
Unveiling the role of RARs in stomach adenocarcinoma: clinical implications and prognostic biomarkers
Highlight box
Key findings
• Retinoic acid receptors (RARs) family is promising biomarkers for stomach adenocarcinoma (STAD).
What is known and what is new?
• Some studies have shown that RARs participated in the pathogenesis of various tumors including acute promyelocytic leukemia and cholangiocarcinoma.
• RARs are promising biomarkers for STAD.
What is the implication, and what should change now?
• RARs might potentially participate in the carcinogenesis of STAD, which provides a direction for future research on the mechanism of RARs in STAD.
Introduction
Stomach adenocarcinoma (STAD) is the most common histologic form of gastric cancer (GC), which is a highly aggressive and extremely heterogeneous tumor originating from the epithelium of the stomach (1). An estimated of 1 million people were newly diagnosed in 2008, and over 783,000 patients died of this disease worldwide (2). Recently, the incidence and fatality rate of GC ranks fifth and fourth respectively, which is the main global burden of cancer estimated by disability-adjusted life-years (DALYs) (3). In China, GC is also the most burdensome gastrointestinal disease as the cancer with the second highest incidence rate (4). Research reported that the occurrence and development of GC are on account of complicated influencing factors, such as Helicobacter pylori infection, diet, smoking, drinking and genetic factors (5). Due to the atypical symptoms of early GC, most patients are diagnosed at an advanced stage. Unfortunately, despite multidisciplinary approaches including surgical resection, chemotherapy and targeted therapy are employed to treat GC, the prognosis of STAD patients is unacceptable with less than 50% rate of overall survival (OS) (6). As a result, it is necessary to identify effectively predictive biomarkers and molecular mechanism of STAD.
As the member of the steroid hormone receptor superfamily, retinoic acid receptors (RARs) mediate the biological processes (BPs) of retinoids including inhibition of proliferation, induction of differentiation, and regulation of apoptosis (7). RARs consist of three subtypes: α, β and γ, which are encoded by RARA, RARB and RARG, respectively (8). It is noteworthy that RARs are widely distributed in various tissues and cell types. For example, chromosomal translocations involving the RARα locus probably indicate the malignant initiating events in acute promyelocytic leukemia (APL) (9). Oncogenic activity of RARγ is exhibited through activation of the Akt/NF-κB and Wnt/β-catenin pathways in cholangiocarcinoma (CCA) (10). On the contrary, RARβ is believed to play a role as a tumor suppressor gene (11). Our previous research has demonstrated that RARα is frequently elevated in GC and exerts oncogenic properties (12). However, the clinical significance and potential function of the RARs family in STAD based on large cohort data are poorly ascertained.
In the current study, the expression profile and clinical value of the RARs family in STAD were comprehensive evaluated using a series of online databases. We hope that this study will give insight into the potential molecular mechanism of RARs in the pathogenesis and clinical prognosis of STAD. We present this article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2154/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene expression profiling interactive analysis (GEPIA)
RARs expressions and correlations in STAD were analyzed using the GEPIA database (13) (http://gepia.cancer-pku.cn/index.html). Furthermore, the relationships between RARs and clinical stage as well as the prognosis of STAD patients were obtained from the GEPIA database. In data analysis, the median RARs expressions were used as a cutoff value to classify groups.
The University of Alabama at Birmingham cancer data (UALCAN)
The expression and methylation of RARs in STAD were examined using the UALCAN database (14) (http://ualcan.path.uab.edu). In addition, we used UALCAN database to evaluate the relationship between RARs expression or methylation status and clinicopathological parameters of STAD patients. All results were analyzed on line and the difference at a P value <0.05 was considered significant.
Human Protein Atlas (HPA)
HPA (15) (https://www.proteinatlas.org) was performed to collect representative immunohistochemistry (IHC) images of RARs in patients with STAD and normal tissues. Images available at https://www.proteinatlas.org/ENSG00000077092-RARB/pathology/stomach+cancer#img and https://www.proteinatlas.org/ENSG00000172819-RARG/pathology/stomach+cancer#img (accessed on 22 May 2023).
Survival analysis
The predictive significance of RARs messenger ribonucleic acid (mRNA) in OS and relapse free survival (RFS) of STAD patients was examined using the Kaplan-Meier plotter (16) (http://kmplot.com). In addition, multivariate cox regression analysis was used to construct a prognostic model, which was analyzed from the Home for Researchers platform (https://www.home-for-researchers.com/). P values and hazard ratio (HR) with 95% confidence interval (CI) were generated by log-rank tests and univariate cox proportional hazards regression. All the analysis methods and R packages were implemented by R (foundation for statistical computing 2020) version 4.0.3. P value <0.05 was considered statistically significant.
Tumor immune estimation resource (TIMER)
TIMER (http://timer.cistrome.org/) is a comprehensive resource for systematical analysis of immune infiltrates across diverse cancer types (17). We used TIMER database to assess the expression and immune cell infiltration of RARs in patients with STAD. The median RARs expressions were severed as cutoff values, respectively. And P value cutoff was set at 0.05.
Gene set cancer analysis (GSCA)
In this study, we used GSCA database (18) (http://bioinfo.life.hust.edu.cn/GSCA) to evaluate the relationship between immune cell infiltration of RARs and STAD. The color and size of the bubbles in the visualization were used to represent the degree of correlation and significance, respectively. Specifically, darker shades of red or blue denoted positive or negative correlation, while the size of the bubble indicated the level of significance. Additionally, the presence of a black contour coil highlighted a false discovery rate (FDR) value of less than 0.05.
cBioPortal
cBioPortal (https://www.cbioportal.org/) is a database that retrieves, downloads, analyzes, and visualizes cancer genomics data across a wide range of genomic data types (19). We used cBioPortal tool to analyze the data of genetic alterations of RARs in STAD. Moreover, the OS and disease-free survival (DFS) of STAD patients were compared with or without RARs genetic alteration.
