
HNF1A, KRT81, and CYP3A5: three more straws on the back of pancreatic cancer?
In this post-genomic era of cancer research, our ability to molecularly define various subtypes of cancer raises tremendous promise for the concepts of patient targeted therapies and precision medicine. A particular bright spot in the utility of cancer subtyping is the example of breast cancer, which has been well defined at the molecular level: tumor subtypes are currently classified based upon the expression of estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2, and current targeted treatment strategies based on subtype assessment have increased clinical responses in the last decade (1). In stark contrast to the subtype success achieved in breast cancer analysis is the present state of pancreatic ductal adenocarcinoma (PDAC). PDAC is one of the most lethal malignancies and the fourth leading cause of cancer deaths, associated with a dismal 5-year survival rate of only ~5%. This poor outlook results from late-stage diagnosis of disease, as non-invasive early detection methods remain elusive in the clinic, and current treatment modalities extend patient lifespan by only several weeks to months (2).
Typically PDAC has been treated as a single aggressive form of cancer, because disease is detected almost universally at advanced stages. The identification of common druggable molecular targets, especially those representing specific subgroups of PDACs, will allow therapy to be optimized to increase patient survival. In recent years genomic expression profiling has begun to define separate PDAC subtypes (3,4), a promising discovery for our efforts in treating PDAC. The next major step will therefore be to determine how these subtypes differ in clinically relevant ways and use these findings to target PDACs in a more focused and accurate approach. This year Noll et al. reported studies identifying new diagnostic markers for PDAC subtypes and demonstrating the first putative subtype-specific drug resistance mechanism (5). These findings highlight molecular differences among PDAC subtypes and cumulatively represent a substantial step forward in understanding the pathogenesis of pancreatic cancer. This work is an important starting point for translating molecular studies to the clinic and the development of new therapeutics with the potential to achieve a significant improvement in patient outcome.
Historically subtyping PDAC has been difficult, in large part due to the complex tumor microenvironment. In 2011 Collisson et al. (3) reported the existence of three separate subtypes—“classical”, “quasimesenchymal”, (QM-PDA), and “exocrine-like”—defined by differences in gene expression signatures, KRAS addiction, and drug response. Due to the absence of exocrine-like cell lines in vitro, this study only evaluated features of the classical and QM-PDA subtypes; the nature of the exocrine-like subtype, and whether this population is a true subgroup, has therefore remained an open question. In another study, Moffitt et al. (4) employed an approach that subtracted mRNA transcripts associated with normal pancreatic tissue that defined only two PDAC subtypes, “classical” and “basal,” the latter subtype showing some overlap with the previously defined QM-PDA group (6). In the recent work of Noll et al. (5), patient tumor specimens were implanted into mice used to derive pancreatic adenocarcinoma (PACO) cell lines representative of each of Collisson’s subtypes, accompanied by histological and RNA profile verification, which confirms the existence of all three subtypes defined by Collisson et al. The establishment of cell lines of each subtype is an important accomplishment, as the absence of exocrine-like cells in vitro has in the past posed an unidentified challenge to accurate assessment of disease representative of all PDAC patients. Identifying molecular and functional differences among these cell types, such as metabolism (7), tumor microenvironment (8), drug resistance (3,5), and immune evasion and immunotherapy (9), in future studies will be highly valuable.
