
Effect and mechanism of curcumin on colon cancer cell senescence through early growth response 1 (EGR1)
Highlight box
Key findings
• Curcumin facilitates the senescence of colon cancer (CC) cells by modulating the transcriptional activity of early growth response 1 (EGR1).
What is known and what is new?
• Curcumin, an extract derived from traditional Chinese medicine, exhibits inhibitory effects on multi-cancer growth.
• Curcumin facilitates CC cell senescence and impedes tumor progression via modulation of EGR1.
What is the implication, and what should change now?
• Curcumin holds promise as a potential therapeutic agent for CC, with EGR1 identified as its molecular target.
• Further validation through animal and clinical trials is warranted to corroborate these findings.
Introduction
Colon cancer (CC) is a prevalent malignancy worldwide, ranking as the third most common cancer and causing nearly 900,000 deaths annually (1). The pathogenesis of CC is a multifaceted and protracted process, with increasing morbidity and mortality rates observed in aging populations and high-income countries primarily due to dietary factors (2). Over the past few decades, treatment strategies for CC have advanced from simple surgical excision to the introduction of radiotherapy, chemotherapy, and targeted therapy, resulting in improved treatment outcomes. However, the 5-year survival rate for CC remains below 40% (3). Consequently, there is an urgent moment to explore novel therapeutic approaches and investigate the molecular mechanisms underlying the occurrence and progression of CC.
In recent years, Chinese herbal medicine has gained global research attention as a potential anti-cancer treatment (4). Curcumin, a diketone compound extracted from Gingeraceae and Araceae, has attracted significant attention due to its antioxidant, anti-inflammatory, anti-angiogenic, and anti-tumor pharmacological effects (5). Curcumin exhibits diverse anti-tumor properties, including the inhibition of tumor cell proliferation, metastasis, and invasion, rendering it a promising candidate for the prevention and treatment of multiple cancers (6,7). However, further investigation is required to elucidate the precise mechanisms by which curcumin exerts its anti-tumor effects and establish a solid scientific foundation for its clinical application.
Cell senescence is characterized by a stable state of cell stasis accompanied by morphological, biochemical, and epigenetic changes (8). Senescence cells are often detected in tumor tissues following radiotherapy and chemotherapy, suggesting their potential anti-tumor effects (9).
Early growth response 1 (EGR1) is a gene located on chromosome 5 that encodes a protein belonging to the EGR family of C2H2 zinc finger proteins (10). EGR1 is a nuclear protein with transcriptional regulatory functions, and its target genes play crucial roles in differentiation and mitosis (11,12). Elevated expression levels of EGR1 have been observed in CC cell lines and are associated with poor prognosis in CC tissues. However, the role of EGR1 as a transcription factor (TF) influencing cell senescence in the progression of CC has been less explored.
To enhance the survival rates of CC, it is imperative to explore new treatment options and improve existing ones. In this study, we elucidated that EGR1 is a TF that inhibits cell senescence. Curcumin reduces the transcriptional activity of EGR1 and increases the expression of EGR1 target genes TERT and SIRT6, thereby promoting the senescence of CC and exerting a therapeutic effect. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-26/rc).
Methods
Materials
The human CC cell line (LoVo) was derived from American Type Culture Collection (ATCC) and has been identified by short tandem repeat (STR). Details of the materials used were as follows: Fetal bovine serum (Hyclone, Logan, Utah, USA); F-12K cell culture medium (Thermo Fisher Scientific, Massachusetts, USA); Purinomycin (Shanghai Biyuntian Biotechnology Co., LTD., Shanghai, China); PrimeScriptTM RT Master Mix Kit (TaKaRa Company, Osaka, Japan); AceQ qPCR SYBR Green Master Mix (Shanghai Xinyu Biotechnology Co., LTD., Shanghai, China); Cell senescence β-galactosidase staining kit (Shanghai Biyuntian Biotechnology Co., LTD.); Antibody (Wuhan Sanying Biotechnology Co., LTD., Wuhan, China); Transwell Chamber (Corning Corporation, New York, USA); EGR1 eukaryotic expression and short hairpin RNA (shRNA) vector (Shanghai Jikai Jiin Medical Technology Co., LTD., Shanghai, China). The primers were synthesized by Jinsilui Biotechnology Co., LTD. (Shanghai, China).
Cell culture
The cell lines were cultured in 5% CO2 at 37 ℃ using F-12K medium containing 10% fetal bovine serum and 100 IU/mL cyanine and streptomycin.