MethSurv
MethSurv database (https://biit.cs.ut.ee/methsurv/) is a web tool to perform multivariable survival analysis using DNA methylation data (20). In this study, we used MethSurv database to analyze the methylation sites of RARs in STAD and the effect of RARs methylation sites on the prognosis of patients with STAD.
GeneMANIA
The GeneMANIA database (http://genemania.org/) is a user-friendly and interactive online platform designed for constructing protein-protein interaction (PPI) networks, generating hypotheses related to the prediction of gene functions, and identifying genes with shared activities (21). In this study, we used the GeneMANIA database to assess the interacting genes and potential functions of RARs in STAD.
LinkedOmics
LinkedOmics (http://www.linkedomics.org) provides a unique platform for biologists and clinicians to access, analyze and compare cancer multi-omics data within and across tumor types (22). In this study, we used LinkedOmics platform to locate the co-expressed genes of RARs. The findings were evaluated utilizing the Pearson correlation coefficient and visually represented through heat maps and volcano plots.
Metascape
Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and visualization were obtained from the Metascape network (23) (http://metascape.org). The screening conditions for Min overlap and Min Enrichment were set at 3 and 1.5, respectively. A P value of less than 0.01 was deemed to be statistically significant.
Search tool for the retrieval of interacting genes (STRING)
The PPI networks of RARs in STAD were analyzed using STRING database (24) (https://string-db.org/), and then constructed by Cytoscape software (version 3.9.1) in order to identify the key hub genes. The cluster analysis was conducted using the Cytoscape with Molecular Complex Detection (MCODE) plug-in tools with default parameters, including a K-core of 2, a degree cutoff of 2, a maximum depth of 100, and a node score cutoff of 0.2.
Tumor immune single-cell hub (TISCH)
TISCH (http://tisch.comp-genomics.org/home/) is a single-cell RNA sequencing (scRNA-seq) database focusing on tumor microenvironment (TME). TISCH provides detailed cell-type annotation at the single-cell level, enabling the exploration of TME across different cancer types (25). In this study, single-cell functional analyses of RARs in STAD were obtained from TISCH database.
Cancer cell line encyclopedia (CCLE)
The differential expressions of RARs in STAD cell lines were obtained from the CCLE dataset (26) (https://portals.broadinstitute.org/ccle). The analysis was constructed by the R v4.0.3 software package ggplot2 (v3.3.3), which was analyzed from the Home for Researchers platform (https://www.home-for-researchers.com/).
Statistical analysis
Differences between the two groups were calculated using Student’s t-test. Kaplan-Meier method and log-rank tests were used to construct survival curves and to assess differences between groups, respectively. Correlations were determined using Pearson or Spearman correlation tests, as appropriate. P values less than 0.05 was considered statistically significant.
Results
Transcriptional and protein levels of RARs in STAD
We first explored the expression of RARs in different cancers and normal tissues using TIMER database. As shown in Figure S1A-S1C, significant differences of RARs were discovered in numerous cancers. The GEPIA and UALCAN databases were used to compare the mRNA transcription levels of RARs between STAD tissues and normal gastric tissues (Figure 1A-1F). The results indicated that RARA were more highly expressed in STAD tissues than in normal tissues (P=0.03). In contrast, significant low expression was detected in RARB (P=0.02). We further used HPA database to analyze the protein levels of RARs in STAD. As shown in Figure 1G,1H, RARβ was more lowly expressed in STAD tissues compared with normal tissues. And there were no obvious differences of RARγ between STAD and normal tissues. Hence, these results demonstrated that RARs were significantly differentially expressed in STAD.
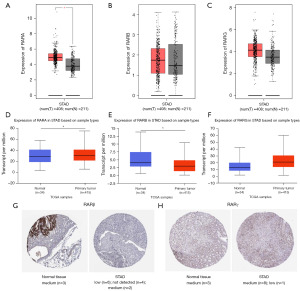
Relationship between the mRNA expressions of RARs and the clinical characteristics of patients with STAD
This study then analyzed the relationship between mRNA expression levels of RARs and clinicopathological parameters of STAD patients using UALCAN database. Compared to normal population, the expression of RARA was significantly increased in STAD among stage 2, Caucasian, male, 61–80 years, intestinal adenocarcinoma tubular, and no regional lymph node metastasis (Figure S2A-S2I). RARB expression was markedly decreased in STAD patients among stage 2, African-American, Asian, male, female, 61–80 years, 81–100 years, grade 1, grade 2, tumors with H. pylori infection, tumors without H. pylori infection, tumors not available, intestinal adenocarcinoma [not otherwise specified (NOS)], intestinal adenocarcinoma tubular, intestinal adenocarcinoma papillary, no regional lymph node metastasis, metastases in 1 to 3 axillary lymph nodes, metastases in 4 to 9 axillary lymph nodes, TP53-mutant, and TP53-nonmutant (Figure S3A-S3I). RARG was significantly increased in STAD among African-American and adenocarcinoma signet ring (Figure S4A-S4I).