To extend molecular characteristics of each subtype into clinical utility, an important step is to determine diagnostic markers for patient stratification. Noll et al. (5) presented surrogate protein markers for the PDAC subtypes, showing that hepatocyte nuclear factor 1 alpha (HNF1A) and cytokeratin 81 (KRT81) are enriched almost exclusively to exocrine-like and QM-PDA subtypes, respectively (Table 1). Levels of HNF1A and KRT81 are inversely correlated in PDAC, and neither protein is frequently found in the classical subtype. Therefore using standard clinical immunohistochemical methods, the classical PDAC subtype can be determined by double negative (KRT81-HNF1A−, DN) status, KRT81+HNF1A− for QM-PDA, and KRT81-HNF1A+ for the exocrine-like group in patients. HNF1A and KRT81 are associated with survival and grade: the KRT81+ subtype correlates with low mean survival and poor differentiation, whereas HNF1A+ tumors show the greatest mean survival and cell differentiation, with DN tumors lying in between for both survival and differentiation (Table 1). Although subtype appears to predict patient prognosis, overall average survival is very low (<5 years), underscoring the need to develop improved approaches to targeting PDACs, likely by identifying and targeting cellular processes unique to each group.

Full table
While classical and QM-PDA tumors are sensitive to cell death by small molecule chemotherapeutics, the exocrine-like subtype is resistant to these agents. Noll et al. go on to show that this drug resistance mechanism stems from elevated expression of cytochrome P450 3A5 (CYP3A5), a heme-thiolate monooxygenase normally expressed in the liver that mediates xenobiotic metabolism to detoxify drugs in the body (10,11). Expression of CYP3A5 correlates positively with HNF1A and inversely with KRT81 in PDAC, and within the exocrine-like subtype CYP3A5 expression is both elevated and drug-inducible relative to normal tissues, QM-PDA, and classical PDAC cells. CYP3A5 mediates resistance against tyrosine kinase inhibitors including erlotinib (EGFR inhibitor) and dasatinib (SRC and ABL1 inhibitor), as well as the CYP3A5 substrate paclitaxel, a cytoskeletal drug that disrupts microtubule breakdown during cell division. Both the pan-CYP inhibitor ketoconazole and shRNA-mediated knockdown of CYP3A5 sensitize exocrine-like cells to treatment, while expression of CYP3A4 or CYP3A7 appear to be neither elevated nor important to PDAC drug resistance, suggesting that CYP3A5 is the lone culprit within the CYP family of enzymes. Furthermore, tumors cannot adapt to the loss of CYP3A5 following sustained treatment, which suggests that CYP3A5 makes an important candidate therapeutic target in exocrine-like tumors. Basal CYP3A5 expression in exocrine-like tumors is mediated by the transcription factor HNF4A, while drug-induced CYP3A5 is controlled by NR1I2, also known as PXR, and the presence of both CYP3A5-regulating proteins appears to have an additive effect in PDAC drug resistance (Figure 1). Therefore targeting these upstream regulators of CYP3A5 may also be therapeutically valuable.
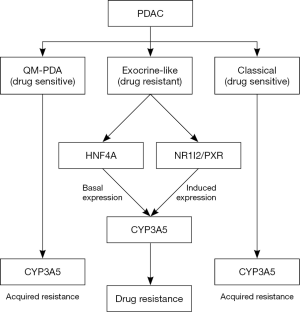
Though CYP3A5 appears to play a predominant role in facilitating cancer cell protection against chemotherapy only in exocrine-like tumors, CYP3A5-mediated drug resistance is not limited solely to exocrine-like PDACs. Indeed, when classical and QM-PDA tumors acquire resistance against therapy, CYP3A5 expression and activity are elevated as an adaptation to sustained treatment, and ectopic expression of CYP3A5 confers protection to non-exocrine-like cells. Tissue microarray analyses suggest that CYP3A5-mediated drug resistance is not only a feature of PDAC but other cancers as well, and these observations were confirmed using cancer cells of non-pancreatic origin. This suggests that CYP3A5 is a potential general target in cancer, and it will be important to determine its pro-tumor activities in other cancer types.
