaling pathways, thus interfering the procedures of the normal cell proliferation and apoptosis, ultimately contributing to the occurrence and progression of CML (2). Tyrosine kinase inhibitor (TKI), represented by imatinib, significantly improves the 5-year overall survival and progression-free survival rate of CML patients to over 90%. Patients who achieve sustained deep molecular response (DMR) after TKI treatment for more than 2 years can try to discontinue the drug, and some patients can achieve long-term treatment free remission, namely, functional cure (3,4). However, TKI resistance contributes to the inability of CML patients to obtain DMR, leading to treatment failure and relapse after remission. Overcoming TKI resistance is still the focus and difficulty of CML clinical research.
Currently, the main causes of TKI resistance are classified into BCR::ABL1 kinase dependent and independent (5). BCR::ABL1 kinase dependent resistance is usually caused by the mutation in the ABL1 gene that prevents TKI from binding effectively to p210bcr-abl, and such resistance can be overcome by developing a new generation of TKI. Whereas, the main cause of BCR::ABL1 kinase independent resistance is that the activation of CML cell proliferation and/or self-renewal related signaling pathways is independent of p210bcr-abl tyrosine kinase activity (6), and the new generation of TKI fails to defeat such resistance. In clinical practice, it is difficult for CML patients with BCR::ABL1 independent drug resistance to achieve optimal efficacy. Thus, it is of great significance to investigate the BCR::ABL1 kinase independent mechanism of TKI resistance in CML patients to develop targeted therapeutic strategy.
Long non-coding RNA (lncRNA) is a class of non-protein-coding RNA sequences with a length of more thanw 200 nucleotides. Studies have shown that abnormally expressed lncRNAs are associated with pathogenesis and drug resistance of various human diseases, including CML (7,8). As reported by Guo and colleagues, served as a major regulatory molecule in BCR::ABL1 mediated CML, lncRNA BGL3 promotes apoptosis of CML cells through positive regulation of PTEN expression, and ultimately contributes to the progression of CML through (9). In addition, previous studies have demonstrated that both lncRNA OIP5-AS1 and lncRNA MALAT1 are involved in the regulation of TKI resistance in CML patients through sponging microRNAs (10,11). LncRNA MEG3 regulates TKI resistance in CML cells by inhibiting the expression of miR-21/miR-184 (12). Collectively, these studies mainly focused on investigating the role of lncRNA in BCR::ABL1 dependent TKI resistance and pathogenesis of CML patients.
Herein, we aimed to explore the function of lncRNA in BCR::ABL1 independent TKI resistance of CML patients, demonstrating that CBR3-AS1 might be involved in BCR::ABL1 independent TKI resistance of CML patients through targeting KCNA6. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-281/rc).
Methods
Patients
CML patients diagnosed according to the guidelines for diagnosis and treatment of chronic myelogenous leukemia in China (2020 edition) (4) were enrolled at Department of Hematology, the First Hospital of Putian City in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the ethics committee of the First Hospital of Putian City (approval No. 2022-027). All eligible participants provided written informed consent. Patients receiving regular and continuous oral administration of TKI for more than 3 months and with BCR::ABLIS ≤0.01% (MR4.0) were included in the DMR group. Patients receiving regular and continuous oral administration of TKI for more than 2 years, previously received or being treated with at least one type of second-generation TKI, and with 0.01%< BCR::ABLIS ≤1% were included in the non-DMR group. All enrolled patients had no BCR::ABL kinase mutations and had not received other drugs that affect the efficacy of TKI. Patients receiving drugs that affect the efficacy of TKI, in combination with other hematological disease, malignant tumor, and severe systemic disease, and with pregnant were excluded. Twelve health individuals were enrolled as control. Once the patients were determined for DMR or non-DMR, the peripheral blood mononuclear cell (PBMC) was collected for analysis.