Screening of stable cell lines
Lentiviral vectors EGR1-CV186 and EGR1-GV298 were transfected into LoVo cell in the logarithmic growth phase, and puromycin was added after culture for 72 h. Monoclonal cells were selected by limited dilution method to obtain stable cell lines.
Real-time fluorescence quantitative polymerase chain reaction (RT-qPCR)
Trizol reagent was used to extract total RNA from cells, which was subsequently reverse-transcribed into cDNA. RT-qPCR samples were collected using SYBR Green PCR Master Mix and primers, and transcription levels were measured by 2–ΔΔCt method. Primers sequences were as follows: EGR1, forward: 5'-GGTCAGTGGCCTAGTGAGC-3'; reverse: 5'-GTGCCGCTGAGTAAATGGGA-3'. TERT, forward: 5'-AAATGCGGCCCCTGTTTCT-3'; reverse: 5'-CAGTGCGTCTTGAGGAGCA-3'. SIRT6, forward: 5'-CCCACGGAGTCTGGACCAT-3'; reverse: 5'-CTCTGCCAGTTTGTCCCTG-3'. β-actin, forward: 5'-CATGTACGTTGCTATCCAGGC-3'; reverse: 5'-CTCCTTAATGTCACGCACGAT-3'.
Western blot (WB)
Total protein was extracted from cells cell lysis buffer, and protein concentration was measured by bicinchoninic acid (BCA) to prepare protein sample. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE gel) was prepared, protein samples were taken from each hole at 30 µg, and separated into protein Marker bands by electrophoresis at constant pressure of 80 V. Cutted and transferred the gel, the membrane transfer rack was placed in ice water and the membrane was transferred at 300 mA for 1 h (the transfer time was determined according to the size of the target protein). After the skim milk was sealed for 1 h, the primary antibody was incubated at 4 ℃ overnight, the secondary antibody was incubated at room temperature for 1 h, and the TBS + Tween (TBST) solution was rinsed and the gel imaging system was used for imaging.
Senescence β-galactosidase (SA-β-gal) staining
The adherent cells were planted in the pore plate, with staining fixative added, and fixed at room temperature. The dyeing solution was prepared according to the instructions, added to the orifice plate, incubated at 37 ℃ overnight (note: incubation at 37 ℃ cannot be carried out in a carbon dioxide incubator), rinsed with phosphate buffer saline (PBS), observed under a microscope and photographed (this kit uses X-Gal as a substrate, catalyzed by senescence-specific β-galactosidase to produce dark blue product, which represents positive senescence).
Matrigel invasion assays
Added 500 µL of medium containing 10% serum to the lower layer of 24-well plate, gently place the Transwell chamber coated with matrix glue, it was important to make sure that bubbles were not produced in the step here. After cell counting, 1×105 cells were added to each upper well, and a serum-free medium of 200 µL was added to each of the multiple Wells to ensure the same pressure, and the cells were infiltrated for 36 h. The cells in the small chamber were gently wiped with cotton swabs, fixed with methanol, stained with 0.1% crystal violet for 2 h, and counted under a microscope.
Methylthiazolyldiphenyl-tetrazolium bromide (MTT)
The cell growth was detected by inverted microscope and recorded by 10× photography. 10 µL MTT solution was added to each well, resulting the final concentration was 0.5 mg/mL. the solution was cultured under conditions of 37 ℃, 5% CO2 and 90% humidity for 4 hours. All the supernatant in the wells was carefully absorbed. Subsequently, 100 µL formazan solubilizing solution was added to each well. The board was placed on an oscillator and shaken at 300 rpm/min for 10 minutes to ensure complete dissolution. Finally, the absorbance was measured at 570 nm with an enzyme-labeled instrument.
Molecular docking
The curcumin PDB format file was obtained by DrugBank (https://go.drugbank.com/), the EGR1 protein PDB format file was obtained by pdbj (https://pdbj.org/), then pretreated by PyMoL 2.5, hydrogen was added, partial charge and protonation state were assigned, and water molecules were removed. HDOCK database (http://hdock.phys.hust.edu.cn/) was used to predict molecular docking, binding sites were attached in Glide standard precision (SP) mode, and visualized in PyMoL 2.5.
Statistical analysis
Descriptive statistics were performed using SPSS 26.0. The measurement data is expressed as the mean ± standard deviation. Comparisons of two groups were performed by t-test. P<0.05 was considered statistically significant.