In addition, we also used Kaplan-Meier plotter database to verify the above relationship. As shown in Table 1, elevated RARA expression was markedly related with the shorter OS of STAD among female (HR =2.08, 95% CI: 1.13–3.83, P=0.02), Asian (HR =6.31, 95% CI: 1.82–21.86, P=0.001), stage 1 (HR =3.62, 95% CI: 0.96–13.71, P=0.04), stage 3 (HR =1.73, 95% CI: 1.03–2.91, P=0.04), grade 3 (HR =1.71, 95% CI: 1.11–2.64, P=0.01), and high mutation burden (HR =2.07, 95% CI: 1.25–3.42, P=0.004). Low RARB expression was significantly associated with the longer OS of STAD among male (HR =1.75, 95% CI: 1.18–2.59, P=0.005), White (HR =1.67, 95% CI: 1.12–2.46, P=0.01), Asian (HR =2.74, 95% CI: 1.07–7.01, P=0.03), stage 2 (HR =2.37, 95% CI: 1.18–4.76, P=0.01), grade 3 (HR =1.81, 95% CI: 1.18–2.77, P=0.006), and low mutation burden (HR =2.04, 95% CI: 1.26–3.29, P=0.003). High RARG expression was obviously correlated with the longer OS of STAD among male (HR =0.58, 95% CI: 0.39–0.88, P=0.009), White (HR =0.52, 95% CI: 0.33–0.81, P=0.003), stage 3 (HR = 0.58, 95% CI: 0.35–0.96, P=0.03), grade 2 (HR =0.54, 95% CI: 0.30–0.98, P=0.04), and high mutation burden (HR =0.50, 95% CI: 0.30–0.84, P=0.007). Additionally high RARG expression was significantly associated with the worse OS of STAD among Asian (HR =3.25, 95% CI: 1.25–8.43, P=0.01). However, the results of GEPIA database indicated that there were no obvious associations between RARs and clinical stage of STAD patients (Figure S5A-S5C).
Table 1
| Subtypes | RARA | RARB | RARG | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High (n) | Low (n) | Log-rank P | HR (95% CI) | High (n) | Low (n) | Log-rank P | HR (95% CI) | High (n) | Low (n) | Log-rank P | HR (95% CI) | |||
| Gender | ||||||||||||||
| Female | 34 | 99 | 0.02* | 2.08 (1.13–3.83) | 79 | 54 | 0.15 | 1.59 (0.84–2.99) | 57 | 76 | 0.12 | 0.61 (0.33–1.13) | ||
| Male | 125 | 113 | 0.051 | 1.48 (1.00–2.21) | 88 | 150 | 0.005* | 1.75 (1.18–2.59) | 167 | 71 | 0.009* | 0.58 (0.39–0.88) | ||
| Race | ||||||||||||||
| White | 64 | 173 | 0.20 | 1.33 (0.86–2.05) | 113 | 124 | 0.01* | 1.67 (1.12–2.46) | 95 | 142 | 0.003* | 0.52 (0.33–0.81) | ||
| Asian | 36 | 37 | 0.001* | 6.31 (1.82–21.86) | 23 | 50 | 0.03* | 2.74 (1.07–7.01) | 19 | 54 | 0.01* | 3.25 (1.25–8.43) | ||
| Black/African America | – | – | – | – | – | – | – | – | – | – | – | – | ||
| Stage | ||||||||||||||
| 1 | 22 | 28 | 0.04* | 3.62 (0.96–13.71) | 32 | 18 | 0.17 | 0.43 (0.12–1.50) | 23 | 27 | 0.23 | 0.48 (0.14–1.63) | ||
| 2 | 29 | 82 | 0.08 | 1.92 (0.92–3.98) | 42 | 69 | 0.01* | 2.37 (1.18–4.76) | 54 | 57 | 0.20 | 0.64 (0.32–1.28) | ||
| 3 | 42 | 107 | 0.04* | 1.73 (1.03–2.91) | 71 | 78 | 0.13 | 1.45 (0.90–2.33) | 59 | 90 | 0.03* | 0.58 (0.35–0.96) | ||
| 4 | 24 | 14 | 0.08 | 2.26 (0.88–5.81) | 18 | 20 | 0.11 | 1.96 (0.85–4.52) | 12 | 26 | 0.07 | 0.41 (0.15–1.11) | ||
| Grade | ||||||||||||||
| 1 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 2 | 91 | 43 | 0.26 | 1.46 (0.76–2.81) | 66 | 68 | 0.27 | 0.72 (0.41–1.28) | 55 | 79 | 0.04* | 0.54 (0.30–0.98) | ||
| 3 | 61 | 157 | 0.01* | 1.71 (1.11–2.64) | 123 | 95 | 0.006* | 1.81 (1.18–2.77) | 92 | 126 | 0.05 | 0.65 (0.42–1.01) | ||
| 4 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| Mutation burden | ||||||||||||||
| High | 47 | 139 | 0.004* | 2.07 (1.25–3.42) | 61 | 125 | 0.12 | 1.48 (0.90–2.42) | 80 | 106 | 0.007* | 0.50 (0.30–0.84) | ||
| Low | 120 | 62 | 0.16 | 1.44 (0.86–2.40) | 104 | 78 | 0.003* | 2.04 (1.26–3.29) | 72 | 110 | 0.11 | 0.68 (0.43–1.09) | ||
| Neoantigen load | ||||||||||||||
| High | 20 | 54 | 0.09 | 1.85 (0.91–3.76) | 30 | 44 | 0.15 | 1.63 (0.83–3.20) | 30 | 44 | 0.07 | 0.51 (0.24–1.07) | ||
| Low | – | – | – | – | – | – | – | – | – | – | – | – | ||
*, a P value of less than 0.05 was considered statistically significant; –, not available. CI, confidence interval; HR, hazard rate; RAR, retinoic acid receptor.
Association between RARs mRNA expressions and STAD prognosis
The correlation between RARs mRNA expressions and STAD prognosis was analyzed by using GEPIA and Kaplan-Meier plotter databases, with the follow-up threshold of 120 months (10 years). As shown in Figure 2A-2F, the results of GEPIA database revealed that higher expressions of RARA and RARB correlated with shorter OS and DFS for STAD patients. For RARG, the survival curve analysis showed that lower expression of RARG was connected to shorter OS in STAD patients. And there was no obvious association between RARG expression and DFS of STAD patients. Similarly, the results of Kaplan-Meier plotter databases (Figure 2G-2L) revealed that high expression of RARA was related with shorter OS (HR =1.51, 95% CI: 1.09–2.09, P=0.01) and relapse free survival (RFS) (HR = 2.16, 95% CI: 1.02–4.59, P=0.04) for STAD patients. And high expression of RARB was correlated with worse OS (HR =1.55, 95% CI: 1.12–2.15, P=0.007) and RFS (HR =3.83, 95% CI: 2–7.34, P<0.001) for STAD patients. Moreover, low expression of RARG was associated with worse OS (HR =0.63, 95% CI: 0.44–0.88, P=0.007), and not related with RFS (HR =1.66, 95% CI: 0.86–3.19, P=0.13) of STAD patients.
