
GGT5 as a promising prognostic biomarker and its effects on tumor cell progression in gastric cancer
Highlight box
Key findings
• Knockdown of gamma-glutamyl transferase 5 (GGT5) reduced proliferation and migration in gastric cancer (GC) cells by modulating the PI3K/AKT-MAPK-MMPs pathway.
What is known and what is new?
• GGT5 is highly expressed in GC.
• GGT5 mediated proliferation and migration of GC cells by directly targeting PI3K/AKT-MAPK-MMPs pathways.
What is the implication, and what should change now?
• GGT5 might be a new target for GC and can be used for the diagnosis, prognosis and treatment of GC.
Introduction
Gastric cancer (GC) is a common cancer worldwide, ranking fifth in incidence and fourth in mortality. In 2020, there were more than 1 million new cases of GC and 769,000 deaths worldwide (1). At present, GC is difficult to diagnose in the early stage, and surgery, endoscopic resection, chemotherapy, biological agents and immunotherapy have been included in the treatment of GC, which reduces the mortality of GC to a certain extent, but the effect is not satisfactory. Therefore, new diagnosis or treatment methods are urgently needed to improve the clinical outcome of GC.
Gamma-glutamyl transferase (GGT) is a membrane-bound extracellular enzyme (2) that serves as a novel non-invasive marker that can be detected in the blood and used to indicate various diseases. Studies have shown that elevated serum GGT levels predict chronic diseases such as liver injury and insulin resistance (3,4). Apart from this, most studies have shown that GGT expression is significantly increased in human malignant tumors. For example, high GGT levels are associated with poor prognosis in cancers such as gallbladder cancer, pancreatic cancer, and epithelial ovarian cancer (5-7). GGT genes belong to a multigene family with at least seven genes to adapt to high physiological variations and expression in different physiological states and encode enzymes involved in glutathione (GSH) metabolism and amino acid transpeptidation which include GGT1, GGT2, GGT3P, GGT4P, GGT5, GGT6, GGT7 and GGT8P (8). GGT1 and GGT5 are the only two extracellular enzymes that cleave γ-glutamyl bonds (9,10).
Gamma-glutamyl transferase 5 (GGT5) belongs to the GGT family and has been shown to be the only GGT enzyme of having catalytic activity to date (10). GGT5 protein is expressed by local macrophages in multiple organs, including Kupffer cells in the liver and macrophages. GGT5 has been implicated in the control of re-oxidation (11), drug metabolism, immunological function (12), and other body functions, and there has been evidence that GGT5 may induce inflammation. Wei et al. had demonstrated that high levels of GGT5 in cancer-associated fibroblasts (CAFs) correlated with cancer cell survival and treatment resistance in lung adenocarcinoma (13). With the development of bioinformatics and the deepening of GC research (14-16), some papers have shown that GGT5 may play an important role in the pathogenesis and development of GC. However, the mechanism of GGT5 in GC remains unclear.
We hypothesized that GGT5 was related to tumor cell growth. In this study, bioinformatics analysis was used to explore the expression and function of GGT5 in GC. In addition, we conducted in vitro experiments to evaluate the impact of GGT5 knockdown on GC cell proliferation and migration. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2222/rc).
Methods
Bioinformatics analysis
A harmonized pan-cancer dataset was downloaded from the UCSC (http://xenabrowser.net/) datasets: TCGA Pan-Cancer, further extracted ENSG00000099998 (GGT5) gene expression data from each sample, log2(x+0.001) transformation was performed for each expression value. GGT5 expression in different tumors and the relationship between GGT5 and prognosis, immune infiltration, clinical stage were analyzed in Sangerbox (17) (http://www.sangerbox.com/tool). GSE27342, GSE29272 and GSE118916 expression profiles were obtained from the Gene Expression Omnibus (GEO) datasets (https://www.ncbi.nlm.nih.gov/geo/). RNAseq data and corresponding clinical information for GC were obtained from the Cancer Genome Atlas (TCGA) datasets (http://portal.gdc.com), and differential mRNA expression was investigated using the R software Limma package. The LinkedOmic datasets (18) (http://www.linkedomics.org) and cBioPortal (19,20) (https://www.cbioportal.org/) were used to conduct a correlation study between GGT5 and other GC genes. In order to further determine the potential function of GGT5, ClusterProfiler (21) package in R software was used to analyze the GGT5 Gene Ontology (GO) functions and enrich Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, as well as to use LinkedOmic datasets for gene set enrichment analysis (GSEA) analysis.
Clinical information
A total of 124 primary GC cases (all patients underwent surgical resection by endoscope, laparotomy or laparoscopy, and had not received any treatment for GC before surgery. No epilepsy drugs. No other malignant tumor within 5 years) diagnosed and treated in The First Affiliated Hospital of Zhejiang Chinese Medical University in the past three years were selected. A total of 287 subjects (no abnormal blood lipids, normal tumor markers, and no other disease diagnosed within 1 year) who underwent a physical examination in the last year were selected as healthy controls. Biochemical indexes of alanine aminotransferase (ALT), aspartate aminotransferase (AST) and GGT in GC patients prior to surgery and healthy controls (excluding abnormal liver function. ALT ranged from 5 to 40 U/L in males and 5 to 35 U/L in females, and AST 8 to 40 U/L.) were collected, and differences in GGT were analyzed. This research only collected the serum test data of the patients’ clinical cases. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Institutional Review Board (IRB) of The First Affiliated Hospital of Zhejiang Chinese Medical University (IRB: 2022-KL-150-01) and was exempted from informed consent by the IRB.