Results
TF EGR1 regulates cell senescence and promotes the progression of CC
In the Gene Ontology Resource, we obtained the list of genes related to cell senescence, and predicted their TF through ChIP-X. The eminent TF, EGR1, was discovered to govern the largest number of genes implicated in the process of cellular senescence (Figure 1A and Tables S1,S2). The motif sequence for EGR1 was obtained from the JASPAR website: https://jaspar.elixir.no/ (Figure 1B). An analysis on the UCSC website (https://genome.ucsc.edu/) found that the key Cellular Senescence genes—telomerase reverse transcriptase (TERT) and sirtuin 6 (SIRT6) promoter regions were highly enriched with EGR1. In addition, a large number of binding sites of EGR1 exist in the promoter sequence. This suggests that EGR1 may be directly involved in the regulation of TERT and SIRT6 gene expression. On the other hand, in the public colon cancer database (GSE17538), high expression levels of EGR1 in 232 CC tissue specimens were associated with poor prognosis. Simultaneously, high expression of cell senescence-related genes, TERT and SIRT6, predicted a better prognosis for CC patients (Figure 1C). Hence, we hypothesize that EGR1 may exert a negative regulatory influence on the expression levels of TERT and SIRT6, consequently impacting the advancement of CC.

EGR1 suppresses the expression of TERT and SIRT6 genes associated with cell senescence
To further substantiate the regulatory impact of the TF EGR1 on genes associated with cell senescence, we established stable LoVo cell lines with either overexpression or knockdown of EGR1. RT-qPCR and WB analyses revealed an inverse relationship between EGR1 expression and the levels of senescence-related genes TERT and SIRT6 (Figure 2A,2B). Consistently, diminished EGR1 expression levels facilitated the upregulation of TERT and SIRT6 in CC cells (Figure 2C,2D). These experiments provide compelling evidence supporting the notion that EGR1 exerts a negative regulatory influence on the expression of cell senescence-related genes TERT and SIRT6 in CC cells.

EGR1 promotes CC progression by suppressing cell senescence
SA-β-Gal senescence staining is widely regarded as one of the most effective methods for detecting cell senescence (13). The proportion of senescent cells was observed to decrease or increase in LoVo cells with stable overexpression or knockdown of EGR1, respectively (Figure 3A). Furthermore, the results from MTT and Matrigel invasion assays indicated that modulation of EGR1 expression in LoVo cells led to alterations in both invasive cell numbers and cell viability (Figure 3B,3C). Additionally, H2O2 was used as a cell senescence inducer to verify the effect of EGR1 when exploring whether EGR1 inhibits cell senescence. As we suspected, the results illustrated that EGR1 could mitigate H2O2-induced senescence while enhancing cell viability and invasion capacity (Figure 3D-3F). In summary, these results suggest that EGR1 promotes tumor progression by suppressing cell senescence in CC cells.
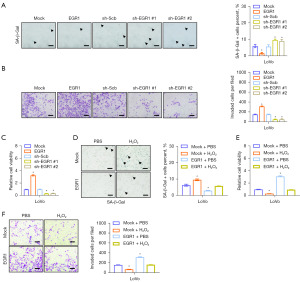
Curcumin regulates EGR1 transcriptional activity via directly binding to EGR1 in CC
In traditional Chinese medicine, CC is thought to be related to freezing cold, Qi Stagnation, and blood stasis. In recent years, the treatment of CC with traditional Chinese medicine has aroused the increasing interest of many clinical medical workers. Curcumin, a traditional Chinese medicine derived primarily from the rhizome of turmeric (14), has been identified by modern pharmacological studies as having multiple benefits for the human body, including its anti-inflammatory, anti-tumor, anti-angiogenesis, anti-metastasis, and anti-multidrug resistance activities (15). We predicted that curcumin directly bound to EGR1 protein by docking the molecular model of curcumin and EGR1 secondary structure (Figure 4A). This prediction was subsequently verified by the curcumin-pull down assays (Figure 4B). In addition, we found that curcumin inhibited transcriptional activity and transcription levels of EGR1 in CC cells (Figure 4C,4D, Figure S1 and Appendix 1). Further studies found that curcumin increased the cell senescence rate of CC cells and decreased cell viability and invasion (Figure 4E-4G). In summary, we propose that curcumin induce cell senescence and impeding the progression of CC.

Curcumin promotes CC cell senescence through EGR1
To further elucidate the mechanism of curcumin in CC cells, we subjected CC cells to treatment with curcumin and EGR1 individually. Our findings revealed that curcumin administration significantly elevated the transcription and protein expression levels of TERT and SIRT6 in LoVo cells compared to dimethyl sulfoxide (DMSO) group. Conversely, overexpression of EGR1 led to decreased transcription and protein expression levels of TERT and SIRT6 (Figure 5A,5B). Additionally, SA-β-Gal staining demonstrated that EGR1 overexpression reversed curcumin-induced cell senescence (Figure 5C). Based on these observations, we delineated a mechanistic pathway illustrating how curcumin enhances the transcription and translation of target genes TERT and SIRT6 by inhibiting the activity of the EGR1 (Figure 5D). This compelling evidence underscores the pivotal role of EGR1 in mediating the effects of curcumin.