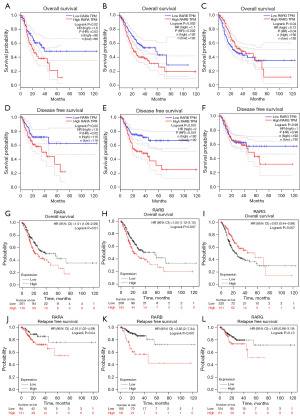
Prognostic signatures of RARs in STAD patients
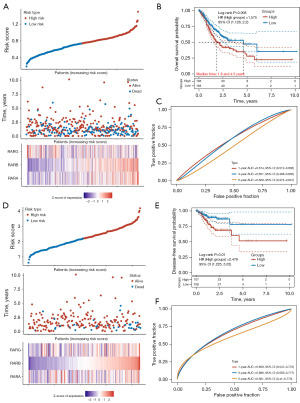
To further identify the prognostic signatures of RARs in STAD patients, multivariate Cox regression analysis was performed. The prognostic model was constructed to predict OS based on the Cox coefficients as follows: risk score = 0.1394 × RARA + (0.2122) × RARB + (−0.1036) × RARG. According to the risk scores, the STAD patients were divided into a high-risk group (n=185) and a low-risk group (n=185) (Figure 3A). The Kaplan-Meier survival curves indicated that the high-risk group had shorter OS compared with the low-risk group (Figure 3B). In addition, receiver operating characteristic (ROC) curves were constructed to evaluate diagnostic value of prognostic model. As shown in Figure 3C, the area under the ROC curve (AUC) of The Cancer Genome Atlas (TCGA) cohort was 0.574, 0.551, and 0.509 at 1, 3, and 5 years, respectively. Similarly, we also constructed prognostic model to predict DFS of STAD patients based on the risk score = (0.0695) × RARA + (0.7558) × RARB + (0.0349) × RARG. The STAD patients were divided into a high-risk group (n=107) and a low-risk group (n=108) (Figure 3D). The Kaplan-Meier survival curves indicated that the high-risk group had worse DFS than the low-risk group (Figure 3E). The AUC of DFS in TCGA cohort was 0.659, 0.666, and 0.591 at 1, 3, and 5 years, respectively (Figure 3F).
Immune cells infiltration of RARs and their prognostic values of STAD patients
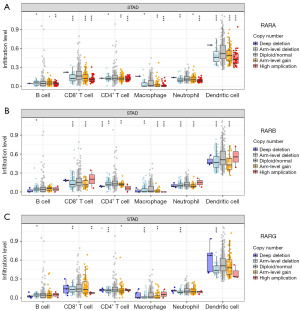
The correlations of RARs in STAD were analyzed using the GEPIA database. As shown in Figure 4A, the results were as follows: RARA-RARB (R=−0.017), RARA-RARG (R=−0.0063), and RARB-RARG (R=0.22). Then, we analyzed the relationship between RARs and immune cell infiltration of STAD patients using the GSCA. As shown in Figure 4B, RARA was positively associated with CD4_T, NK, NKT, Tfh, Th2, CD8_naive, and Gamma_delta. In contrast, there were consistent negative correlations between the expression of RARA and Central_memory, cytotoxic, macrophage, DC, Effector_memory, exhausted, monocyte, Th1, nTreg, as well as neutrophil. For RARB, persistent positive correlations were observed in CD4_T, NK, NKT, Tfh, Th2, B cell, CD4_naive, MAIT, Tr1, and Central_memory. Moreover, constant negative correlations between RARB expression and DC, Effector_memory, exhausted, monocyte, Th1, nTreg, as well as neutrophil were observed in STAD. For RARG, a persistent positive correlation was observed in Th17, and a constant negative correlation was observed in neutrophil. The data of six immune infiltrating cells in STAD were also retrieved from the TIMER database. As shown in Figure 4C, RARA expression level was statistically correlated with CD4+ T cell, macrophage and neutrophil in STAD. The expression of RARB in STAD was markedly related with six immune infiltrating cells including B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil and dendritic cell (Figure 4D). For RARG, there were no obvious associations between RARs and the above six immune infiltrating cells in STAD (Figure 4E). The detail cox proportional hazard model of RARs and six tumor-infiltrating immune cells in STAD performed using TIMER is shown in Table S1.