RNA-sequencing and data analysis
Total RNA was extracted from PBMC using TRIzol (Invitrogen, CA, USA; 15596026). After ribosomal RNA removal, RNA was purified by RNA purification magnetic beads and interrupted using metal ion solution. The first strand of complementary DNA (cDNA) was synthesized in the reverse transcriptase system species by using the fragmented RNA as the template and the random oligonucleotides as the primers. Thereafter, the second stranded of cDNA was synthesized, repaired and A tail was added to the 3’ end. After purification and fragment selection (300–500 bp), the products were amplified and enriched by polymerase chain reaction (PCR) to construct library. The obtained raw data in FASTQ format was filtered using fastp (v0.21.0) software (https://github.com/OpenGene/fastp) to collect clean data through removing reads containing adapter, containing ploy, and with low quality. Thereafter, the filtered reads were mapped to the reference genome using HISAT2 (v2.1.0) (13). FeatureCounts (v2.0.0) (14) statistics was utilized to calculate the expression of genes, and fragments per kilobase of exon model per million mapped fragments (FPKM) was then used to standardized the expression. Differentially expressed genes were identified using DESeq2 (v1.30.1) with screened conditions as follows: expression difference multiple |log2FoldChange| >1, significant Padj ≤0.05. Online database genemania was used to analyze the major interacted proteins of KCNA6 (https://genemania.org/search/homo-sapiens/KCNA6). Gene Ontology (GO) analysis within the molecular function (MF), cellular component (CC), and biological process (BP) categories, as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the major interacted genes of KCNA6 were performed using R software (v 4.2.1). LncRNAs-messenger RNAs (mRNAs) co-expression pairs were obtained using the weighted gene co-expression network analysis.
Real-time quantitative PCR (RT-qPCR)
RT-qPCR was performed to validate the expression of lncRNA CBR3-AS1 in PBMC of CML patients. Total RNA was extracted from peripheral blood using Trizol (Ambion, CA, USA; 15596-026) and reverse transcribed into cDNA using HiScript® II Q RT SuperMix for qPCR (+gDNA wiper) (VAZYME, Nanjing, China; R223-01), followed by qPCR amplification using SYBR Green Master Mix (VAZYME, Q111-02). The primer sequences were as follows: lncRNA CBR3-AS1, forward, 5'-TCAAGAAGGTGGTGAAGCAGG-3', reverse, 5'-TCAAAGGTGGAGGAGTGGGT-3'; GAPDH, forward, 5'-CCTCGATAAGACCGTGAC-3', reverse, 5'-TCTCCCTCTGCTTCCCTA-3'. The relative expression of lncRNA CBR3-AS1 was calculated using the method of 2−ΔΔCt.
Statistical analysis
Enumeration data were described using the number of cases (percentage, %), and measurement data were described as the mean ± standard deviation or mean (range). The chi-squared test was performed to comparison baseline data between groups. Comparisons of lncRNAs expression among DMR, non-DMR, and health control groups were analyzed using the Mann-Whitney U test for non-parametric variables. As for data analysis of RT-qPCR, comparisons between groups were performed using the one-way analysis of variance followed by Tukey’s post-hoc test. Statistical significance was defined as P value <0.05.
Results
Baseline characteristics of enrolled CML patients
As shown in Table 1, a total of 27 CML patients were enrolled in this study, 15 received DMR after TKI treatment, with mean age of 42 years (range, 31–67 years), and 12 divided in non-DMR group, with mage age of 45 (range, 24–56 years). Most patients received imatinib as first-line therapy and subsequently switched to second-generation TKI including dasatinib, flumatinib, and nilotinib. All the enrolled patients had elevated white blood cell, and most patients showed decreased hemoglobin. Ten (67%) patients in DMR group showed low-risk, whereas 10 (83%) patients in non-DMR group showed high-risk according to European Treatment and Outcome Study (EUTOS) long-term survival (ELTS). Most patients in both DMR and non-DMR groups demonstrated intermediate risk according to SOKAL score. The results of statistical difference analysis suggested that there was no statistical difference of included baseline data between two groups (age, P=0.31; gender, P>0.99; first treatment drugs, P=0.78; BCR::ABLIS, P=0.66; hemoglobin, P=0.47; platelet, P=0.29; SOKAL score, P=0.12) except for ELTS (P=0.001), indicating that patients with low-risk are easier to obtain DMR.