Discussion
CC poses a formidable global health challenge, characterized by its complexity and heterogeneity. Despite advancements in treatment modalities, there persists an unmet need for efficacious and safe therapeutic interventions (16). Curcumin, a natural compound derived from the turmeric plant, has garnered attention for its potential anticancer properties (17). This study endeavors to explore the role of curcumin in promoting cell senescence, particularly its capacity to impede the proliferation and metastasis of CC cells by regulating target genes TERT and SIRT6 via the TF EGR1.
Cell senescence, an irreversible growth arrest state, plays a critical role in tumor suppression (18). Prior investigations have underscored curcumin’s induction of cell senescence across various cancer types, including CC (19). Our research extends these findings by delineating the mechanism through which curcumin orchestrates the transcription and expression of key genes such as TERT and SIRT6, mediated by EGR1, thereby contributing to cell senescence.
TERT, recognized as a cyclin-dependent kinase inhibitor, stands out as a prominent mediator of cell cycle arrest and senescence (20). It plays a crucial role in telomere synthesis, thereby preserving chromosome stability and cellular division capacity (21,22). Our investigations unveil that curcumin elevates the expression of TERT, prompting cell cycle arrest and subsequent senescence in CC. This effect is, in part, mediated by the TF EGR1, which binds to the promoter region of TERT, thereby amplifying its transcription.
SIRT6, often hailed as the “Guardian of the genome”, emerges as a tumor suppressor gene crucial for regulating cell cycle arrest, DNA repair, and apoptosis (23-25). Our experimental endeavors unveil that curcumin activates SIRT6, bolstering its transcriptional activity and culminating in heightened expression of downstream target genes associated with senescence induction. Notably, the TF EGR1 also plays a role in mediating the regulation of SIRT6 by curcumin.
The capacity of curcumin to induce cell senescence bears significant implications for treating CC. Senescence cells lose their proliferative potential, thereby impeding tumor growth. Moreover, curcumin has been reported to curb the metastatic propensity of CC cells by stifling epithelial-mesenchymal transition (EMT), a pivotal process in initiating metastasis (5). Our study sheds light on the novel role of EGR1 as a TF in cell senescence. Curcumin dampens EGR1 activity and promotes senescence, consequently hindering tumor progression.
However, it’s important to note that the conclusions of the study are predominantly based on findings from public databases and in vitro experiments. Further validation through in vivo studies and clinical trials is essential to substantiate these findings. Additionally, more research and clinical investigation are required to fully assess the efficacy and safety of curcumin in CC treatment.
Conclusions
In conclusion, curcumin mitigates the activity of the TF EGR1, augments the expression of target genes TERT and SIRT6, and accelerates tumor cell senescence. This positions curcumin a potential candidate for CC therapy, offering an alternative approach to clinical treatment. Its ability to inhibit cell proliferation and metastasis underscores its therapeutic potential in CC. Therefore, strategies targeting EGR1 with curcumin may represent a viable option for untreated CC.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-26/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-26/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-26/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-26/coif). L.L. reports funding from the Hubei Provincial Health Commission Project (No. WJ2019M240) to this study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liu L, Mo M, Chen X, et al. Targeting inhibition of prognosis-related lipid metabolism genes including CYP19A1 enhances immunotherapeutic response in colon cancer. J Exp Clin Cancer Res 2023;42:85. [Crossref] [PubMed]
- Strating E, Verhagen MP, Wensink E, et al. Co-cultures of colon cancer cells and cancer-associated fibroblasts recapitulate the aggressive features of mesenchymal-like colon cancer. Front Immunol 2023;14:1053920. [Crossref] [PubMed]