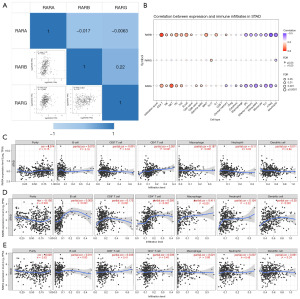
We further detected the effect of somatic copy-number alterations (SCNA) for RARs on the infiltration levels of various immune cells. As showcased in Figure 5A-5C, the variations of RARA, RARB and RARG were significantly correlated to the infiltrating levels of six immune related cells including B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil and dendritic cell. In addition, we also investigated the effect of immune cell infiltration on the association between RARs mRNA expressions and STAD prognosis using Kaplan-Meier plotter database. Our results revealed that the infiltrating levels of decreased CD4+ memory T cells (HR =0.61, 95% CI: 0.35–1.07, P=0.08), enriched CD4+ memory T cells (HR =1.74, 95% CI: 1.05–2.88, P=0.03), decreased macrophages (HR =2.28, 95% CI: 1.39–3.72, P<0.001) and enriched macrophages (HR =1.48, 95% CI: 0.85–2.57, P=0.16) markedly effected the relationship between RARA and STAD prognosis (Figure S6A-S6D). The effect of RARB on STAD prognosis also depended on the infiltrating levels of decreased B cells (HR =2.09, 95% CI: 1.15–3.79, P=0.01), enriched B cells (HR =1.79, 95% CI: 1.16–2.76, P=0.008), decreased CD8+ T cells (HR =1.75, 95% CI: 1.05–2.9, P=0.03), enriched CD8+ T cells (HR =1.69, 95% CI: 1.09–2.63, P=0.02), decreased CD4+ memory T cells (HR =1.9, 95% CI: 1.05–3.43, P=0.03), enriched CD4+ memory T cells (HR =1.49, 95% CI: 0.96–2.32, P=0.08, decreased macrophages (HR =1.82, 95% CI: 1.07–3.1, P=0.02) and enriched macrophages (HR =1.69, 95% CI: 1.07–2.66, P=0.02) (Figure S6E-S6L).
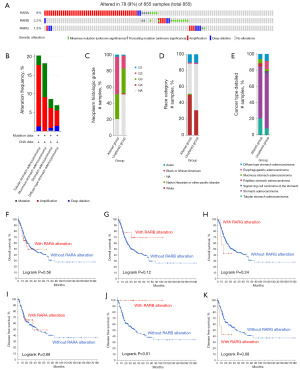
Genetic alterations of RARs and their prognostic values of STAD patients
The genetic alterations of RARs family members in STAD patients were explored using the cBioPortal database. As showcased in Figure 6A, RARs were altered in 78 samples of 855 patients with STAD, and the overall mutation frequency was 9%. RARA, RARB and RARG were altered at about 6%, 2.2% and 1.3%, respectively. Moreover, the most common gene alteration patterns of RARs in STAD were mutation, amplification, and deep deletion (Figure 6B). We further investigated the relationship between genetic alterations in RARs and the clinical characteristics of STAD. As shown in Figure 6C-6E, RARs mutations were more frequently detected in patients with neoplasm histological grade 2, White and STAD. Next, we analyzed the correlation between genetic alterations of RARs and outcomes in STAD patients. As shown in Figure 6F-6K, our results revealed that genetic alteration of RARB was significantly associated with the longer DFS of STAD patients. In contrast, there were no obvious differences in the OS and DFS of STAD patients with genetic alterations of RARA and RARG.
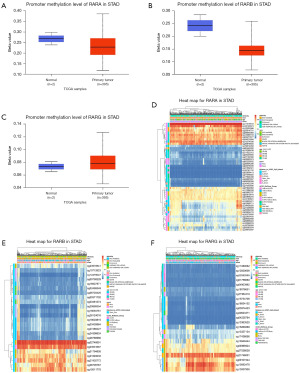
RARs methylation sites and their prognostic significances in STAD patients
To clarify the underlying mechanisms of RARs abnormal expression in STAD, we explored the correlation between RARs and methylation status using UALCAN database. Due to the number of normal groups, there were no statistically significant differences between methylation and expression of RARs (Figure 7A-7C). However, the promoter methylation level of RARA in STAD was increased at the stage 2 and stage 3 comparing with stage 1 (Figure S7A). The methylation level of RARA in Asian, grade 3, N3, and TP53 nonmutant groups was higher than in African-American, grade 2, N2, and TP53 mutant groups, respectively (Figure S7B-S7G). As shown in Figure S8A-S8G, the methylation level of RARB in 21–40 years, grade 3 and TP53 nonmutant groups was higher than in 61–80 years, grade 2 and TP53 mutant groups, respectively. And the methylation level of RARG in Asian, female, grade 3 and TP53 nonmutant groups was higher than in African-American, male, grade 2 and TP53 mutant groups, respectively (Figure S9A-S9G).
Next, we used MethSurv database to perform multivariable survival analysis to demonstrate the relationship between RARs methylation sites and prognosis in STAD patients. As shown in Figure 7D-7F and Table 2, nine RARA CpG sites (Body-Open_Sea-cg02477677; TSS200-Open_Sea-cg09858022; 5'UTR-N_Shore-cg00442282; TSS200-Open_Sea-cg03053374; Body-Island-cg11522012; Body-Open_Sea-cg11316510; Body-S_Shore-cg10062919; TSS200-Open_Sea-cg22935450; Body-Island-cg08446900) were significantly associated with the prognosis of STAD. And nine RARB CpG sites were markedly related to the prognosis of STAD patients, including TSS1500-Open_Sea-cg00371702, 5'UTR;1stExon-Open_Sea-cg19003815, TSS1500-Open_Sea-cg18094781, 5'UTR;Body-Open_Sea-cg01794805, 5'UTR;1stExon-Open_Sea-cg03428864, 5'UTR;1stExon-Open_Sea-cg27486427, TSS200-Open_Sea-cg06720425, TSS1500-Open_Sea-cg26124016, and TSS1500-Open_Sea-cg20899354. Moreover, five RARG CpG sites (Body-N_Shelf-cg21746001; 1stExon;5'UTR;Body-Island-cg15260268; Body-Open_Sea-cg27036638; TSS1500-S_Shore-cg12820608; Body-N_Shore-cg13937905) were markedly corelated to the prognosis of STAD patients.
