
Systematic analysis of TREM2 and its carcinogenesis in pancreatic cancer
Highlight box
Key findings
• Overexpression of Triggering receptor expressed on myeloid cells 2 (TREM2) promotes migration and metastasis of pancreatic cancer cells. The expression of TREM2 in cancer cells was increased by intraperitoneal culture, and then knockdown experiments were performed to demonstrate that knockdown of TREM2 could inhibit the progression of pancreatic cancer.
What is known and what is new?
• TREM2 is mostly expressed in monocytes/macrophages of pan-cancer.
• TREM2 upregulation was found to be associated with lymph node metastasis and poor prognosis in pancreatic cancer, and TREM2, expressed on pancreatic cancer malignant cells, promoted tumor progression.
What is the implication, and what should change now?
• TREM2 is a bridge between the tumor microenvironment and malignant cells and has a carcinogenesis effect. Guidance for future mechanistic exploration must be provided.
Introduction
Cancer is a significant public health problem worldwide. Cancer mortality rates have been declining steadily since 1991, with an overall decrease of 33 percent (1). The third leading cause of cancer death is pancreatic cancer. To reduce mortality from pancreatic cancer, the main measures include the promotion of early screening and improvements in treatment, such as with adjuvant chemotherapies, targeted treatment, and immunotherapy. However, the treatment of cancer has always been a tricky puzzle. The major obstacles in limiting the effectiveness of conventional cancer therapies are cancer heterogeneity, the dynamic nature of cancer (2), and the complexity of the tumor microenvironment (TME) (3). Therefore, it is essential to analyze the tumor characteristics from all aspects.
Triggering receptor expressed on myeloid cells 2 (TREM2) is mainly expressed on cells such as macrophages (including osteoclasts and microglia) and dendritic cells (DCs) (4). TREM2 associates with the tyrosine kinase binding protein (TYROBP) to form a receptor signaling complex, which recruits intracellular signal transduction machinery and mediates cell activation (5). TREM2 signaling induces changes in cellular phenotype and function in a variety of contexts that involve induction of phagocytosis (6) and lipid metabolism (7), promotion of cell survival (8) and counteracting inflammatory activation (9). In addition, TREM2 is a key signalling hub for Alzheimer’s disease (AD), metabolic syndrome, and cancer. TREM2 plays a central role in AD. In a mouse model of AD, targeting the overexpression of TREM2 in microglia improved pathological phenotypes, which include plaque load and cognitive impairment (10). In addition, TREM2 signaling in macrophages has been found to be linked to metabolic diseases like obesity, fatty liver, and atherosclerosis. Recently, emerging roles of TREM2 in cancer have become hotspots of research. TREM2 expression is restricted within most normal tissue, whereas approximately 75% of cancer types have been shown to express TREM2 (11). TREM2 is abnormally expressed in various tumors, including breast cancer, lung cancer, and gastric cancer (12). In various cancers, there is growing evidence for a role of TREM2 in tumor-associated macrophages (TAMs). For example, TREM2+ macrophages are poorly represented in healthy breast tissues but increase with tumor development (13). TREM2 macrophages promote a pro-tumorigenic monocyte-derived macrophages state and drive natural killer (NK) cell paucity and dysfunction in lung cancer (14). However, the expression and function of TREM2 in pancreatic cancer is unknown, especially in pancreatic cancer cells.
We used multiple databases to explore the potential value of TREM2 in different cancers in a comprehensive and in-depth manner. In this study, we clarified the different expression levels of TREM2 in pan-cancer and particularly cell type of cancer, and we explored the correlation between TREM2 and the level of immune infiltration in tumor tissue. In addition, we also focused on analyzing the relationship between TREM2 expressed in cancer cells and pancreatic cancer progression using in vitro experiments. The results of this study are expected to lay the foundation for future research. We present this article in accordance with the MDAR and the ARRIVE reporting checklists (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-201/rc).
Methods
Data collection
The correlation between TREM2 expression and the molecular subtypes and immune subtypes of tumors based on the Tumor-Immune System Interactions Database (TISIDB, http://cis.hku.hk/TISIDB/) was investigated. Moreover, Tumor Immune Single-cell Hub 2 (TISCH2, http://tisch.comp-genomics.org/home/) was used to clarify the expression level of TREM2 in each cell type in different cancers. Furthermore, Tumor IMmune Estimation Resource 2.0 (TIMER2.0, http://timer.cistrome.org/) was also used to conduct an immune correlation assessment between the TREM2 and immune cell infiltration in each tumor. The cBio Cancer Genomics Portal (cBioPortal, http://www.cbioportal.org/) was used to obtain information on the types and frequencies of genomic alterations of TREM2 in pan-cancer. Additionally, we used the Human Protein Atlas database (HPA, https://www.proteinatlas.org/) to reveal the immunohistochemical staining of TREM2 in pancreatic cancer. Furthermore, clinical features and transcriptome profiling of The Cancer Genome Atlas-pancreatic adenocarcinoma (TCGA-PAAD) were retrieved from the Genomic Data Commons (GDC) data portal by TCGAbiolinks (2.30.0), which was used to probe the expression of TREM2 in cancer tissues of patients with lymph node metastasis and non-lymph node metastasis of pancreatic cancer and explore the relationship between TREM2 and tumor progression-related pathways. For the differential expression analysis between pancreatic cancer and normal pancreas, expression profile of TCGA-PAAD and Genotype-Tissue Expression (GTEx) pancreas was extracted from University of California Santa Cruz (UCSC) Xena (https://xena.ucsc.edu/). All the differential expression analysis was performed by limma (3.56.2) using limma-trend method. We used the Kaplan-Meier plotter (https://kmplot.com/analysis) to assess the relationship between TREM2 expression levels and overall survival (OS) and recurrence-free survival (RFS) in pancreatic cancer. Gene expression profiles of pancreatic cancer cell lines and corresponding half-maximal inhibitory concentration (IC50) of the drug were obtained from the Genomics of Drug Sensitivity in Cancer (GDSC) database (https://www.cancerrxgene.org/) (15). Finally, the dataset (GSE154778), which can be found in the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/), was used for showing the expression of TREM2 in different pancreatic cancer cell lines. In Table S1, we list the abbreviation for each tumor type.