Table 1
| Characteristics | DMR (n=15) | Non-DMR (n=12) | χ2 | P value |
|---|---|---|---|---|
| Age, years | 42 [31–67] | 45 [24–56] | 1.025 | 0.31 |
| Gender | 0.000 | >0.99 | ||
| Male | 11 [73] | 9 [75] | ||
| Female | 4 [27] | 3 [25] | ||
| First treatment drugs | 0.077 | 0.78 | ||
| Imatinib | 13 [87] | 9 [75] | ||
| Replaced with a second generation TKI | 1.081 | 0.58 | ||
| Dasatinib | 7 [47] | 6 [50] | ||
| Flumatinib | 4 [27] | 5 [42] | ||
| Nilotinib | 3 [20] | 1 [8] | ||
| BCR::ABLIS | 93% [50–271%] | 92% [53–191%] | 0.439 | 0.66 |
| Elevated white blood cell | 15 [100] | 12 [100] | 0.000 | >0.99 |
| Hemoglobin | 0.519 | 0.47 | ||
| Normal | 4 [27] | 1 [8] | ||
| Decreased | 11 [73] | 11 [92] | ||
| Platelet | 2.466 | 0.29 | ||
| Elevated | 10 [67] | 6 [50] | ||
| Normal | 4 [27] | 6 [50] | ||
| Decreased | 1 [7] | 0 | ||
| ELTS | 15.082 | 0.001 | ||
| Low risk | 10 [67] | 1 [8] | ||
| Intermediate risk | 3 [20] | 1 [8] | ||
| High risk | 2 [13] | 10 [83] | ||
| SOKAL score | 4.182 | 0.12 | ||
| Low risk | 1 [7] | 4 [33] | ||
| Intermediate risk | 13 [87] | 8 [67] | ||
| High risk | 1 [7] | 0 |
Data are presented as mean [range] or n [%]. CML, chronic myeloid leukemia; DMR, deep molecular response; TKI, tyrosine kinase inhibitor; ELTS, EUTOS long-term survival; EUTOS, European Treatment and Outcome Study.
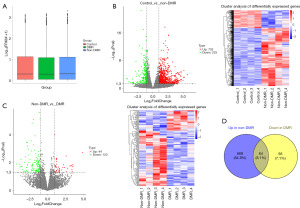
Overview of lncRNA data
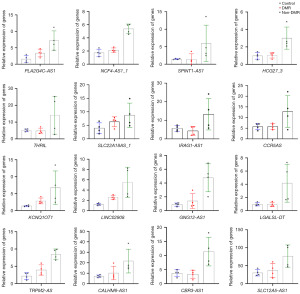
As shown in Figure 1A, FPKM box plot revealed the close value of the log10(FPKM+1) among control, DMR, and non-DMR groups, indicating the reliability of the data. Differential expression analysis suggested that, compared with healthy controls, 733 lncRNAs were up-regulated and 225 lncRNAs were down-regulated in CML patients with non-DMR. Heatmap clustering clearly revealed a separation of the differentially expressed lncRNAs between control and non-DMR groups (Figure 1B). Additionally, compared with patients in non-DMR group, 44 lncRNAs were up-regulated and 120 lncRNAs were down-regulated of patients in DMR groups. Heatmap clustering revealed a separation of the differentially expressed lncRNAs between DMR and non-DMR groups (Figure 1C). Venn plot identified 64 overlapped lncRNAs between up-regulated lncRNAs in non-DMR group (compared with healthy control) and down-regulated lncRNAs in DMR group (compared with non-DMR group) (Figure 1D), which was considered as the resistance related genes. Excluding novel genes or informal genes, a total of 16 lncRNAs were obtained, containing, PLA2G4C-AS1, NCF4-AS1_1, SPINT1-AS1, HCG27_3, THRIL, SLC22A18AS_1, IRAG1-AS1, CCR5AS, KCNQ1OT1, LINC02908, GNG12-AS1, LGALSL-DT, TRPM2-AS, CALHM6-AS1, CBR3-AS1, SLC12A5-AS1 (Figure 2).