Table 2
| CpG | HR | 95% CI | P value |
|---|---|---|---|
| RARA-Body-Open_Sea-cg02477677 | 0.522 | 0.341–0.798 | 0.001 |
| RARA-TSS200-Open_Sea-cg09858022 | 0.610 | 0.411–0.907 | 0.01 |
| RARA-5'UTR- N_Shore-cg00442282 | 1.515 | 1.072–2.141 | 0.02 |
| RARA-TSS200-Open_Sea-cg03053374 | 0.677 | 0.483–0.949 | 0.02 |
| RARA-Body-Island-cg11522012 | 1.416 | 1.025–1.957 | 0.03 |
| RARA-Body-Open_Sea-cg11316510 | 0.707 | 0.509–0.982 | 0.04 |
| RARA-Body-S_Shore-cg10062919 | 1.542 | 1.022–2.325 | 0.03 |
| RARA-TSS200-Open_Sea-cg22935450 | 0.717 | 0.520–0.989 | 0.04 |
| RARA-Body-Island-cg08446900 | 1.492 | 1.000–2.226 | 0.04 |
| RARB-TSS1500-Open_Sea-cg00371702 | 0.607 | 0.430–0.858 | 0.006 |
| RARB-5'UTR;1stExon-Open_Sea-cg19003815 | 0.585 | 0.399–0.858 | 0.004 |
| RARB-TSS1500-Open_Sea-cg18094781 | 0.584 | 0.384–0.886 | 0.008 |
| RARB-5'UTR;Body-Open_Sea-cg01794805 | 1.723 | 1.127–2.633 | 0.008 |
| RARB-5'UTR;1stExon-Open_Sea-cg03428864 | 0.580 | 0.377–0.893 | 0.009 |
| RARB-5'UTR;1stExon-Open_Sea-cg27486427 | 0.594 | 0.387–0.914 | 0.01 |
| RARB-TSS200-Open_Sea-cg06720425 | 0.670 | 0.472–0.949 | 0.03 |
| RARB-TSS1500-Open_Sea-cg26124016 | 0.620 | 0.403–0.953 | 0.02 |
| RARB-TSS1500-Open_Sea-cg20899354 | 0.634 | 0.412–0.975 | 0.03 |
| RARG-Body-N_Shelf-cg21746001 | 0.617 | 0.445–0.854 | 0.004 |
| RARG-1stExon;5'UTR;Body-Island-cg15260268 | 0.699 | 0.505–0.969 | 0.03 |
| RARG-Body-Open_Sea-cg27036638 | 0.653 | 0.435–0.980 | 0.04 |
| RARG-TSS1500-S_Shore-cg12820608 | 1.540 | 1.009–2.351 | 0.046 |
| RARG-Body-N_Shore-cg13937905 | 0.658 | 0.436–0.992 | 0.046 |
CI, confidence interval; HR, hazard rate; RARs, retinoic acid receptors; STAD, stomach adenocarcinoma.
Co-expressed genes, associated enrichment pathways and protein-protein networks of RARs in STAD
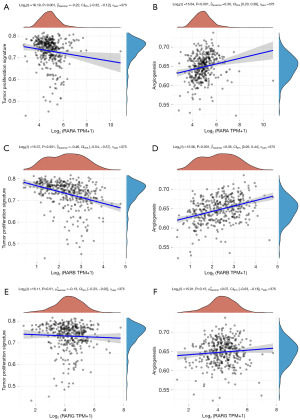
We analyzed the correlations between RARs and STAD related signaling pathways using TCGA database. As shown in Figure 8A-8F, three RARs family members were significantly associated with tumor proliferation in STAD. And RARA and RARB were also markedly related with angiogenesis of STAD. In addition, there were significant correlations between RARs and epithelial mesenchymal transition (EMT) markers, extracellular matrix (ECM) related genes and apoptosis of STAD (Figures S10-S12).
Next, we investigated the interacting genes and potential functions of RARs using GeneMANIA database. As shown in Figure 9A, the network indicated 20 genes with close relational-functional relationships between RARs members, including RXRA, MED1 and TADA3 so on. These genes were physical interactions, co-expression, predicted, co-localization, genetic interactions, pathway and shared protein domains, which were consistent with the biological role of RARs.
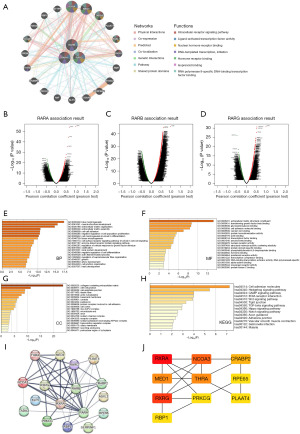
We further screened 500 co-expressed genes of RARs in STAD using LinkedOmics (Figure 9B-9D). The top 50 positively and negatively related genes were shown in Figures S13-S15. GO enrichment and KEGG pathway analyses were performed using Metascape based on the above 500 co-expressed genes. The GO enrichment analyses were divided into three functional groups: BP, cellular composition (CC), and molecular function (MF). As shown in Figure 9E-9G, RARs and its associated genes were mainly enriched for tube morphogenesis, ECM structural constituent and collagen-containing ECM in the BP, MF and CC category, respectively. As for KEGG pathway analysis, cell adhesion molecules were related to RARs and its associated genes (Figure 9H). In addition, we used the STRING database to establish the PPI networks analysis of RARs in patients with STAD (Figure 9I). Then, the CytoScape was used to assess the top 10 hub targets including RXRA, RXRG, NCOA3, MED1, THRA, CRABP2, RPE65, PRKCG, RBP1 and PLAAT4 (Figure 9J).