The pre-clinical characterization of all three PDAC subtypes in vivo is an important step forward in pancreatic cancer treatment. The establishment of PACO cell lines will be useful in determining further differences among subtypes, and the application of the finding that CYP3A5 is a protector of exocrine-like cells will be valuable clinically. Given the known physiological roles of the CYPs, it is not surprising that CYP3A5 protects cancer cells from treatment, but its now confirmed role as a mediator of exocrine-like tumor drug resistance and its regulatory mechanisms in PDAC highlight its value as a potential target. We now have a mechanistic basis for differences in drug resistance among the PDAC subtypes, and in light of these in vivo studies, development of CYP3A5 drugs is warranted. CYP3A5 is dispensable to normal tissues (12,13), but central to chemoresistance in PDAC. Despite high levels of CYP3A5 in tumors relative to healthy tissues, inhibiting the drug clearance activities of CYPs in noncancerous tissues presents the possible clinical pitfall of elevating the systemic toxicity of other chemotherapies. Therefore the significance of achieving high chemical specificity for CYP3A5, but not other CYPs, is worth noting. An alternative approach to CYP3A5 inhibition might instead be to seek out anti-tumor agents that are not metabolized by CYPs and therefore circumvent CYP3A5-mediated resistance. Another possibility is the development of small molecules that serve as substrates for CYP3A5 (such as a modified form of paclitaxel), with the oxidation product functioning as an active form of the drug, thus turning the tumor’s resistance mechanism against itself.
Immunohistological detection of HNF1A and KRT81 may prove important to revealing which patients are likely to benefit from such therapy. Evaluating the subtype and context specificity of CYP3A5 in other cancer types will also be an important goal for therapeutic development, as well as determining if the correlation of HNF1A and KRT81 with CYP3A5 extends to other cancers. Altogether the work of Noll et al. demonstrated a substantial body of clinically useful findings, yet the “smoking gun” for PDAC treatment remains elusive. We can only hope that these targets represent three additional straws on the back of PDAC, and may one day bring us that much closer to improved therapeutic intervention.
Acknowledgments
Funding: This work was supported by grant 1R15CA206067-01 (to J.C.W.) from the National Institutes of Health.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Xiaotian Sun (Department of Internal Medicine, Clinic of August First Film Studio, Beijing, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.08.12). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kelly CM, Buzdar AU. Using multiple targeted therapies in oncology: considerations for use, and progress to date in breast cancer. Drugs 2013;73:505-15. [Crossref] [PubMed]
- Cid-Arregui A, Juarez V. Perspectives in the treatment of pancreatic adenocarcinoma. World J Gastroenterol 2015;21:9297-316. [Crossref] [PubMed]
- Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011;17:500-3. [Crossref] [PubMed]
- Moffitt RA, Marayati R, Flate EL, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47:1168-78. [Crossref] [PubMed]
- Noll EM, Eisen C, Stenzinger A, et al. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat Med 2016;22:278-87. [Crossref] [PubMed]
- Biankin AV, Maitra A. Subtyping Pancreatic Cancer. Cancer Cell 2015;28:411-3. [Crossref] [PubMed]
- Daemen A, Peterson D, Sahu N, et al. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc Natl Acad Sci U S A 2015;112:E4410-7. [Crossref] [PubMed]
- Carr RM, Fernandez-Zapico ME. Pancreatic cancer microenvironment, to target or not to target? EMBO Mol Med 2016;8:80-2. [Crossref] [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Wrighton SA, Brian WR, Sari MA, et al. Studies on the expression and metabolic capabilities of human liver cytochrome P450IIIA5 (HLp3). Mol Pharmacol 1990;38:207-13. [PubMed]
- MacPhee IA. Pharmacogenetic biomarkers: cytochrome P450 3A5. Clin Chim Acta 2012;413:1312-7. [Crossref] [PubMed]
- Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 2001;27:383-91. [Crossref] [PubMed]
- Walsky RL, Obach RS, Hyland R, et al. Selective mechanism-based inactivation of CYP3A4 by CYP3cide (PF-04981517) and its utility as an in vitro tool for delineating the relative roles of CYP3A4 versus CYP3A5 in the metabolism of drugs. Drug Metab Dispos 2012;40:1686-97. [Crossref] [PubMed]


