
TSPAN1 overexpression as an indicator of poor prognosis in estrogen receptor-positive breast cancer
Highlight box
Key findings
• Tetraspanin 1 (TSPAN1) modulates downstream extracellular matrix receptor signaling cascades and promotes cellular proliferation and invasion in breast cancer.
What is known and what is new?
• TSPAN1 is involved in the progression of many types of cancer.
• The TSPAN1 level was highly elevated in breast cancer tissue and was associated with local invasion/distant metastasis.
What is the implication, and what should change now?
• TSPAN1 inhibition may be a potential new treatment strategy for breast cancer.
Introduction
Breast cancer is one of the most common malignancies in women (1). Accurate data related to breast cancer incidence and mortality have been available since 2012. In the five-year period from 2008 to 2012, the average incidence was highest among white women, followed by black women (2,3). Currently, the 5-year survival rate for primary breast cancer is relatively high, ranging from 80–92% in different populations (1). However, when metastasis occurs, it drops to less than 25% (1). Estrogen receptor-positive (ER+) tumors account for the most common form of breast cancer and are the leading cause of death from the disease (4).
Patient survival rates can be improved by identifying the most effective treatment, which is guided by the characteristics of the tumor cells (5). Once metastatic lesions are identified, accurate characterization of tumor cells must be obtained at the outset of treatment (6,7) and one possible approach is the use of biomarkers (8). Currently, a series of various biomarkers, such as tissue markers, genetic markers, serum markers, and noncoding RNAs, have been discovered, but assessing the effectiveness of targeted therapies or prognosis of the disease still poses significant challenges (9,10). Hence, it is essential to identify more biomarkers and determine their clinical utility in future studies.
Tetrameric protein 1 (tetraspanin 1 or TSPAN1) is a member of the tetrameric protein (TSPAN/TM4SF) superfamily characterized by tetraspanin-enriched microdomains (TEMs) that are reported to be involved in many fundamental biological processes, including cell proliferation, adhesion, and migration (11). Recent research has shown that TSPAN1 is involved in the progression of many types of cancer, such as cholangiocarcinoma, ovarian, gastric, and colorectal cancers (11-13). However, further research is needed to uncover the mechanisms underlying this activity.
In this study, we demonstrated that higher TSPAN1 expression is associated with the overall survival (OS) of breast cancer and that TSPAN1 downregulation inhibits breast cancer cell proliferation. We also highlighted the interaction of TSPAN1 with integrin beta 4 (ITGB4) in promoting proliferation, invasion, and angiogenesis. Furthermore, we demonstrated that TSPAN1 can be modulated by the extracellular matrix (ECM) receptor signaling pathway. In addition, we used in vitro and in vivo studies to explore the role of TSPAN1 in breast cancer cell proliferation. We present this article in accordance with the MDAR and ARRIVE reporting checklists (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-409/rc).
Methods
UALCAN database analysis
UALCAN (https://ualcan.path.uab.edu/) is a user-friendly, interactive web resource that allows researchers to explore cancer transcriptome data from The Cancer Genome Atlas (TCGA). It provides easy access to gene expression profiles, survival analysis, and other relevant information across various cancer types. Researchers can input a gene of interest and obtain comprehensive data on its expression levels, clinical relevance, and potential associations with patient survival and other clinical parameters. The UALCAN database contains grade 3 RNA-seq and clinical data for 31 kinds of cancer selected from TCGA (14). It is an interactive web resource for in-depth analysis of RNA-Seq expression. We used the functional module of TCGA analysis in the UALCAN database for pan-cancer analysis of TSPAN1 expression.
Gene Expression Profiling Interactive Analysis (GEPIA) database analysis
GEPIA (http://gepia2.cancer-pku.cn/#index), is another web-based tool for analyzing RNA sequencing expression data from TCGA and the Genotype-Tissue Expression (GTEx) project. It enables users to perform gene expression analysis, including differential expression, survival analysis, correlation analysis, and pathway analysis. GEPIA offers an intuitive interface and provides interactive visualizations to facilitate the interpretation of gene expression patterns and their implications in cancer biology and other research areas. The GEPIA database is a comprehensive resource for gene expression systemic analysis (15). The database includes 9,736 tumor samples and 8,587 normal tissue samples from the TCGA and GTEx programs. We used the single-gene analysis in the functional module to plot the expression map of TSPAN1 in various cancers and normal tissues. We selected a threshold as the default.
