
Immuno-molecular characterization of colorectal cancer tumors and its clinical implications
We thank Oh, Lee and Lockhart for their well-informed comment. In addition to an accurate summary of our work, it provides a relevant discussion in light of the recent immunotherapy trials in colorectal cancer (CRC). The authors notably cite the work of Le and colleagues who have shown the association between response to Pembrolizumab and the microsatellite-instable (MSI) status of CRC tumors (1). Indeed, the immune contexture of MSI CRC tumors supports PD-1 blockade as a potential immunotherapeutic modality (2). This susceptibility is likely due to the high mutational burden of these tumors, which in turns leads to the expression of many potentially immunogenic peptides. Consistent observations have been reported in lung adenocarcinoma, where mutational burden has been shown to be associated with response to anti-PD1 treatment (3). Our work suggests that consensus molecular subgroup (CMS) 1, or CMS1 tumors, would likely respond to checkpoint blockade treatments, as we observed a high abundance of cytotoxic lymphocytes in these tumors, as well as the expression of the PD-1 ligands’ genes (CD274 and PDCD1LG2). Strikingly, although MSI has been known to be associated with high immune infiltration, our data showed that the CMS classification was more predictive of the infiltration by cytotoxic lymphocytes than MSI status (Figure 1). We can thus hypothesize that the benefit of anti-PD1 treatments may extend to CMS1-microsatellite stable (MSS) tumors, while MSI non CMS1 tumors may not respond. Mutational burden is another possible marker of response that has not been yet completely addressed in CRC. It is highly correlated with MSI (which represent about 75% of hypermutated CRC tumors) (4), but other molecular phenotypes may lead to an hypermutated tumor genome, notably the POLE mutations (4). Mutational load is also correlated with CMS subtypes, with CMS1 showing the highest mutational burden (5). Carefully-designed clinical trials including CMS annotations, MSI status and mutational burden will thus be required to really pinpoint the best marker of response to these therapies.
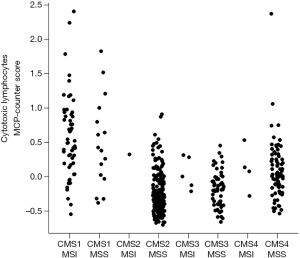
Our data have shown that CMS4 tumors had higher infiltration by CD8 T cells than CMS2 and CMS3 subgroups (6). It is thus tempting to postulate that these patients may respond to checkpoint-blockade therapies. Given the abysmal response rate reported by Le et al. in MSS CRC for Pembrolizumab (1), it is however unlikely that CMS4 tumors are good responders to PD-1-blockade alone. However, our work revealed that, aside from cytotoxic lymphocytes, CMS4 tumors are highly inflammatory and angiogenic. These additional features have been shown to promote immunosuppression through mechanisms that may be distinct from checkpoint-related immunosuppression, notably through the promotion regulatory phenotypes for T cells and myeloid cells (7). It may thus be necessary to use combination of immunotherapies in CMS4 tumors, targeting the redundant immunosuppressive mechanisms involved (7).
Our data reveal that CRC immune-oncology may be more complex than previously-thought. We have shown a decade ago that CD8 infiltration is associated with favourable outcome in CRC (8), and that it was associated with expression of CXCL9, CXCL10 and IFNG (9). These markers almost perfectly correlate with the CMS1 subtype of favourable outcome. However, our work showed that the poor-prognosis CMS4 subgroup has an intermediate infiltration with CD8 T cells, challenging the idea of a monotonic relationship between CD8 T cells abundance and overall survival, and suggesting that multiple cellular and molecular markers need to be measured simultaneously to accurately describe the microenvironment of a given tumour subgroup. This problem is even more apparent in exploratory research, where a large number of potential markers have to be measured. In that regard, we and others (6,10) have shown that leveraging high-dimensional data, such as tumor transcriptomes, provide a rapid estimate of the abundances of multiple cellular populations. The large amount of publically-available transcriptomic data can thus help formulate hypothesis on which restricted set of cellular markers may be associated with disease subtypes or response to treatments, and study the association between immunological, genomic and clinical features of tumors on the same datasets.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Zhen-Yu Lin, MD (Cancer center, Union hospital, Huazhong University of Science and Technology, Wuhan, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.07.50). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43-51. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350-6. [Crossref] [PubMed]
- Becht E, de Reyniès A, Giraldo NA, et al. Immune and Stromal Classification of Colorectal Cancer Is Associated with Molecular Subtypes and Relevant for Precision Immunotherapy. Clin Cancer Res 2016;22:4057-4066. [Crossref] [PubMed]
- Becht E, Giraldo NA, Dieu-Nosjean MC, et al. Cancer immune contexture and immunotherapy. Curr Opin Immunol 2016;39:7-13. [Crossref] [PubMed]
- Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960-4. [Crossref] [PubMed]
- Mlecnik B, Tosolini M, Charoentong P, et al. Biomolecular network reconstruction identifies T-cell homing factors associated with survival in colorectal cancer. Gastroenterology 2010;138:1429-40. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]