Single-cell functional analyses of RARs in STAD
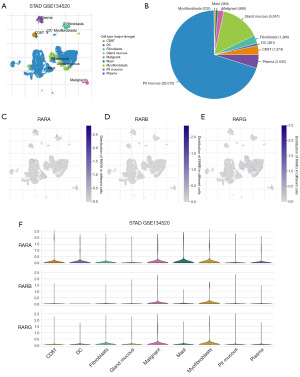
We used the TISCH database to evaluate RARs expression at the single-cell level. Two datasets (STAD_GSE134520, STAD_GSE167297) were used to interpret RARs expression in tumor microenvironment-related immune cells. As shown in Figure S16A-S16C, the results indicated that RARA and RARG showed the highest degree of infiltration in mast cells and malignant cells, respectively. RARB expression was higher in myofibroblast and malignant cells. Then, the analysis was performed on the STAD_GSE134520, which comprised 9 types of cells (Figure 10A). The results indicated that RARs were mainly expressed in pit mucous, gland mucous and plasma cells (Figure 10B). As shown in Figure 10C-10F, we demonstrated that RARs expression in STAD existed some extent infiltration of tumor microenvironment-related cells, which is consistent with Figure S16.
Differential expressions of RARs in STAD cell lines
We further used CCLE dataset to assess the RARs expression in different cell lines of STAD, which might be beneficial to select suitable cell lines for subsequent analysis or validation. As shown in Figure S17A-S17C, RARA, RARB and RARG expressions were highest in NCI-N87, NCC-StC-KI40 and HGC-27 cells, respectively.
Discussion
As vitamin A derivatives, RARs were discovered in the landmark 1987 (27), which uncovered a genomic kinship between the fields of vitamin A biology and steroid receptors. The presence of an ancestral RAR has been postulated in the genome of the most recent common ancestor of protostomes and deuterostomes. Nonetheless, in protostomes, this RAR gene appears to have been lost in the lineages that gave rise to nematode worms and insects (28). Vertebrates exhibit three paralogous RARs that have arisen from genome duplication (29). Nowadays, RARs are well known to have the ability to participate in the process of various tumors (7). Interestingly, RARA, RARB and RARG show the different roles in the development of malignant cancers. Our previous study has reported that RARα exerts oncogenic properties in GC (12). However, the clinical significance and potential function of the RARs family in STAD based on large cohort data are not identified.
In this study, we discovered dramatically increased expression of RARA and decreased expression of RARB in STAD tissues. Additionally, many clinical variables are closely related to RARs. Contrary to our expectations, there were no associations between RARs expression and the cancer stages of STAD patients. Notably, higher expressions of RARA and RARB correlated with worse OS, DFS and RFS for STAD patients. In contrast, lower expression of RARG was connected to shorter OS in STAD patients. Moreover, the clinical value of prognostic model indicated that RARs were identified to be potential prognostic biomarkers for STAD patients. Among RARs family members, RARB was closely related to immune cell infiltration, which had effect on the role of RARB in STAD prognosis. And the genetic alteration of RARB was significantly associated with the longer DFS of STAD patients.
Increasing evidence suggests that abnormal gene methylation and transcription level can lead to carcinogenesis, and accumulated genetic mutations result in cancer progression (30). In gastrointestinal cancers, epigenetic alterations have effect on cell cycle control, DNA repair, apoptosis, and tumorigenic-specific signaling pathways (31). Epigenetic age acceleration of STAD is related with tumor stemness, immunoactivation, and favorable prognosis (32). In the present study, we found that some CpG sites of the RARs family were associated with the prognosis of STAD patients.
The potential molecular mechanism of abnormal RARs expression in cancers have not yet been clarified. On the one hand, these three subtypes of RARs (RARα, RARβ, and RARγ) function as dependent transcription factors that mediate retinoic acid signaling through the formation of heterodimers with the retinoid X receptors (RXRα, RXRβ, and RXRγ) (33). Heterodimers play a crucial role in enhancing affinity, specificity, and diversity of binding sites from a structural standpoint. Additionally, these heterodimers govern cellular processes such as growth, differentiation, and apoptosis through interactions with ligands (such as retinoids), transcriptional co-regulators, and DNA (34).
On the other hand, RARs have been shown to have non-genomic effects and are capable of activating kinase signaling pathways, thereby modulating the transcription of retinoic acid target genes. Research has shown that inhibition of RARα phosphorylation repressed the progression of triple-negative breast cancer through transactivating miR-3074-5p (35). In GC, overexpression of RARα might promote the progression of GC via forming the positive feedback of IL-1β/Akt/RARα/Akt signaling (12). Moreover, upregulation of RARβ significantly decreased drug resistance of CCA cells to chemotherapeutic agents by increasing the susceptibility of these cells to caspasedependent apoptosis (36). In GC, all-trans retinoic acid (ATRA) could inhibit the role of LncHOXA10 in gastric tumorigenesis by the regulation of RARβ (37). In addition, RARγ promoted the proliferation, metastasis, and chemoresistance of CCA through simultaneous activation of the Akt/NF-κB and Wnt/β-catenin pathways (10).
In the present study, we identified that RARs family members were significantly associated with tumor proliferation EMT markers, ECM-related genes and apoptosis of STAD. GO enrichment and KEGG pathway analyses indicated that RARs and its associated genes were mainly enriched for tube morphogenesis, ECM structural constituent, collagen-containing ECM and cell adhesion molecules. In addition, STRING database was used to establish the PPI networks analysis of RARs in STAD patients. The top 10 hub targets included RXRA, RXRG, NCOA3, MED1, THRA, CRABP2, RPE65, PRKCG, RBP1 and PLAAT4. At the single-cell level, there was some extent of infiltration of tumor microenvironment-related cells in the RARs expression in STAD. Furthermore, RARA, RARB and RARG expressions were highest in NCI-N87, NCC-StC-KI40 and HGC-27 cells, respectively.
Although we have found interesting results in the current study, there are still some flaws. First, all of the data analyzed were uncovered by the online databases, hence the biological experiments and clinical trials are needed to validate the results. Second, an in-depth study requires a larger sample size to verify our conclusions due to the online database’s sample size limitation. Finally, the potential mechanism of RARs in STAD will be the focus of our research work in the future.