LinkedOmics database analysis
We analyzed co-expressed genes associated with TSPAN1 expression using the LinkedOmics database (https://www.linkedomics.org/) (16). We used the LinkedOmics database, which includes mRNA sequencing data from 173 breast cancer patients from the TCGA database, to determine co-expression of TSPAN1 in breast cancer. In the LinkFinder module of LinkedOmics, we analyzed the data using Pearson’s correlation coefficient and generated a volcano map to visualize the results.
We generated the Kaplan-Meier curve of the OS of breast cancer patients using the Kaplan-Meier plotter, an online resource (17).
Tissue samples
We obtained breast cancer tissue and adjacent non-cancerous tissue samples from patients in our hospital (The Affiliated Hospital of Qingdao University) (n=60). The methodology followed the principles of the Declaration of Helsinki (as revised in 2013). Patients gave their informed consent for tissue collection and use, and all protocols were approved by the ethics committee of the Affiliated Hospital of Qingdao University (Nos. QU20210135ANIMAL and QU2016523PAT). The protocol was prepared before the study, and this protocol was not registered. We collected samples from patients who underwent surgery without adjuvant radiation therapy or chemotherapy. Prior to analysis, the collected tissues were frozen at −80 ℃. The diagnosis of breast cancer was independently confirmed by two experienced pathologists for all isolated patient tissues.
Western blot analysis
Patient tissue samples or cells were lysed using radioactive immunoprecipitation assay (RIPA) buffer containing a mixture of phosphatase and protease inhibitors (Roche, Mannheim, Germany; Beyotime Institute of Biotechnology, Shanghai, China). Protein concentration was measured using Pierce protein assay reagents (Thermo Fisher Scientific, Waltham, MA, USA). Proteins (20 µg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride membrane (Merck, Kenilworth, NJ, USA). After 1 hour of blockade with 5% skim milk, the membrane was incubated overnight at 4 ℃ with the primary antibody dissolved in poly(butylene succinate-co-terephthalate) (PBST) containing 5% body surface area (BSA).
We used the following primary antibodies: anti-TSPAN1 (Cat. SC-376551. Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-ITGB4 (Cat. sc-13543. Santa Cruz Biotechnology), anti-MEK1 (Cat. ab32091, Abcam, Cambridge, UK), anti-pMEK1 (Cat. ab96379, Abcam), anti-ERK1/2 (Cat. ab184699, Abcam), and anti-pERK1/2 (Cat. ab201015, Abcam). Anti-glyceraldehyde-3-phosphate dehydrogenase (Anti-GAPDH, Bioworld, Dublin, OH, USA) was used as the control. In addition, the membrane was incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Bethyl Laboratories, Montgomery, TX, USA) for 90 minutes and then detected using ECL kits and ChemiDoc XRS+ (Bio-Rad, Hercules, CA, USA).
Cell culture and chemicals used
For the in vitro study, we purchased three human breast cancer cell lines, namely, T47D, MCF7, and BT474, from the Cell Bank Culture Library of the Chinese Academy of Sciences. We obtained MCF10A, human normal breast cells, from the cell bank culture library of the Chinese Academy of Sciences. All cell lines were cultured with RPMI 1640 medium (Hyclone Laboratory, Logan, UT, USA) containing 10% fetal bovine serum (FBS) (Corning, Woodland, CA, USA) and 1% streptomycin/penicillin (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) in a humidified incubator at 37 ℃ with 5% CO2. We purchased FR180204 from Baiyang Times (Beijing, China). FR180204 (10 mM) solution was added to the medium and serially diluted to achieve the desired concentration.
siRNA transfection and plasmid construction
We transfected the human TSPAN1 siRNA and TSPAN1 plasmids using either lipid essence RNAiMAX reagent or lipogen 2000 reagent (Invitrogen, Carlsbad, CA, USA) as per the protocol detailed by the manufacturer. Two TSPAN1-specific siRNAs were purchased from GenePharma (Shanghai GenePharma Co., Ltd., Shanghai, China); human TSPAN1 was cloned using GV141-TSPAN1. We obtained the TSPAN1 clone from Genechem Co., Ltd. (Shanghai, China). The sequences used in the reaction are described in Table S1.