Signature score computation
The transcriptome profiling of PAAD in TCGA was acquired in the same way as described above. To explore the correlations between the expression levels of TREM2 and tumor microenvironment, we evaluated the related gene signatures, including genes related to glycolysis, the p53 pathway, angiogenesis, hypoxia, and KRAS signaling up (16) as well as pan-fibroblast TGF-β response signature (pan_f_tbrs) (17), which were first z-score transformed. Then, a principal component analysis was performed, and principal component 1 was extracted to serve as a gene signature score (17).
Cell culture and acquisition
Mouse Panc02 cells were purchased from Procell (Wuhan, China), and as-Panc02 cells were acquired by injecting Panc02 cells into the abdomens of C57BL/6J mice. The cell lines were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and kept at 37 ℃ in a 5% CO2 atmosphere.
Acquisition of ascites cancer cells in mice
Nine 6- to 8-week-old male C57BL/6J mice (weighing 20–25 g) were purchased from the Experimental Animal Centre of Soochow University. All animals were fed with standard diets under a controlled environment with a 12-hour light/dark cycle, 40–60% relative humidity, and room temperature of 24–26 ℃. Panc02 cells (1×106) were injected into the abdomens of three mice to generate Panc02 cells randomly. The prepared Panc02 cell suspension was absorbed with a 1-mL sterile syringe, and the gas was emptied after the abdominal skin of the mice was sterilized with 75% ethanol. The mice were fixed in the supine position, and then the needle was inserted at an angle into the left lower abdomen of the mice. After feeling a breakthrough, 0.1 mL of Panc02 cell suspension (1×106) was injected into each mouse intraperitoneally. The whole inoculation process was guaranteed to be clean, and it is completed within 30 minutes. The activity state of the mice was observed after inoculation, and sufficient water and food were provided. After the mice produced ascites, they were killed by cervical dislocation and then soaked with 75% alcohol for 5 minutes. The ascites of the mice were collected on a biological ultra-clean table. Specific methods were as follows: a skin incision of approximately 2 cm in the median line of the abdomen of each mouse was made to expose the muscle layer; then, the muscle layer was opened with tissue scissors to expose the peritoneum. After that, the peritoneum was lifted with forceps in the left hand, a small incision was made on the peritoneum with scissors in the right hand, and then the abdominal ascites of the mice were absorbed with a syringe without a needle (18). The ascites fluid was centrifuged, and then the obtained cells were cultured. The suspended cells were removed after cell adhesion, and the cells were verified by flow cytometry to exclude the interference of ascites-derived macrophages. Finally, the as-Panc02 was obtained. Animal experiments were performed under a project license (No. SUDA202304A0754) granted by the Animal Ethics Committee of Soochow University, in compliance with the Animal Ethics Committee of Soochow University guidelines for the care and use of animals.
Patients and tissue samples
Human PAAD and their adjacent normal pancreas tissues (eleven pairs) were collected from PAAD patients who underwent resection at The First Affiliated Hospital of Soochow University. All tissue samples were obtained from patients who did not receive any anti-tumour therapy and tissue samples were stored in liquid nitrogen before use. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of The First Affiliated Hospital of Soochow University (No. 2023302) and informed consent was obtained from all individual participants.
Cell transfection
Vector (pLVX-mCherry-N1) and pLVX-mCherry-N1-TREM2 were purchased from Genewiz (80-1294071107-CDS-Trem2, Genewiz, Suzhou, China). Sh-negative control (sh-NC) and sh-Trem2 were designed by Thermo Fisher Scientific (Massachusetts, USA) and synthesized by the Genewiz Company. Table S2 lists the sequences of the sh-RNAs. The plasmid was transfected into 293T cells using Lipomaster 2000 Transfection Reagent (TL201, Vazyme, Nanjing, China). Next, viral supernatant was used to infect pancreatic cancer cells in 8 µg/mL polybrene. Finally, puromycin was used to screen and establish pancreatic cancer cell lines stably transfected with the plasmids mentioned above.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples and cells using an RNA-Quick Purification Kit (RN001, ESscience, Shanghai, China) according to the manufacturer’s instructions. RNA (1 µg) was reverse-transcribed into complementary DNA (cDNA) using 5× All-In-One RT MasterMix (G490, Abm, Richmond, Canada). qPCR was performed using 2× SYBR Green qPCR Master Mix (B21202, Selleck, Shanghai, China). Relative gene expression was quantified using the 2−ΔΔCt method. Table S3 lists the sequences of all PCR primers used.