Expression validation of lncRNA CBR3-AS1 and lncRNA TRPM2-AS in clinical samples
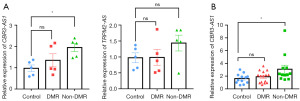
We first excluded genes that had not been reported in the literature and whose functions were unclear. Genes associated with a variety of tumors but not studied in CML were retained. Then, combined with the expression profiles of genes in the GEPIA database, we finally identified CBR3-AS1 and TRPM2-AS as targets of interest. We first evaluated the expression of lncRNA CBR3-AS1 and lncRNA TRPM2-AS using small samples (five each group) as preliminary experiment, demonstrating that, compared with control group, there was no statistical difference of CBR3-AS1 expression in DMR group (P=0.50), whereas, CBR3-AS1 was up-regulated in non-DMR group (P=0.03) (Figure 3A). However, compared with control group, there was no statistical difference in the expression of TRPM2-AS in both DMR (P=0.99) and non-DMR group (P=0.33) (Figure 3A). Thus, only CBR3-AS1 was chosen for the further validation in an expended cohort, demonstrating the similar the results of preliminary experiment (Figure 3B). Collectively, these results suggest that CBR3-AS1 may be associated with BCR::ABL1 independent TKI resistance of CML patients.
Overview of mRNA data
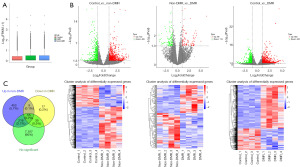
Furthermore, we analyzed the data of mRNA, showing the close value of the log10(FPKM+1) among control, DMR, and non-DMR groups, indicating the reliability of the mRNA data (Figure 4A). Differential expression analysis showed that, compared with control group, 773 mRNAs were up-regulated and 405 mRNA were down-regulated in non-DMR group, whereas 853 mRNAs were up-regulated and 426 mRNA were down-regulated in DMR group; compared with non-DMR group, 89 mRNAs were up-regulated and 113 mRNA were down-regulated in DMR group (Figure 4B). Heatmap clustering showed a separation of the differentially expressed mRNAs between groups (Figure 4B). We considered genes that up-regulated in non-DMR group (compared with control group), down-regulated in DMR group (compared with non-DMR group), showed no statistical difference between control and DMR group as the potential genes that are associated with TKI resistance of CML patients. As shown in Figure 4C, 55 corresponding mRNAs were obtained including KCNA6, a target gene of CBR3-AS1 (available online: https://cdn.amegroups.cn/static/public/10.21037tcr-24-281-1.xls). Thus, KCNA6 was selected for the subsequent analysis.
LncRNA CBR3-AS1 might be involved in BCR::ABL1 independent TKI resistance of CML patients through targeting KCNA6
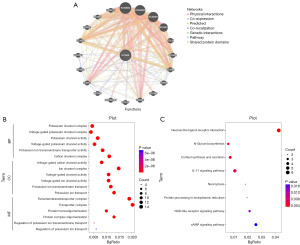
Online database genemania was used to analyze the major interacted proteins of KCNA6, as shown in Figure 5A. Furthermore, functional enrichment and pathway enrichment analysis of the major interacted proteins were performed using R software. GO functional enrichment analysis demonstrated that the genes were mainly enriched in potassium channel complex term of BP category, voltage-gated cation channel activity of CC category, and transmembrane transporter complex of MF category (Figure 5B). KEGG pathway analysis revealed that the major interacted genes were enriched in several resistance-associated pathways including interleukin (IL)-17 signaling pathway and cyclic adenosine monophosphate (cAMP) signaling pathway (Figure 5C).