RNA isolation and real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNA was isolated from human breast cancer cell lines using the RNAiso reagent (Takara, Shiga Prefecture, Japan) as per the manufacturer’s protocol. The cDNA was synthesized using a reverse transcription system (Takara), and we used the SYBR Premix EX Taq Kit (Takara) for performing the RT-qPCR. The primers used in the reaction are shown in Table S1.
5-ethynyl-2'-deoxyuridine (EdU) staining
We used the EdU detection kit (China Beyotime Institute of Biotechnology) as per the instructions provided by the manufacturer. We incubated 1×106 breast cancer cells with 50 µM EdU-labeled medium (provided with the kit) for 2 hours at 37 ℃ with different treatments. After fixation, the cells were stained with Azide 555 solution (provided with the kit) for 30 minutes at 37 ℃ and Hoechst 33342 solution (provided with the kit) for 30 minutes at 37 ℃. We used a fluorescence microscope (Olympus, Tokyo, Japan) for imaging.
Colony formation
Cells were seeded into 6-well plates with a density of 1.0×105 cells/well. After 16 hours of inoculation, we replaced the medium with fresh medium with different treatments, allocating replicates to each group. After 48 hours of treatment, cells were detached with 0.25% trypsin and resuspended in 1640 medium containing 10% FBS. Cells were seeded into 6-well plates with a density of 1×103 cells per well and then incubated at 37 ℃ with 5% CO2. After the incubation period, the medium was discarded, and the cells were fixed with 4% acetal and stained with 0.5% crystal violet staining solution for 10 minutes. After that, the cells were given a mild wash and air-dried. The colony formation rate was determined by visually counting the cloned clusters.
Transwell invasion detection
We transfected human breast cancer cells with siRNA (siNC or TSPAN1 siRNA) and vector plasmids (GV141 or GV141/TSPAN1). A total of 2×104 cells/200 µL of FBS-free medium was added onto the filter coated with matrix gel (BD Biosciences, New Jersey, USA) in the transwell upper chamber (Corning Costar, Cambridge, MA, USA) 24 hours after transfection. RPMI 1640 medium supplemented with 10% FBS and 1% antibiotics was added to the lower chamber, and the plates were incubated for 24 hours. The invading cells were visualized with 1% crystal violet staining. For quantification, we counted cells in five randomly selected regions in each well using a wide-field microscope. Data were expressed as mean ± standard deviation (SD) from three independent experiments.
Tube formation assay
To assess tube formation, matrigel was incubated in a plate containing 48 wells with 150 µL/well (BD Biosciences) for 30 minutes at 37 ℃. Then 4×104 cells/well were seeded and cultured for 4–6 hours. The plate was monitored using an inverted microscope, and we calculated the tube length using ImageJ software (Washington, USA).
Co-immunoprecipitation (co-IP) and western blotting
Cells were lysed in RIPA buffer (Beyotime Institute of Biotechnology) containing a mixture of phosphatase and protease inhibitors (Roche) for 1 hour with brief vortexing every 15 minutes. The cells were then centrifuged at 12,000 ×g for 30 minutes at 4 ℃ to remove cell debris. We retained 5% of the cell lysate as input. The remainder was pre-clarified with protein A- or protein-G-coupled agarose (Life Technologies, Shanghai, China) for 2 hours at 4 ℃, followed by an overnight immunoprecipitation with the corresponding antibody at 4 ℃. The immunoprecipitate was washed four times with RIPA buffer and then boiled in SDS loading buffer for western blot analysis. For western blotting, we analyzed protein samples using SDS-PAGE and transferred these to a nitrocellulose membrane, followed by blockade and probing with the indicated antibody for detection.