Western blot
Dissected tissue samples were mechanically lysed in ice-cold RIPA buffer (P0013C, Beyotime, Shanghai, China) supplemented with 1% phenylmethylsulfonyl fluoride (PMSF) on ice for 30–60 minutes. The samples were then centrifuged at 12,000 rpm for 5 minutes at 4 ℃ and the supernatant was collected. The concentration of the proteins was quantified using a bicinchoninic acid (BCA) protein assay (P0012, Beyotime) in accordance with the manufacturer’s instructions. Samples (30 µg) were subjected to 10–12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) separation and subsequently transferred onto polyvinylidene difluoride (PVDF) membranes. Following the blocking of the membranes with 5% bovine serum albumin (BSA) for a period of 2 hours at room temperature, the membranes were incubated with the indicated antibodies overnight at 4 ℃. Following three washes with phosphate-buffered saline with Tween 20 (PBST) solution, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 hour at room temperature. The blots were then subjected to enhanced chemiluminescence (ECL) detection (E411-04, Vazyme). The following antibodies were used: rabbit anti-TREM2 (1:1,000) (MAB17291, R&D, Minneapolis, USA) and GAPDH (diluted 1:5,000; 10494-1-AP, Proteintech, Wuhan, China). The original image of the full western blot can be found in Appendix 1.
Flow cytometry assay
The cell cycle of cancer cells and the identification of as-Panc02 cells were determined using flow cytometry. A cell cycle assay kit (E-CK-A352, Elabscience, Wuhan, China) was used following the manufacturer’s protocol for analysis of the cell cycle distribution of the cancer cells. Moreover, the steps for identifying as-Panc02 cells are described below. The as-Panc02 cells were harvested and equally divided into 1×106 cells/100 µL fluorescence-activated cell sorting (FACS) tubes. Subsequently, the cells were blocked with blocking immunoglobulin G (IgG) (156603, BioLegend, California, USA) for 10 minutes on ice, and then incubated with phycoerythrin (PE)-conjugated anti-mouse F4/80 antibody (123109, BioLegend) for 30 minutes at room temperature in the dark. Unbound antibody was removed by washing the cells in 2 mL of PBS. The cells were centrifuged at 1,200 rpm for 5 minutes, and this wash step was repeated two times. After washing, cells were suspended in PBS and then analyzed by flow cytometry (Beckman Coulter, Gallios, USA).
Immunofluorescence
For fluorescent multiplex immunohistochemistry (mIHC) analysis, a three-color fluorescence kit based on tyramide signal amplification (TSA) was employed by the manufacturer’s instructions (abs50014, Absin Bioscience, Shanghai, China) (19). In summary, tissue sections were incubated with primary antibodies by the protocol for two sequential cycles of IHC, after which the corresponding secondary antibodies and TSA solution at 650 and 520, respectively, were applied. The following primary antibodies were used: anti-PANCK (1:400; 123684, Absin Bioscience), anti-CD68 (1:200; 76437, Cell Signaling Technology, Shanghai, China) and anti-TREM2 (1:400; AF8229, Beyotime). After the last TSA cycle, the sections were counterstained with DAPI at a dilution of 1:1,000 for 10 minutes. Fluorescence images were obtained under a confocal microscope (Olympus, FV1000, Tokyo, Japan).
Colony formation assay
In brief, 800/1,600 transfected cells were cultured in 6-well plates for 7 days. The medium was replaced on a 3-day basis. Following a 7-day incubation period, the colonies were fixed with 4% paraformaldehyde at room temperature for 20 minutes and then stained with crystal violet (C0121-100 mL, Beyotime) at room temperature for 20 minutes. The number of colonies was counted under a light microscope.
Transwell migration and invasion assays
Transwell migration and invasion assays were used to determine cell metastasis capacities. Briefly, [6–10] ×104 cells were placed into the upper chamber with an 8 µm pore size (354576, Corning, New York, USA) with 200 µL of serum-free RPMI 1640 medium, while 600 µL of RPMI 1640 supplemented with 10% FBS was added to the lower chamber. Following the transfection period of 24 or 48 hours, the cells in the upper chamber were removed using a cotton swab, and the cells in the lower chamber were then fixed and stained. Finally, the number of invading cells was quantified and imaged under a light microscope. Different from the transwell migration assay, the transwell upper chamber was coated with 50 µL of a 1:8 dilution of Matrigel (356234, Corning) and incubated at 37 ℃ for 2 hours before culturing the cells in the transwell invasion assay.
Statistical analysis
Each independent experiment was repeated at least three times. All the data are expressed as the mean ± standard error of the mean and were analyzed by using GraphPad Prism 8.0 (GraphPad, La Jolla, CA, USA). Student’s t-tests and two-tailed distributions were used to determine significant differences. Furthermore, a two-sample t-test was employed to assess the differential expression of TREM2 in tumor versus normal paired tissues. Differences were considered to indicate statistical significance at P<0.05.