Discussion
Our current findings demonstrated the diversity lncRNA profiles among healthy individuals, CML patients with non-DMR, and CML patients with DMR. Differential expression analysis and Veen plot of up-regulated lncRNAs in patients with non-DMR and down-regulated lncRNAs in patients with DMR revealed that lncRNA CBR3-AS1 overexpression might be related to BCR::ABL1 independent TKI resistance of CML patients. CBR3-AS1 was initially found to be up-regulated in prostate cancer, which promotes proliferation and suppresses apoptosis of prostate cancer cells (15-17). Subsequent studies discovered the carcinogenesis of CBR3-AS1 in several human cancers including cervical cancer, osteosarcoma, non-small cell lung cancer, lung adenocarcinoma, and breast cancer (18-22). Recently, the role of CBR3-AS1 on drug resistance has been uncovered. As reported by Zhang et al., CBR3-AS1 was obviously overexpressed in breast cancer tissues, which contributed to poor prognosis. CBR3-AS1 upregulation resulted in chemotherapy resistance of breast cancer by regulating miR-25-3p and MEK4/JNK1 signaling pathway (23). Furthermore, we verified the expression of CBR3-AS1 in an expended cohort, demonstrating that, compared with control group, there was no statistical difference of CBR3-AS1 expression in DMR group, whereas, CBR3-AS1 was up-regulated in non-DMR group, which strengthen our conclusion that CBR3-AS1 overexpression is associated with BCR::ABL1 independent TKI resistance of CML patients. However, further functional verification experiments in vitro and in vivo are needed in the follow up study.
To further explore the BCR::ABL1 kinase independent mechanism of TKI resistance in CML patients, we analyzed the mRNA data obtained for RNA-sequencing, indicating the different mRNA profiles among healthy individuals, CML patients with non-DMR, and CML patients with DMR. Subsequent analysis identified 55 mRNAs that may be correlated with TKI resistance in CML patients, including KCNA6, a target gene of CBR3-AS1. Thus, we hypothesized that CBR3-AS1 is involved in BCR::ABL1 independent TKI resistance of CML patients through targeting KCNA6. Moreover, we performed bioinformatics analysis, and the results showed that the major interacted genes of KCNA6 were enriched in several resistance-associated pathways including IL-17 signaling pathway and cAMP signaling pathway, which confirmed our hypothesis. IL-17 is a proinflammatory factor that shows an important function in the immune regulation and tumor progression, including tumorigenesis, proliferation, and metastasis (24,25). In addition, IL-17 pathway has been discovered to be involved in chemotherapy resistance in breast cancer (26), and immune checkpoint blockade sensitivity in pancreatic cancer (27). cAMP is a nucleotide that influences multiple signal transduction pathways, thereby involving in the regulation of numerous cellular BP, including proliferation, differentiation, apoptosis, gene transcription, and protein translation (28). Accumulating evidences also demonstrate the drug resistance modulation of cAMP signaling pathway. As reported by Mishra et al., cAMP signaling inactivation alleviated tamoxifen resistance in estrogen receptor -positive breast cancer (29). Xian et al. found that KCNQ1OT1 knockdown enhanced chemosensitivity towards methotrexate in colorectal cancer through regulating cAMP signaling pathway (30).