Immunofluorescence and confocal microscopy analysis
The target cells on the chamber slides were washed twice with PBS, fixed with 4% paraformaldehyde for 30 minutes at room temperature, and washed with 100% ethanol for 10 minutes. The fixed cells were washed three times with PBS and blocked with 5% normal serum from the same species as the secondary antibody for 1 hour with 1% BSA/0.2% Triton X-100/PBS at room temperature. Cells were washed with primary antibodies (anti-TSPAN1 or anti-ITGB4; Santa Cruz, CA, USA) at 37 ℃ for 1 hour three times and incubated with secondary antibodies for 1 hour at 37 ℃. These were then washed three times, mounted on coverslips with fade-resistant mounting medium [with 4',6-diamidino-2-phenylindole (DAPI)], and examined under a Leica TCS SP5 confocal microscope (Leica Microsystems, Wetzlar, Germany).
In vivo growth studies
All animal experiments in this study were approved by the Institutional Animal Care and Use Committee (IACUC) of our hospital and performed in specific pathogen-free facilities in accordance with the IACUC guidelines (IACUC 2018-A0015). We purchased pathogen-free 6-week-old female Balb/c nude mice from the Beijing Weta River Laboratory Animal Technology Co., Ltd. (Beijing Weta River, Beijing, China). A total of 2×106 stable BT474 cells (two types: LV-shNC control and LV-shTSPAN1) were injected subcutaneously onto the left side of the mice. The tumor size was measured every two days using calipers from the time palpable tumors were formed until they were removed. We calculated tumor volume using the following formula: length × width × width × 0.5. The mice were euthanized in a 7.5% CO2 chamber, and tumors were harvested for further analysis.
Hematoxylin-eosin (H&E) and immunohistochemical (IHC) analysis
Tumors were harvested, immediately fixed in formalin, and embedded in paraffin. We subjected the tissues to H&E staining as per the routine protocol. For immunohistochemistry, tumor sections were dewaxed, hydrated, and heated in an active antigen retrieval solution (10 mM citric acid buffer, pH 6.0) in a microwave for 20 minutes. Endogenous peroxidase was inactivated with a 3% hydrogen peroxide solution. The slides were rinsed with distilled water, washed twice with PBS for 3 minutes, and incubated with Superblock (Scytek Labs, Burlingame, CA, USA) for 1 hour at room temperature in a humidified chamber. After rinsing with PBS, the slides were incubated with avidin (Vector Laboratories, Inc., Burlingame, CA, USA) for 15 minutes, washed with PBS, and blocked with biotin in a humidified chamber for 15 minutes at room temperature. The slides were incubated with the primary antibody overnight in a humidified chamber at 4 ℃. After rinsing with PBS, biotinylated secondary antibodies were added to the slides and incubated for one hour. After washing with PBS, the slides were incubated with avidin-peroxidase complex reagents (Vector Laboratories, Inc.) for 30 minutes, washed once with PBS, stained with a freshly prepared diaminobenzidine solution for 15 minutes, and counterstained with hematoxylin.
Statistical analysis
The experiments were conducted in triplicate, and results were expressed as the mean ± SD of at least three independent experiments. A two-tailed t-test was used for statistical analysis. We used the regression curve test for survival analysis. P<0.05 was considered statistically significant.
Results
Overexpression of TSPAN1 in breast cancer and survival rate
ER+ breast cancer is the most common type and is the leading cause of death (4). Patients with ER+ cancer also require long-term hormone therapy. We tested the mRNA expression of TSPAN1 in different human tumor samples using the UALCAN and GEPIA databases. The mRNA expression levels of TSPAN1 were the highest in most types of human cancer (Figure S1A,S1B). The transcription levels of TSPAN1 in cancer were then compared to those in normal samples using the GEPIA database. The results showed that TSPAN1 was significantly upregulated in patients with breast cancer (Figure 1A).
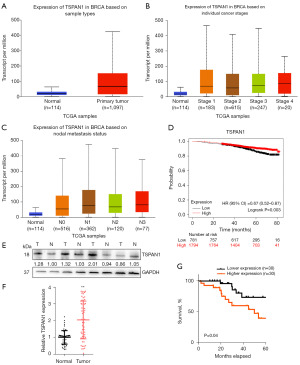
Further analysis from the UALCAN database showed that mRNA levels of TSPAN1 were relatively high in advanced and high-grade breast cancer tissues and relatively low in early- and low-grade breast cancer tissues (Figure 1B,1C). Next, we used the Kaplan-Meier plotter to generate survival curves from the data of ER+ breast cancer patients (Figure 1D). However, the 5-year OS demonstrated a superior survival rate in the high TSPAN1 expression group (n=1,794) when compared to the low TSPAN1 expression group (n=781) (Figure 1D).