Results
Relationship between expression of TREM2 and TME in pan-cancer
The tumor immune microenvironment (TIME) plays an important role in the development of tumors. To investigate the differential expression of TREM2 in various tumor subtypes, we used the TISIDB database to classify tumors based on immune subtypes. We found that the expression of TREM2 in the C6 (TGF-β dominant) subtype is the highest in most tumors like BRCA, COAD, LUSC, PAAD, and STAD, not to mention higher than in the C2 (IFN-γ dominant) subtype (Figure 1A). Similarly, by classifying tumors according to molecular subtype via the TISIDB database, we found that TREM2 expression varied among molecular subtypes such as BRCA, COAD, and OV. However, there was no significant difference in ACC, GBM, and UCEC (Figure S1). To further discuss the specific cells of TREM2 expression in pan-cancer, we found that TREM2 was expressed in monocytes/macrophages mostly by analyzing single-cell sequencing data from various cancers. Interestingly, TREM2 was also found to be expressed in malignant cells in several databases (Figure 1B). In order to elucidate the relationship between TREM2 and immune cell infiltration, we employed the using Estimate the Proportion of Immune and Cancer cells (EPIC) algorithms based on the TIMER 2.0 database. The results showed that the expression of TREM2 was significantly associated with macrophages among the various types of cancer (Figure 1C). Subsequently, we utilized the database to confirm this finding based on other algorithms, such as TIMER and XCELL. Using CIBERSORT algorithms, we clearly showed that the M2 macrophage subtype was strongly correlated with the expression of TREM2 in most cancers (Figure 1D). These findings indicate that TREM2 is associated with anti-inflammatory and immunosuppressive effects in most cancers.
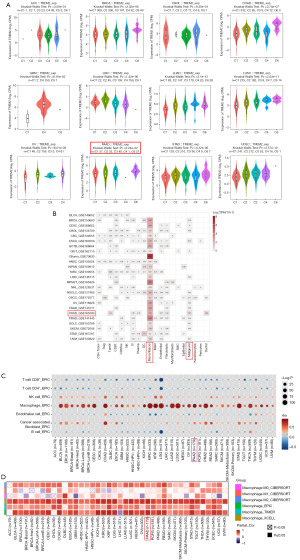
Somatic mutations and drug sensitivity analysis of TREM2 in pancreatic cancer
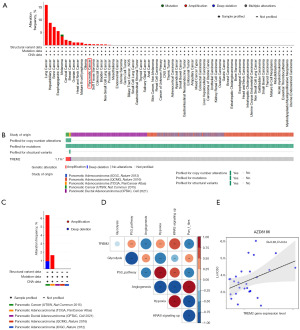
Given the differential expression of TREM2 observed in tumors (Figure 1A), we proceeded to analyze its genetic alterations utilizing the online resource cBioPortal. As illustrated in Figure 2A, the predominant genetic alteration of TREM2 was “amplification”, among which lung cancer (15.79%), hepatobiliary cancer (8.76%), and bone cancer (7.14%) were the most common. “Mutation” was mainly observed in melanoma (0.66%) and breast cancer (0.16%). Furthermore, five databases of PAAD were selected. The overall probability of a TREM2 mutation was 1.1% in the samples (Figure 2B). The mutations are predominantly manifested as amplifications in pancreatic cancer (Figure 2C). By performing transcriptome profiling of TCGA-PAAD data retrieved from the GDC data portal, we found that TREM2 was positively associated with hypoxia and the P53 pathway, especially the KRAS signaling pathway. Moreover, to measure TGF-β pathway activity specifically in fibroblasts, we generated pan_f_tbrs (17). However, TREM2 was negatively associated with both pan_f_tbrs and angiogenesis (Figure 2D). We further investigated the potential correlation analysis between the corresponding IC50 of the drug and TREM2 expression in pancreatic cancer cell lines using the GDSC database. Notably, our results exhibited that TREM2 expression was positively associated with the IC50 of AZD8186 (Figure 2E). AZD8186, a potent and selective inhibitor of PI3K, targets the PI3K/mTOR signaling pathway. It was worth mentioning that TREM2 might not be associated with the sensitivity of certain chemotherapeutic agents, such as gemcitabine and paclitaxel, which are commonly used in pancreatic cancer (Figure S2).