Conclusions
In conclusion, the present findings identified lncRNA CBR3-AS1 as a regulatory gene of BCR::ABL1 independent TKI resistance in CML patients using RNA-seq. The expression of CBR3-AS1 was then validated using clinical samples. Further mRNA data analysis and bioinformatics analysis demonstrated that CBR3-AS1 might be involved in BCR::ABL1 independent TKI resistance of CML patients through targeting KCNA6. The limitation of our current study is that we only verified the expression of CBR3-AS1 in clinical samples, further in vitro and in vivo will be designed to validate the function of CBR3-AS1-KCNA6, as well as IL-17, in BCR::ABL1 independent TKI resistance of CML patients. Collectively, this work investigates the mechanism of TKI resistance in CML patients, providing a new target for the synergy therapy of CML patients.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-281/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-281/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-281/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-281/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the ethics committee of the First Hospital of Putian City (approval No. 2022-027). All eligible participants provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yılmaz U, Bulan B, Belli Ç, et al. Management of chronic myeloid leukemia in myeloid blastic phase with novel therapies: a systematic literature review. Expert Rev Hematol 2022;15:423-9. [Crossref] [PubMed]
- Patnaik MM, Tefferi A. Chronic myelomonocytic leukemia: 2022 update on diagnosis, risk stratification, and management. Am J Hematol 2022;97:352-72. [Crossref] [PubMed]
- Hughes TP, Ross DM. Moving treatment-free remission into mainstream clinical practice in CML. Blood 2016;128:17-23. [Crossref] [PubMed]
- Chinese Society of Hematology, Chinese Medical Association. The guidelines for diagnosis and treatment of chronic myelogenous leukemia in China (2020 edition). Zhonghua Xue Ye Xue Za Zhi 2020;41:E001. [PubMed]
- Braun TP, Eide CA, Druker BJ. Response and Resistance to BCR-ABL1-Targeted Therapies. Cancer Cell 2020;37:530-42. [Crossref] [PubMed]
- Perrotti D, Silvestri G, Stramucci L, et al. Cellular and Molecular Networks in Chronic Myeloid Leukemia: The Leukemic Stem, Progenitor and Stromal Cell Interplay. Curr Drug Targets 2017;18:377-88. [Crossref] [PubMed]
- Feng X, Yang L, Liu X, et al. Long non-coding RNA small nucleolar RNA host gene 29 drives chronic myeloid leukemia progression via microRNA-483-3p/Casitas B-lineage Lymphoma axis-mediated activation of the phosphoinositide 3-kinase/Akt pathway. Med Oncol 2024;41:60. [Crossref] [PubMed]
- Gasic V, Karan-Djurasevic T, Pavlovic D, et al. Diagnostic and Therapeutic Implications of Long Non-Coding RNAs in Leukemia. Life (Basel) 2022;12:1770. [Crossref] [PubMed]
- Guo G, Kang Q, Zhu X, et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene 2015;34:1768-79. [Crossref] [PubMed]
- Dai H, Wang J, Huang Z, et al. LncRNA OIP5-AS1 Promotes the Autophagy-Related Imatinib Resistance in Chronic Myeloid Leukemia Cells by Regulating miR-30e-5p/ATG12 Axis. Technol Cancer Res Treat 2021;20:15330338211052150. [Crossref] [PubMed]
- Wen F, Cao YX, Luo ZY, et al. LncRNA MALAT1 promotes cell proliferation and imatinib resistance by sponging miR-328 in chronic myelogenous leukemia. Biochem Biophys Res Commun 2018;507:1-8. [Crossref] [PubMed]