We then evaluated TSPAN1 expression in 60 pairs of breast cancer tissues obtained from patients who had surgery at our hospital. A panel of experienced pathologists confirmed the diagnosis of breast cancer in all samples. Western blot analysis further revealed that the total level of TSPAN1 expression in tumor tissues was higher than that in the normal tissues (T) (Figure 1E and Figure S1C).
To investigate the frequency of TSPAN1 regulation in breast cancer, we examined its expression in 60 freshly frozen breast cancer tissues using RT-qPCR. We observed a significant upregulation of TSPAN1 in breast cancer tissues (Figure 1F). The Kaplan-Meier survival curve showed that OS was significantly longer in patients with low TSPAN1 expression than in patients with high TSPAN1 expression (Figure 1G, P=0.04). Overall, these results suggested that upregulation of TSPAN1 in patients with primary breast cancer was associated with a lower survival rate.
TSPAN1 downregulation inhibited breast cancer cell growth in vitro
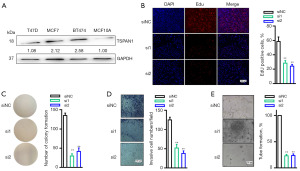
Next, we assessed the levels of TSPAN1 in three separate ER+ breast cancer cell lines and normal breast cell lines using western blotting analysis. TSPAN1 expression assessed using western blotting and RT-qPCR. The expression of TSPAN1 evaluated through western blotting (Figure S2A) and RT-qPCR (Figure S2B) in BT474 cancer cell lines with TSPAN1 knockdown. As seen in Figure 2A, TSPAN1 expression was upregulated in all sampled breast cancer cells. Next, we performed the TSPAN1 downregulation experiment in BT474 (Figure 2B). After the downregulation of TSPAN1, when compared to the control group, BT474 cells experienced a significant decrease in cell growth (P<0.001, Figures 2B) and viability (P<0.001, Figures 2C). The downregulation of TSPAN1 significantly reduced cell invasion compared to the siNC group (P<0.001, Figure 2D). Further, we evaluated tube formation in umbilical vein endothelial human cells, which were co-cultured with the supernatant of BT474 cells to downregulate TSPAN1. Our results showed that the BT474 cell supernatant inhibited endothelial tube formation by downregulating TSPAN1 (P<0.001, Figure 2E). In summary, TSPAN1 could be a potential therapeutic target for breast cancer.
TSPAN1 co-expression network and Gene Ontology/Kyoto Encyclopedia of Genes and Genomes (GO/KEGG) bioprocess enrichment
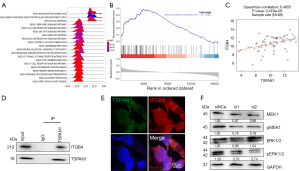
We used LinkedOmics to study the TSPAN1 co-expression gene in the breast cancer cohort to further explore the mechanism by which TSPAN1 promotes breast cancer proliferation and invasion. The top 50 significant genes that were positively and negatively correlated with TSPAN1 were visualized in a heat map (Figure S3A-S3C). Next, we used Gene Set Enrichment Analysis (GSEA) to analyze the enrichment functions of GO annotation and KEGG pathways. The most prominent biological processes identified were bioregulatory and metabolic processes (Figure S3D), with cellular components enriched in membranes and nuclei (Figure S3D). The first two molecular functional terms were protein binding and ion binding (Figure S3D). KEGG pathway analysis showed ECM receptor enrichment as well as focal adhesion signaling pathways, among others (Figure 3A,3B). These results suggested that TSPAN1 has an extensive role in the progression of breast cancer and that ITGB4 is positively correlated with TSPAN1 expression (Figure 3C).
TSPAN1 promoted proliferation through the ECM receptor signaling pathway
TSPAN1 has been found to promote colon cancer metastasis by activating the ECM receptor signaling pathway, as bioinformatics analyses have revealed (18). In this study, using the Co-IP assay, we confirmed this mechanism in breast cancer by observing the interaction of TSPAN1 and ITGB4 in BT474 cells (Figure 3D).