Identification of TREM2 in PAAD
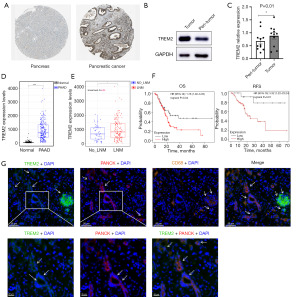
Many studies have focused on the function of TREM2 in TAMs in cancer (13,14). However, it is worth noting that malignant cells also express TREM2 in some PAAD (Figure 1B). Consequently, the study was designed to examine the potential oncogenic effect of TREM2 on malignant cells of PAAD. Firstly, analysis of the HPA database revealed that the intensity of immunohistochemical staining for TREM2 was significantly higher in PAAD tissues than in normal pancreatic tissues (Figure 3A). In addition, we determined the expression of TREM2 in PAAD tissues and adjacent normal tissues from eleven patients via western blotting. Compared with that in adjacent normal tissues, the expression of TREM2 in tumor tissues was elevated (Figure 3B,3C). Moreover, the expression profile of TCGA-PAAD and GTEx pancreas provided further evidence to support the aforementioned points (Figure 3D). Subsequently, we found that TREM2 was highly expressed in cancer tissues with lymph node metastasis compared to those without lymph node metastasis by utilizing data from the TCGA-PAAD (Figure 3E). Furthermore, the prognostic value of TREM2 was evaluated in white pancreatic cancer patients using the Kaplan-Meier plotter database, which illustrated that the OS and RFS of patients with low TREM2 expression were significantly longer than those of patients with the high TREM2 expression (Figure 3F). Finally, based on the GSE154778 single-cell sequencing dataset, TREM2 was expressed on various types of cells in pancreatic cancer tissue, and some TREM2 was expressed in malignant tumor cells (Figure S3), consistent with what was shown before (Figure 1B). We examined the relationship between TREM2 and pancreatic cancer cells in tumor tissues from PAAD patients. Results showed that TREM2 was mainly colocalized with macrophage marker CD68, and a percentage of TREM2 was colocalized with the tumor cell marker PANCK according to mIHC staining (Figure 3G). Therefore, we speculated that the effect of TREM2 on cancer cells is associated with tumor progression.
Elevated expression of TREM2 in pancreatic cells promotes PAAD progression
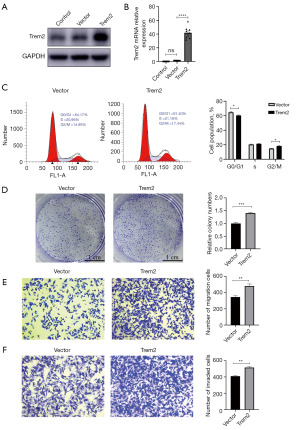
According to several studies, TREM2 is positively associated with metastasis of cancer both on macrophages and tumor cells (20,21). However, it is still unknown whether this effect occurs in pancreatic cancer cells. To investigate the role of TREM2 in PAAD, TREM2 was overexpressed in Panc02 cells using the pLVX-mCherry-N1-TREM2 plasmid. The success of the transfection was determined by means of western blot assays and qPCR (Figure 4A,4B). Subsequently, cell cycle assays and colony formation assays were performed to determine the effect of TREM2 on cell proliferation. Compared with that in the vector group, the proportion of Panc02 cells in the G2/M phase was higher in the TREM2-overexpressing group (17.44% vs. 14.86%, Figure 4C), and the proportion of Panc02 cells in the G0/G1 phase was lower (61.40% vs. 64.17%, Figure 4C). Moreover, compared to those formed by the vector group, the colony formation assay results demonstrated that the overexpression of TREM2 increased the number of colonies (Figure 4D). To further decipher the detailed mechanism of TREM2 in PAAD, transwell migration and invasion assays were performed. The results of the transwell migration and invasion assays demonstrated that the overexpression of TREM2 increased the number of migrating cells (Figure 4E) and invaded cells (Figure 4F), indicating that the migratory and invaded capabilities of PAAD cells were enhanced following TREM2 overexpression. In summary, elevated expression of TREM2 in pancreatic cells promotes the progression of cancer.
Silencing the expression of TREM2 in pancreatic cells inhibits PAAD progression
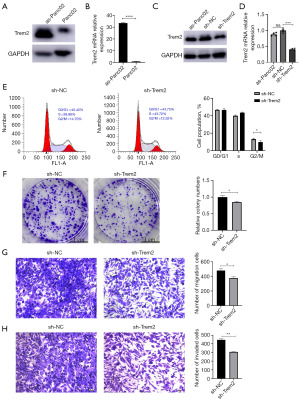
A previous study found that the migration and invasion ability of pancreatic cancer cells were promoted through injecting Panc02 into the mouse abdominal cavity (18). We found that this phenomenon does did exist (Figure S4). To rule out the effect of mouse macrophages, we detected the mouse macrophage marker F4/80 by flow cytometry and found that as-Panc02 cells did not express this marker (Figure S5). Furthermore, we found that the expression of TREM2 in Panc02 cells increased after abdominal culture in C57BL/6J mice (as-Panc02) (Figure 5A,5B). Subsequently, we employed RNAi technology to inhibit the expression of TREM2 effectively (Figure 5C,5D). After that, we performed cell cycle assays and colony formation assays to determine the effect of TREM2 on cell proliferation, as usual. Flow cytometry revealed that TREM2 interference blocked the cell cycle at the G2/M-phase transition in the as-Panc02 cells (14.70% vs. 12.52%, Figure 5E). In addition, by performing colony formation assays, we found that interfering with TREM2 expression inhibited the viability of the as-Panc02 cells (Figure 5F). Furthermore, transwell migration and invasion assays were employed to investigate the potential involvement of TREM2 in the metastasis of as-Panc02 cells. As shown in Figure 5G,5H, the migration of as-Panc02 cells with disrupted TREM2 expression was significantly inhibited. Based on this study, we can see that it was suggested that TREM2 on pancreatic cancer cells promoted the progression of PAAD.