Conclusions
In this study, we focused on the comprehensive analysis of the role of RAR family members in STAD using a series of online databases, which suggested that RARs might be able to serve as potential therapeutic targets and prognostic markers for patients with STAD. These results would be beneficial for understanding of the potential molecular mechanism of RARs in the pathogenesis and clinical prognosis of STAD.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2154/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2154/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2154/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- López MJ, Carbajal J, Alfaro AL, et al. Characteristics of gastric cancer around the world. Crit Rev Oncol Hematol 2023;181:103841. [Crossref] [PubMed]
- Zhou D, Xiong S, Xiong J, et al. Integrated analysis of the microbiome and transcriptome in stomach adenocarcinoma. Open Life Sci 2023;18:20220528. [Crossref] [PubMed]
- Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229-63. [Crossref] [PubMed]
- Xie W, Yang T, Zuo J, et al. Chinese and Global Burdens of Gastrointestinal Cancers From 1990 to 2019. Front Public Health 2022;10:941284. [Crossref] [PubMed]
- Machlowska J, Baj J, Sitarz M, et al. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci 2020;21:4012. [Crossref] [PubMed]
- Johnston FM, Beckman M. Updates on Management of Gastric Cancer. Curr Oncol Rep 2019;21:67. [Crossref] [PubMed]
- di Masi A, Leboffe L, De Marinis E, et al. Retinoic acid receptors: from molecular mechanisms to cancer therapy. Mol Aspects Med 2015;41:1-115. [Crossref] [PubMed]
- Giguère V, Evans RM. Chronicle of a discovery: the retinoic acid receptor. J Mol Endocrinol 2022;69:T1-T11. [Crossref] [PubMed]
- Parrado A, Chomienne C, Padua RA. Retinoic acid receptor alpha (RAralpha) Mutations in Human Leukemia. Leuk Lymphoma 2000;39:271-82. [Crossref] [PubMed]
- Huang GL, Luo Q, Rui G, et al. Oncogenic activity of retinoic acid receptor γ is exhibited through activation of the Akt/NF-κB and Wnt/β-catenin pathways in cholangiocarcinoma. Mol Cell Biol 2013;33:3416-25. [Crossref] [PubMed]
- Alvarez S, Germain P, Alvarez R, et al. Structure, function and modulation of retinoic acid receptor beta, a tumor suppressor. Int J Biochem Cell Biol 2007;39:1406-15. [Crossref] [PubMed]
- Ren HY, Liu F, Huang GL, et al. Positive feedback loop of IL-1β/Akt/RARα/Akt signaling mediates oncogenic property of RARα in gastric carcinoma. Oncotarget 2017;8:6718-29. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Chandrashekar DS, Karthikeyan SK, Korla PK, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022;25:18-27. [Crossref] [PubMed]
- Thul PJ, Åkesson L, Wiking M, et al. A subcellular map of the human proteome. Science 2017;356:eaal3321. [Crossref] [PubMed]
- Győrffy B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience 2023;45:1889-98. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Liu CJ, Hu FF, Xie GY, et al. GSCA: an integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Brief Bioinform 2023;24:bbac558. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018;10:277-88. [Crossref] [PubMed]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214-20. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023;51:D638-46. [Crossref] [PubMed]
- Sun D, Wang J, Han Y, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res 2021;49:D1420-30. [Crossref] [PubMed]
- Ghandi M, Huang FW, Jané-Valbuena J, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019;569:503-8. [Crossref] [PubMed]
- Giguere V, Ong ES, Segui P, et al. Identification of a receptor for the morphogen retinoic acid. Nature 1987;330:624-9. [Crossref] [PubMed]
- Bertrand S, Brunet FG, Escriva H, et al. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol 2004;21:1923-37. [Crossref] [PubMed]
- Escriva H, Bertrand S, Germain P, et al. Neofunctionalization in vertebrates: the example of retinoic acid receptors. PLoS Genet 2006;2:e102. [Crossref] [PubMed]
- Hu Y, Wang M, Wang K, et al. A potential role for metastasis-associated in colon cancer 1 (MACC1) as a pan-cancer prognostic and immunological biomarker. Math Biosci Eng 2021;18:8331-53. [Crossref] [PubMed]
- Heydari Z, Moeinvaziri F, Mirazimi SMA, et al. Alteration in DNA methylation patterns: Epigenetic signatures in gastrointestinal cancers. Eur J Pharmacol 2024;973:176563. [Crossref] [PubMed]
- Hong C, Yang S, Wang Q, et al. Epigenetic Age Acceleration of Stomach Adenocarcinoma Associated With Tumor Stemness Features, Immunoactivation, and Favorable Prognosis. Front Genet 2021;12:563051. [Crossref] [PubMed]
- le Maire A, Teyssier C, Balaguer P, et al. Regulation of RXR-RAR Heterodimers by RXR- and RAR-Specific Ligands and Their Combinations. Cells 2019;8:1392. [Crossref] [PubMed]
- le Maire A, Germain P, Bourguet W. Protein-protein interactions in the regulation of RAR-RXR heterodimers transcriptional activity. Methods Enzymol 2020;637:175-207. [Crossref] [PubMed]
- Lou S, Gao H, Hong H, et al. Inhibition of retinoic acid receptor α phosphorylation represses the progression of triple-negative breast cancer via transactivating miR-3074-5p to target DHRS3. J Exp Clin Cancer Res 2021;40:141. [Crossref] [PubMed]
- Ren HY, Chen B, Huang GL, et al. Upregulation of retinoic acid receptor-β reverses drug resistance in cholangiocarcinoma cells by enhancing susceptibility to apoptosis. Mol Med Rep 2016;14:3602-8. [Crossref] [PubMed]
- Wang C, Zhao D, Wang K, et al. All-Trans Retinoic Acid Rescues the Tumor Suppressive Role of RAR-β by Inhibiting LncHOXA10 Expression in Gastric Tumorigenesis. Nutr Cancer 2021;73:2065-77. [Crossref] [PubMed]