Immunofluorescence staining of BT474 cells also demonstrated the ability of TSPAN1 (green) and ITGB4 (red) to colocalize each other (Figure 3E). To further elucidate its mechanism, we used western blotting to detect protein expressions of MEK1, pMEK1, ERK1/2, and pERK1/2 in breast cancer cells after TSPAN1 siRNA transfection. We found that downregulation of TSPAN1 expression significantly reduced the expression levels of MEK1, pMEK1, and pERK1/2 in breast cancer cells (Figure 3F), which indicates that TSPAN1 is involved in the MEK-ERK1/2 signal cascade. This result showed that TSPAN1 promotes metastasis through the ECM receptor signaling pathway.
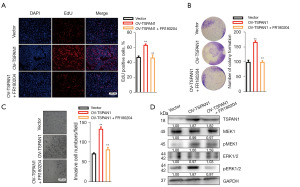
We performed salvage functional experiments to detect whether TSPAN1 affected cancer cell proliferation and motility through the ECM receptor signaling pathway. To explore the role of ERK1/2 activation in breast cancer, we used FR180204, a potent and selective adenosine triphosphate (ATP) inhibitor of ERK1 and ERK2 that inhibits the activity of ERK1 and ERK2 kinases (19). We tested the TSPAN1-overexpressing cell line using the breast cancer cell line T47D with TSPAN1 overexpression.
Efficiency was detected using western blotting (Figure 4). EdU staining and colony formation assays showed that the upregulation of TSPAN1 expression somewhat restored the impaired cell proliferation capacity of FR180204 (Figure 4A,4B). Using the Transwell assay to detect the invasion capability of TSPAN1-overexpressed cells, we found that treatment with FR180204 salvaged the invasion capability (Figure 4C). Transfection of T47D with the TSPAN1-expressed plasmid enhanced the protein expression of TSPAN1 (Figure 4D). Notably, treatment with TSPAN1 resulted in a significant increase in the expression of pERK1/2 in T47D (Figure 4D), and treatment with FR180204 reversed these effects (Figure 4D). Overall, these results suggest that TSPAN1 regulates proliferation and invasion by modulating ECM receptor signaling.
TSPAN1 shRNA inhibited primary tumor growth and metastasis in the nude mouse model
We established primary tumors using breast cancer cells and their TSPAN1 knockdown counterparts (Figure S4A,S4B). We injected stable TSPAN1 knockdown or control cells into left-sided nude mice. Twenty-four days after tumor inoculation, we found a significant reduction in the growth rate of TSPAN1-silencing tumor cells (Figure 5A,5B). On the last day, tumor volumes in the LV-shTSPAN1 group were 166.35±69.15 mm3, while those in the LV-shNC group were 416.44±104.43 mm3. The mean tumor weight in the LV-shTSPAN1 group was significantly lower than in the LV-shNC group (P<0.01, Figure 5C; LV-shTSPAN1, 0.29±0.08 g; LV-shNC, 0.51±0.14 g).
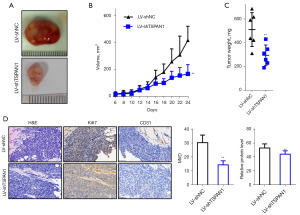
We also found that the expression of Ki67 protein in TSPAN1-silencing tumor cells was reduced after IHC staining (Figure 5D). Using light microscopy, we detected tumor cell invasion capability on H&E-stained tissue xenografts. The microvascular density of TSPAN1-silencing tumors was significantly lower than that of controls (Figure 5D). These findings suggest that TSPAN1 plays a vital role in vivo in the regulation of tumor growth, invasion, and angiogenesis.
Discussion
The TSPAN1 gene is located on chromosome 1p34.1 and encodes a 26 kDa protein. However, the molecular weight of the wild-type TSPAN1 protein expressed in ovarian cancer cells is observed to be higher than the value calculated by N-glycosylation (20). Band shifts of TSPAN1 have also been consistently confirmed in breast cancer cells. Recent research on TSPAN1 has focused on its ability to promote cancer proliferation and invasion. However, the mechanism behind TSPAN1 activity remains unclear.