Discussion
At the beginning of the 21st century, triggering receptors expressed on myeloid cells (TREMs) as a new class of receptors were identified (22,23). The TREMs of humans and mouse are encoded by gene clusters on human chromosome 6p21.1 and mouse chromosome 17C, respectively. The human cluster includes TREM1, TREM2, TREML1 (encoding TREM-like 1), TREML2, TREML4, and NCR2 (encoding NKp44) (24). TREMs are comprised of three domains: an extracellular immunoglobulin-like domain, a transmembrane domain, and a short cytoplasmic tail (5). These receptors are expressed on the plasma membrane of a wide range of myeloid cells, including granulocytes, monocytes, macrophages, and DCs (25,26). It has been demonstrated that alterations in TREMs expression are associated with a variety of infectious and inflammatory diseases (27), with a particular association observed in neurodegenerative disorders such as AD (28).
TREM2, a member of the TREMs, is a lipid-binding receptor associated with DAP12 and DAP10 and is expressed by tissue macrophages, including microglia in the central nervous system, osteoclasts in bone, adipose tissue, pulmonary alveoli, and tumors. In recent years, studies on TREM2 have been extraordinarily broad. It has been suggested that TREM2 is a double-edged sword. In neurodegenerative disorders, researchers have focused on the function of TREM2 in microglial phagocytosis, which plays a protective role in preventing the onset of AD (29). In infectious diseases, TREM2-resident macrophages protect the septic heart by keeping the self-renewal ability and maintaining cardiomyocyte homeostasis (30). In addition, in metabolic diseases, a prolonged period of hypernutrition has been demonstrated to impair TREM2-dependent macrophage efferocytosis, which in turn has been shown to cause the development of chronic liver inflammation and nonalcoholic steatohepatitis (NASH) development (31). Paradoxically, in terms of tumor immunosuppression, TREM2+ mo-macs suppress NK cell accumulation and reduce NK cell activity in lung cancer (14). TREM2+ macrophages suppress CD8+ T-cell infiltration after transarterial chemoembolization in hepatocellular carcinoma. TREM2 deficiency dramatically increases CD8+ T cell infiltration and augments the therapeutic efficacy of anti-programmed cell death 1 ligand 1 (anti-PD-L1) blockade (32). In conclusion, TREM2 may be involved in the pathogenesis of various cancers in different ways. Nevertheless, there have been few reports exploring the role of TREM2 in pancreatic cancer. In this study, we conducted a comprehensive analysis of TREM2 expression profiles and genetic alterations in pan-cancer. Furthermore, we analyzed the correlation between TREM2 expression and TME immune cell infiltration. The study also elucidated the oncogenic role of TREM2 in PAAD by employing functional experiments.
In this study, bioinformatics analysis demonstrated that TREM2 expression varies across distinct molecular and immune subtypes in various tumor types. We found that the C6 (TGF-β dominant) subtype had higher TREM2 expression than the C2 (IFN-γ dominant) subtype in most tumors, possibly because IFN-γ inhibits TREM2 expression in macrophages, while TGF-β promotes TREM2 expression (33). A more detailed examination of these correlations may assist in identifying the underlying mechanisms of TREM2.
Interestingly, TREM2 demonstrated a remarkably consistent positive correlation with TAMs across a range of cancers. In recent years, an increasing number of studies have demonstrated that TAMs can either promote or inhibit tumor progression by influencing tumour growth (34), immune escape (35), and metastasis of tumors (36,37). In this study, we also showed that TREM2 was mainly expressed on macrophages in most cancers by analysis of single-cell sequencing data. Moreover, the EPIC algorithm analysis demonstrated a strong association between TREM2 expression and macrophages and carcinoma-associated fibroblasts (CAFs), while also indicating a negative association between TREM2 expression and CD4+ T cells, CD8+ T cells, and B cells infiltration in the TME, which further showed that TREM2 is associated with immunosuppressive effects (38,39). However, this analysis also showed that TREM2 expression was positively associated with NK cells, which are common tumor-killer cells. TREM2 and TYROBP/DAP12, the intracellular adaptor of TREM2, may be important activating signal transduction elements in NK cells (40,41). In addition, research has shown that Nasu-Hakola disease (NHD) patients carrying biallelic variants in TREM2 or TYROBP may manifest a reduction of the NK cell population (42).
Notably, somatic mutations of TREM2 were mainly amplification in pan-cancer. In PAAD, TREM2 expression was positively correlated with the KRAS signaling up pathway and AZD8186 resistance (a selective inhibitor of PI3K), which can explain that the oncogenic-KRAS mutation is the major event in pancreatic cancer (43), and oncogenic-KRAS reprogramming of pancreatic cancer cells mediate the polarization of TAMs (44) that occurs through a PI3K-AKT pathway (45). In addition, TREM2 appears to be highly expressed in TAMs populations across different subtypes (46).
Previous studies have shown that macrophages in human and mouse pancreatic tumors overexpress TREM2, especially macrophages at the site of liver metastases of pancreatic cancer (47). The present study has demonstrated that TREM2 is highly expressed in pancreatic cancer cells at both mRNA and protein levels. Moreover, TREM2 expression was found to be associated with lymph node metastasis in patients with PAAD. In vitro experiments found that elevated expression of TREM2 in pancreatic cells can promote their proliferation, migration, and invasion. Furthermore, the migratory and invasive ability of pancreatic cancer cells improve through peritoneal culture (18). Interestingly, the expression of TREM2 on malignant cells was also enhanced. Therefore, we reduced the expression of TREM2 and found that the proliferation and migratory capacity of malignant cells were inhibited. These findings verified the oncogenic role of TREM2 in PAAD.