- Zhou X, Yuan P, Liu Q, et al. LncRNA MEG3 Regulates Imatinib Resistance in Chronic Myeloid Leukemia via Suppressing MicroRNA-21. Biomol Ther (Seoul) 2017;25:490-6. [Crossref] [PubMed]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357-60. [Crossref] [PubMed]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923-30. [Crossref] [PubMed]
- Fang Z, Xu C, Li Y, et al. A feed-forward regulatory loop between androgen receptor and PlncRNA-1 promotes prostate cancer progression. Cancer Lett 2016;374:62-74. [Crossref] [PubMed]
- Zhang A, Zhang J, Kaipainen A, et al. Long non-coding RNA: A newly deciphered "code" in prostate cancer. Cancer Lett 2016;375:323-30. [Crossref] [PubMed]
- Cui Z, Ren S, Lu J, et al. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol Oncol 2013;31:1117-23. [Crossref] [PubMed]
- Cai Y, Huang Y, Zhang J, et al. LncRNA CBR3-AS1 predicts a poor prognosis and promotes cervical cancer progression through the miR-3163/LASP1 pathway. Neoplasma 2022;69:1406-17. [Crossref] [PubMed]
- Yao W, Hou J, Liu G, et al. LncRNA CBR3-AS1 promotes osteosarcoma progression through the network of miR-140-5p/DDX54-NUCKS1-mTOR signaling pathway. Mol Ther Oncolytics 2022;25:189-200. [Crossref] [PubMed]
- Liu S, Zhan N, Gao C, et al. Long noncoding RNA CBR3-AS1 mediates tumorigenesis and radiosensitivity of non-small cell lung cancer through redox and DNA repair by CBR3-AS1 /miR-409-3p/SOD1 axis. Cancer Lett 2022;526:1-11. [Crossref] [PubMed]
- Hou M, Wu N, Yao L. LncRNA CBR3-AS1 potentiates Wnt/β-catenin signaling to regulate lung adenocarcinoma cells proliferation, migration and invasion. Cancer Cell Int 2021;21:36. [Crossref] [PubMed]
- Xu L, Zhu H, Gao F, et al. Upregulation of the long non-coding RNA CBR3-AS1 predicts tumor prognosis and contributes to breast cancer progression. Gene 2019;721S:100014. [Crossref] [PubMed]
- Zhang M, Wang Y, Jiang L, et al. LncRNA CBR3-AS1 regulates of breast cancer drug sensitivity as a competing endogenous RNA through the JNK1/MEK4-mediated MAPK signal pathway. J Exp Clin Cancer Res 2021;40:41. [Crossref] [PubMed]
- Huangfu L, Li R, Huang Y, et al. The IL-17 family in diseases: from bench to bedside. Signal Transduct Target Ther 2023;8:402. [Crossref] [PubMed]
- Rodriguez C, Araujo Furlan CL, Tosello Boari J, et al. Interleukin-17 signaling influences CD8(+) T cell immunity and tumor progression according to the IL-17 receptor subunit expression pattern in cancer cells. Oncoimmunology 2023;12:2261326. [Crossref] [PubMed]
- Dai H, Sheng X, Wang Y, et al. HIF1α Regulates IL17 Signaling Pathway Influencing Sensitivity of Taxane-Based Chemotherapy for Breast Cancer. Front Cell Dev Biol 2021;9:729965. [Crossref] [PubMed]
- Zhang Y, Chandra V, Riquelme Sanchez E, et al. Interleukin-17-induced neutrophil extracellular traps mediate resistance to checkpoint blockade in pancreatic cancer. J Exp Med 2020;217:e20190354. [Crossref] [PubMed]
- Yan K, Gao LN, Cui YL, et al. The cyclic AMP signaling pathway: Exploring targets for successful drug discovery Mol Med Rep 2016;13:3715-23. (Review). [Crossref] [PubMed]
- Mishra RR, Belder N, Ansari SA, et al. Reactivation of cAMP Pathway by PDE4D Inhibition Represents a Novel Druggable Axis for Overcoming Tamoxifen Resistance in ER-positive Breast Cancer. Clin Cancer Res 2018;24:1987-2001. [Crossref] [PubMed]
- Xian D, Zhao Y. LncRNA KCNQ1OT1 enhanced the methotrexate resistance of colorectal cancer cells by regulating miR-760/PPP1R1B via the cAMP signalling pathway. J Cell Mol Med 2019;23:3808-23. [Crossref] [PubMed]