In our study, using the UALCAN and GEPIA datasets from Gene Expression Omnibus (GEO), we found that the expression of TSPAN1 was higher in primary breast cancer than in normal breast tissue. Therefore, TSPAN1 may have a role in metastasis. Using UALCAN expression analysis, we found that the expression of TSPAN1 varied significantly among patients based on disease stage and lymph node metastasis status. Using Kaplan-Meier curve analysis, we found that high levels of TSPAN1 expression were associated with a poor prognosis in ER+ breast cancer. These results indicated that TSPAN1 is a biomarker and potential therapeutic target for ER+ breast cancer.
Although basal-like breast cancer and triple-negative breast cancer (TNBC) can predict a poor prognosis, they have low TSPAN1 expression. Furthermore, TSPAN1 could not predict the prognosis of basal-like breast cancer or TNBC. Finally, we conducted a comprehensive genome-wide association study to identify TSPAN1-related genes, and we enriched the GO keywords for these genes. The majority of the enriched GO keywords referred to bioregulatory and metabolic processes, with cellular components enriched in membranes and nuclei. The first two molecular functional terms were protein binding and ion binding. These GO keywords can help direct future studies aiming at elucidating the mechanism of TSPAN1 in breast cancer and developing new therapies for ER+ breast cancer.
Like other members of the tetrameric protein family, TSPAN1 is described as a typical cell surface receptor that activates various signaling cascades through interactions with other tetramerins, integrins, receptors, and/or cytoplasmic proteins called tetrameric protein networks. Interactions between TSPAN1 and integrins have been described in cholangiosarcoma and head and neck squamous cell carcinoma (HNSCC) models (21-24). TSPAN1’s probable dimerization and transfer from cytoplasm to nucleus localization raise the possibility that it activates the ECM receptor pathway by interacting with integrins or other cell surface receptors, and this is worth exploring in the future (25).
Based on these observations, we postulate that TSPAN1 is an oncogenic protein involved in both the development and progression of breast cancer, with the ECM receptor pathway serving as the main downstream effector. TSPAN1 overexpression is associated with tumor progression observed in mouse tumor biopsies and human breast cancer tissues. Our findings lend credence to the hypothesis that activation of the ECM receptor pathway contributes to the spread of breast cancer metastases. There were limitations to this study. The mechanism of TSPAN1 in breast cancer requires further investigation through in vitro and in vivo experiments.
Considering our findings in this study, we anticipate that TSPAN1 suppression may be an effective strategy for targeted therapies for breast cancer and possibly metastatic sites. Inhibition of TSPAN1 has been shown to induce cellular sensitivity to different chemotherapy drugs, making it a promising new target for cancer treatment.
Conclusions
This study established that TSPAN1 played a crucial role in the tumorigenesis of breast cancer. The TSPAN1 level was highly elevated in breast cancer tissue and was associated with local invasion/distant metastasis. In addition, high TSPAN1 expression was inversely correlated with OS in patients. TSPAN1 expression silenced by siRNA inhibited the proliferation and invasion capabilities of breast cancer cells. TSPAN1, via its interaction with ITGB4, participated in the MEK-ERK1/2 signal cascade, as found in a mechanistic analysis. Overall, our study not only revealed the critical role of TSPAN1 in breast cancer tumorigenesis, but also demonstrated the rationale for targeting TSPAN1 as a novel treatment option for breast cancer.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-409/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-409/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-409/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-409/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of The Affiliated Hospital of Qingdao University (Nos. QU20210135ANIMAL and QU2016523PAT). Written informed consent was obtained from all participants. All animal experiments in this study were approved by the Institutional Animal Care and Use Committee (IACUC) of our hospital and performed in specific pathogen-free facilities in accordance with the IACUC guidelines (No. IACUC 2018-A0015).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Borges VF. Options for Endocrine-Refractory, Hormone Receptor-Positive Breast Cancer: Which Target and When? J Clin Oncol 2021;39:3890-6. [Crossref] [PubMed]