However, there are some limitations to this study. Firstly, the role of TREM2 in promoting cancer progression needs to be further investigated in in vivo models which take into account the PAAD immune microenvironment. Secondly, the tumor microenvironment promoted TREM2 expression in pancreatic cancer cells, which increased the immunosuppressive ability of cancer cells and needs further exploration. Therefore, it is imperative to identify potential mechanisms through multi-layer data analysis.
Conclusions
In this study, the expression of TREM2 was found to be aberrant in a number of tumour types, according to different molecular and immune subtypes of pan-cancer. Furthermore, the contribution of TREM2 to TME and the abundance of immune cell infiltration are not to be overlooked. Meanwhile, the functional study of TREM2 indicated that TREM2 may be involved in the progression of PAAD. Further prospective and experimental studies of TREM2 in different cancer populations may provide additional insights into tumor mechanisms, which could then be used to inform the development of treatment strategies.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR and the ARRIVE reporting checklists. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-201/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-201/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-201/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-201/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. For human experiments: the study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the First Affiliated Hospital of Soochow University (No. 2023302) and informed consent was obtained from all individual participants. For animal experiments: animal experiments were performed under a project license (No. SUDA202304A0754) granted by the Animal Ethics Committee of Soochow University, in compliance with the Animal Ethics Committee of Soochow University guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17-48. [Crossref] [PubMed]
- Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018;15:81-94. [Crossref] [PubMed]
- Tiwari A, Trivedi R, Lin SY. Tumor microenvironment: barrier or opportunity towards effective cancer therapy. J Biomed Sci 2022;29:83. [Crossref] [PubMed]
- Kober DL, Brett TJ. TREM2-Ligand Interactions in Health and Disease. J Mol Biol 2017;429:1607-29. [Crossref] [PubMed]
- Deczkowska A, Weiner A, Amit I. The Physiology, Pathology, and Potential Therapeutic Applications of the TREM2 Signaling Pathway. Cell 2020;181:1207-17. [Crossref] [PubMed]
- Kleinberger G, Brendel M, Mracsko E, et al. The FTD-like syndrome causing TREM2 T66M mutation impairs microglia function, brain perfusion, and glucose metabolism. EMBO J 2017;36:1837-53. [Crossref] [PubMed]
- Nugent AA, Lin K, van Lengerich B, et al. TREM2 Regulates Microglial Cholesterol Metabolism upon Chronic Phagocytic Challenge. Neuron 2020;105:837-854.e9. [Crossref] [PubMed]
- Wang Y, Ulland TK, Ulrich JD, et al. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J Exp Med 2016;213:667-75. [Crossref] [PubMed]
- Ito H, Hamerman JA. TREM-2, triggering receptor expressed on myeloid cell-2, negatively regulates TLR responses in dendritic cells. Eur J Immunol 2012;42:176-85. [Crossref] [PubMed]
- Lee CYD, Daggett A, Gu X, et al. Elevated TREM2 Gene Dosage Reprograms Microglia Responsivity and Ameliorates Pathological Phenotypes in Alzheimer's Disease Models. Neuron 2018;97:1032-1048.e5. [Crossref] [PubMed]
- Molgora M, Esaulova E, Vermi W, et al. TREM2 Modulation Remodels the Tumor Myeloid Landscape Enhancing Anti-PD-1 Immunotherapy. Cell 2020;182:886-900.e17. [Crossref] [PubMed]
- Zhang X, Wang W, Li P, et al. High TREM2 expression correlates with poor prognosis in gastric cancer. Hum Pathol 2018;72:91-9. [Crossref] [PubMed]
- Nalio Ramos R, Missolo-Koussou Y, Gerber-Ferder Y, et al. Tissue-resident FOLR2(+) macrophages associate with CD8(+) T cell infiltration in human breast cancer. Cell 2022;185:1189-1207.e25. [Crossref] [PubMed]
- Park MD, Reyes-Torres I, LeBerichel J, et al. TREM2 macrophages drive NK cell paucity and dysfunction in lung cancer. Nat Immunol 2023;24:792-801. [Crossref] [PubMed]
- Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41:D955-61. [Crossref] [PubMed]
- Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417-25. [Crossref] [PubMed]
- Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544-8. [Crossref] [PubMed]
- Lu D. Establishment of a murine of pancreatic cancer ascites and the effects of anti-IL-9 antibody on pancreatic cancer ascites in mice. MA thesis. Guangxi Medical University; 2019.