- Kesireddy M, Tenner L. Colon Cancer Survivorship in Patients Who Have Received Adjuvant Chemotherapy. Clin Colorectal Cancer 2023;22:361-74. [Crossref] [PubMed]
- Wang W, Li M, Wang L, et al. Curcumin in cancer therapy: Exploring molecular mechanisms and overcoming clinical challenges. Cancer Lett 2023;570:216332. [Crossref] [PubMed]
- Zhang X, Zhu L, Wang X, et al. Basic research on curcumin in cervical cancer: Progress and perspectives. Biomed Pharmacother 2023;162:114590. [Crossref] [PubMed]
- de Waure C, Bertola C, Baccarini G, et al. Exploring the Contribution of Curcumin to Cancer Therapy: A Systematic Review of Randomized Controlled Trials. Pharmaceutics 2023;15:1275. [Crossref] [PubMed]
- Bashkeran T, Kamaruddin AH, Ngo TX, et al. Niosomes in cancer treatment: A focus on curcumin encapsulation. Heliyon 2023;9:e18710. [Crossref] [PubMed]
- Zeng W, Zhang W, Tse EHY, et al. Restoration of CPEB4 prevents muscle stem cell senescence during aging. Dev Cell 2023;58:1383-1398.e6. [Crossref] [PubMed]
- Kawamoto S, Uemura K, Hori N, et al. Bacterial induction of B cell senescence promotes age-related changes in the gut microbiota. Nat Cell Biol 2023;25:865-76. [Crossref] [PubMed]
- Nie P, Zhang W, Meng Y, et al. A YAP/TAZ-CD54 axis is required for CXCR2-CD44- tumor-specific neutrophils to suppress gastric cancer. Protein Cell 2023;14:513-31. [PubMed]
- Yang Z, Chen F, Wei D, et al. EGR1 mediates MDR1 transcriptional activity regulating gemcitabine resistance in pancreatic cancer. BMC Cancer 2024;24:268. [Crossref] [PubMed]
- Liu T, Xu X, Li J, et al. ALOX5 deficiency contributes to bladder cancer progression by mediating ferroptosis escape. Cell Death Dis 2023;14:800. [Crossref] [PubMed]
- Lucas V, Cavadas C, Aveleira CA. Cellular Senescence: From Mechanisms to Current Biomarkers and Senotherapies. Pharmacol Rev 2023;75:675-713. [Crossref] [PubMed]
- Zhao C, Zhou X, Cao Z, et al. Curcumin and analogues against head and neck cancer: From drug delivery to molecular mechanisms. Phytomedicine 2023;119:154986. [Crossref] [PubMed]
- Ming T, Tao Q, Tang S, et al. Curcumin: An epigenetic regulator and its application in cancer. Biomed Pharmacother 2022;156:113956. [Crossref] [PubMed]
- Lu L, Mullins CS, Schafmayer C, et al. A global assessment of recent trends in gastrointestinal cancer and lifestyle-associated risk factors. Cancer Commun (Lond) 2021;41:1137-51. [Crossref] [PubMed]
- Joshi P, Bisht A, Paliwal A, et al. Recent updates on clinical developments of curcumin and its derivatives. Phytother Res 2023;37:5109-58. [Crossref] [PubMed]
- Sui J, Boatz JC, Shi J, et al. Loss of ANT1 Increases Fibrosis and Epithelial Cell Senescence in Idiopathic Pulmonary Fibrosis. Am J Respir Cell Mol Biol 2023;69:556-69. [Crossref] [PubMed]
- Liu C, Rokavec M, Huang Z, et al. Curcumin activates a ROS/KEAP1/NRF2/miR-34a/b/c cascade to suppress colorectal cancer metastasis. Cell Death Differ 2023;30:1771-85. [Crossref] [PubMed]
- Dasi D, Nallabelli N, Devalaraju R, et al. Curcumin attenuates replicative senescence in human dental follicle cells and restores their osteogenic differentiation. J Oral Biosci 2023;65:371-8. [Crossref] [PubMed]
- Ma M, Wei N, Yang J, et al. Schisandrin B promotes senescence of activated hepatic stellate cell via NCOA4-mediated ferritinophagy. Pharm Biol 2023;61:621-9. [Crossref] [PubMed]
- Li HY, Wei TT, Zhuang M, et al. Iron derived from NCOA4-mediated ferritinophagy causes cellular senescence via the cGAS-STING pathway. Cell Death Discov 2023;9:419. [Crossref] [PubMed]
- Li X, Liu A, Xie C, et al. The transcription factor GATA6 accelerates vascular smooth muscle cell senescence-related arterial calcification by counteracting the role of anti-aging factor SIRT6 and impeding DNA damage repair. Kidney Int 2024;105:115-31. [Crossref] [PubMed]
- Liang J, Cui J, Cheng J, et al. SIRT6 Knockdown in Buffalo Fetal Fibroblasts Exacerbates Premature Senescence Caused by DNA and Telomere Damage. Cell Reprogram 2023;25:277-87. [Crossref] [PubMed]
- Aobulikasimu A, Liu T, Piao J, et al. SIRT6-PAI-1 axis is a promising therapeutic target in aging-related bone metabolic disruption. Sci Rep 2023;13:7991. [Crossref] [PubMed]