- Xu T, Yu S, Zhang J, et al. Dysregulated tumor-associated macrophages in carcinogenesis, progression and targeted therapy of gynecological and breast cancers. J Hematol Oncol 2021;14:181. [Crossref] [PubMed]
- Reinert T, Barrios CH. Optimal management of hormone receptor positive metastatic breast cancer in 2016. Ther Adv Med Oncol 2015;7:304-20. [Crossref] [PubMed]
- Jiang Y, Jiang Z, Wang M, et al. Current understandings and clinical translation of nanomedicines for breast cancer therapy. Adv Drug Deliv Rev 2022;180:114034. [Crossref] [PubMed]
- Wang L, Zhang S, Wang X. The Metabolic Mechanisms of Breast Cancer Metastasis. Front Oncol 2021;10:602416. [Crossref] [PubMed]
- Yeeravalli R, Das A. Molecular mediators of breast cancer metastasis. Hematol Oncol Stem Cell Ther 2021;14:275-89. [Crossref] [PubMed]
- Barzaman K, Karami J, Zarei Z, et al. Breast cancer: Biology, biomarkers, and treatments. Int Immunopharmacol 2020;84:106535. [Crossref] [PubMed]
- Sukumar J, Gast K, Quiroga D, et al. Triple-negative breast cancer: promising prognostic biomarkers currently in development. Expert Rev Anticancer Ther 2021;21:135-48. [Crossref] [PubMed]
- Zubair M, Wang S, Ali N. Advanced Approaches to Breast Cancer Classification and Diagnosis. Front Pharmacol 2021;11:632079. [Crossref] [PubMed]
- Wang J, Huang J, Huang L. TSPAN1 silencing protects against cerulein-induced pancreatic acinar cell injury via targeting AGR2. Drug Dev Res 2022;83:158-66. [Crossref] [PubMed]
- Han J, Xie C, Liu B, et al. Tetraspanin 1 regulates papillary thyroid tumor growth and metastasis through c-Myc-mediated glycolysis. Cancer Sci 2023;114:4535-47. [Crossref] [PubMed]
- Zhou Y, Huang B, Zhang Q, et al. Modeling of new markers for the diagnosis and prognosis of pancreatic cancer based on the transition from inflammation to cancer. Transl Cancer Res 2024;13:1425-42. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Szász AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016;7:49322-33. [Crossref] [PubMed]
- Qi W, Sun L, Liu N, et al. Tetraspanin family identified as the central genes detected in gastric cancer using bioinformatics analysis. Mol Med Rep 2018;18:3599-610. [Crossref] [PubMed]
- Ohori M, Kinoshita T, Okubo M, et al. Identification of a selective ERK inhibitor and structural determination of the inhibitor-ERK2 complex. Biochem Biophys Res Commun 2005;336:357-63. [Crossref] [PubMed]
- Scholz CJ, Sauer G, Deissler H. Glycosylation of tetraspanin Tspan-1 at four distinct sites promotes its transition through the endoplasmic reticulum. Protein Pept Lett 2009;16:1244-8. [Crossref] [PubMed]
- Wang Y, Liang Y, Yang G, et al. Tetraspanin 1 promotes epithelial-to-mesenchymal transition and metastasis of cholangiocarcinoma via PI3K/AKT signaling. J Exp Clin Cancer Res 2018;37:300. [Crossref] [PubMed]
- Garcia-Mayea Y, Mir C, Carballo L, et al. TSPAN1: A Novel Protein Involved in Head and Neck Squamous Cell Carcinoma Chemoresistance. Cancers (Basel) 2020;12:3269. [Crossref] [PubMed]
- Lu Z, Pang T, Yin X, et al. Delivery of TSPAN1 siRNA by Novel Th17 Targeted Cationic Liposomes for Gastric Cancer Intervention. J Pharm Sci 2020;109:2854-60. [Crossref] [PubMed]
- Zhang X, Shi G, Gao F, et al. TSPAN1 upregulates MMP2 to promote pancreatic cancer cell migration and invasion via PLCγ. Oncol Rep 2019;41:2117-25. [Crossref] [PubMed]
- Ferrero Restelli F, Fontanet PA, De Vincenti AP, et al. Tetraspanin1 promotes NGF signaling by controlling TrkA receptor proteostasis. Cell Mol Life Sci 2020;77:2217-33. [Crossref] [PubMed]