- Zhang W, Hubbard A, Jones T, et al. Fully automated 5-plex fluorescent immunohistochemistry with tyramide signal amplification and same species antibodies. Lab Invest 2017;97:873-85. [Crossref] [PubMed]
- Yofe I, Shami T, Cohen N, et al. Spatial and Temporal Mapping of Breast Cancer Lung Metastases Identify TREM2 Macrophages as Regulators of the Metastatic Boundary. Cancer Discov 2023;13:2610-31. [Crossref] [PubMed]
- Li C, Hou X, Yuan S, et al. High expression of TREM2 promotes EMT via the PI3K/AKT pathway in gastric cancer: bioinformatics analysis and experimental verification. J Cancer 2021;12:3277-90. [Crossref] [PubMed]
- Bouchon A, Dietrich J, Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol 2000;164:4991-5. [Crossref] [PubMed]
- Chung DH, Seaman WE, Daws MR. Characterization of TREM-3, an activating receptor on mouse macrophages: definition of a family of single Ig domain receptors on mouse chromosome 17. Eur J Immunol 2002;32:59-66. [Crossref] [PubMed]
- Colonna M. The biology of TREM receptors. Nat Rev Immunol 2023;23:580-94. [Crossref] [PubMed]
- Bleharski JR, Kiessler V, Buonsanti C, et al. A role for triggering receptor expressed on myeloid cells-1 in host defense during the early-induced and adaptive phases of the immune response. J Immunol 2003;170:3812-8. [Crossref] [PubMed]
- Wang Y, Cella M, Mallinson K, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer's disease model. Cell 2015;160:1061-71. [Crossref] [PubMed]
- Larabi A, Barnich N, Nguyen HTT. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020;16:38-51. [Crossref] [PubMed]
- Li RY, Qin Q, Yang HC, et al. TREM2 in the pathogenesis of AD: a lipid metabolism regulator and potential metabolic therapeutic target. Mol Neurodegener 2022;17:40. [Crossref] [PubMed]
- Ulland TK, Song WM, Huang SC, et al. TREM2 Maintains Microglial Metabolic Fitness in Alzheimer's Disease. Cell 2017;170:649-663.e13. [Crossref] [PubMed]
- Zhang K, Wang Y, Chen S, et al. TREM2(hi) resident macrophages protect the septic heart by maintaining cardiomyocyte homeostasis. Nat Metab 2023;5:129-46. [Crossref] [PubMed]
- Wang X, He Q, Zhou C, et al. Prolonged hypernutrition impairs TREM2-dependent efferocytosis to license chronic liver inflammation and NASH development. Immunity 2023;56:58-77.e11. [Crossref] [PubMed]
- Tan J, Fan W, Liu T, et al. TREM2(+) macrophages suppress CD8(+) T-cell infiltration after transarterial chemoembolisation in hepatocellular carcinoma. J Hepatol 2023;79:126-40. [Crossref] [PubMed]
- Fabre T, Barron AMS, Christensen SM, et al. Identification of a broadly fibrogenic macrophage subset induced by type 3 inflammation. Sci Immunol 2023;8:eadd8945. [Crossref] [PubMed]
- Shirabe K, Mano Y, Muto J, et al. Role of tumor-associated macrophages in the progression of hepatocellular carcinoma. Surg Today 2012;42:1-7. [Crossref] [PubMed]
- Curiel TJ, Coukos G, Zou L, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 2004;10:942-9. [Crossref] [PubMed]
- Chen Y, Song Y, Du W, et al. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci 2019;26:78. [Crossref] [PubMed]
- Baghel KS, Tewari BN, Shrivastava R, et al. Macrophages promote matrix protrusive and invasive function of breast cancer cells via MIP-1β dependent upregulation of MYO3A gene in breast cancer cells. Oncoimmunology 2016;5:e1196299. [Crossref] [PubMed]
- Katzenelenbogen Y, Sheban F, Yalin A, et al. Coupled scRNA-Seq and Intracellular Protein Activity Reveal an Immunosuppressive Role of TREM2 in Cancer. Cell 2020;182:872-885.e19. [Crossref] [PubMed]
- Costa A, Kieffer Y, Scholer-Dahirel A, et al. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell 2018;33:463-479.e10. [Crossref] [PubMed]
- Terme M, Tomasello E, Maruyama K, et al. IL-4 confers NK stimulatory capacity to murine dendritic cells: a signaling pathway involving KARAP/DAP12-triggering receptor expressed on myeloid cell 2 molecules. J Immunol 2004;172:5957-66. [Crossref] [PubMed]
- Paloneva J, Kestilä M, Wu J, et al. Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat Genet 2000;25:357-61. [Crossref] [PubMed]
- Errichiello E, Dardiotis E, Mannino F, et al. Phenotypic Expansion in Nasu-Hakola Disease: Immunological Findings in Three Patients and Proposal of a Unifying Pathogenic Hypothesis. Front Immunol 2019;10:1685. [Crossref] [PubMed]
- Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol 2020;17:153-68. [Crossref] [PubMed]
- Boyer S, Lee HJ, Steele N, et al. Multiomic characterization of pancreatic cancer-associated macrophage polarization reveals deregulated metabolic programs driven by the GM-CSF-PI3K pathway. Elife 2022;11:e73796. [Crossref] [PubMed]
- Van de Velde LA, Subramanian C, Smith AM, et al. T Cells Encountering Myeloid Cells Programmed for Amino Acid-dependent Immunosuppression Use Rictor/mTORC2 Protein for Proliferative Checkpoint Decisions. J Biol Chem 2017;292:15-30. [Crossref] [PubMed]
- Azizi E, Carr AJ, Plitas G, et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 2018;174:1293-1308.e36. [Crossref] [PubMed]
- Kemp SB, Steele NG, Carpenter ES, et al. Pancreatic cancer is marked by complement-high blood monocytes and tumor-associated macrophages. Life Sci Alliance 2021;4:e202000935. [Crossref] [PubMed]