Cell culture
Human GC cell lines AGS, HGC-27 and MKN-45 were used in the experiments. HGC-27 was acquired from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). AGS and MKN-45 were donated by the Key Laboratory of Digestive Pathophysiology of Zhejiang Province and the Department of Cell and Immunology of Zhejiang Chinese Medical University, respectively. HGC-27 and AGS were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, USA) containing 10% fetal bovine serum (FBS, Gibco, USA), MKN-45 was grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) supplemented with 10% FBS. All cells were maintained in 100 U/mL penicillin, and 100 µg/mL streptomycin at a humidified incubator (37 ℃, with 5% CO2).
Cell transfections
The lentiviral vector has the characteristics of green fluorescent protein (GFP) gene and puromycin resistance, that is, the cells can express green fluorescence normally and are resistant to puromycin after successful transfection. According to the lentivirus transfection manual, 24 hours before transfection, target cells with good growth were selected, and 5×104 cells were added to each well of 6-well plates for culture under conventional culture conditions. When the cell confluence rate reached 70% after 24 h, the old medium was removed, and 1 mL of suspended diluted GGT5 knockdown lentivirus (sh-GGT5, Genechem, China) was added to each well (sh-GGT5 sequences were GCTCTTCTTCAACGGGACAGA). At the same time, the target cells were transfected with control negative lentivirus (sh-NC) to establish the control cell line (sh-NC sequences were TTCTCCGAACGTGTCACGT). Cells are routinely cultured in 37 ℃, 5% CO2 incubator. Cell growth status was observed 12 h after transfection. If there was no obvious toxic effect, the conventional medium was replaced about 48 hours later. At 72 h after transfection, expression of GFP in cells could be observed under inverted phase contrast fluorescence microscopy. When the expression of green fluorescent protein was good and the growth was stable, puromycin (Beyotime, China, 2 µg/mL) was added to screen the cells with poor transfection. Polymerase chain reaction (PCR) was used to detect GGT5 expression in transfected cells. The cells with high fluorescence expression and stable growth could be used for subsequent cell experiments.
RNA isolation and PCR
Total RNA isolation was performed from cells using Trizol reagent (Glpbio, USA). After its concentration was determined, RNA was reversed-translated into the cDNA kit based on instructions from the BIO-RAD script cDNA synthesis kit (Bio-Rad, USA). The primer sequences were as follows: forward primer 5'-GTCAGCCTAGTCCTGCTGG-3' and reverse primer 5'-GGATGGCTCGTCCAATATCCG-3' for GGT5; forward primer 5'-ACCACAGTCCATGCCATCAC-3' and reverse primer 5'-TCCACCACCCTGTTGCTGT-3' for Glyceraldehyde 3-phosphate dehydrogenase (GAPDH). GAPDH was the reference gene for GGT5.The date were analyzed via using 2−ΔΔCt method.
Cell Counting Kit-8 (CCK-8)
GC cell proliferation was assessed by CCK-8 assay (Beyotime, China). According to the manufacturer’s instructions, 3000 cells were seeded into 96-well plates. The cells were cultured for 24, 48, and 72 h, CCK-8 reagent was added into each well, after incubation for another 2 h at 37 ℃, absorbance at 450 nm was measured using a multifunctional microplate reader.
5-ethynyl-2’-deoxyuridine (EdU) assays
The 5×105 cell suspension was injected into the 6-well plates. After the cells were cultured overnight and returned to normal, EdU labeling (Beyotime, China), fixation, washing and permeability were performed according to kit instructions. A volume of 500 µL of the Click reaction solution containing Azide 594 dye was added to each well for incubation, and then nuclear staining was conducted by using Hoechst dye, followed by fluorescence detection.
Transwell migration assays and wound healing assays
After the cells were digested by trypsin, 5×104 cell suspension was injected into 1% FBS medium and cultured in the 6-well plates starved for 24 h. Digestive cells again, 200 µL 2×104 cell suspension was inoculated in Transwell chambers (Corning, USA), placed in the 24-well plates, and cultured in the lower chamber of 10% FBS medium for 24 h. Then, after fixation with 70% methanol, crystal violet staining was carried out to stain the cells.
For wound healing assays, 5×105 cell suspension was seeded into 6-well plates. After 24 h, a scratch wound was made using a sterile 1,000 µL pipette tip in a monolayer of cells. The scratch wound was imaged at the 0, 24 and 48 h time points using microscope at the same location of 6-well plates. The wound healing rate was measured by the fracture area of the noncellular regions.
Western blotting
The proteins in cells were extracted by using the Radio Immunoprecipitation Assay (RIPA) lysis buffer containing a mixture of protease and phosphatase inhibitors (Beyotime, China), and were mixed with 5× loading buffer (Beyotime, China) at a ratio of 4:1, boiled for denaturation. The proteins were prepared with sodium dodecyl sulfate polyacrylamide gel electrophoresis, and transferred to the polyvinylidene fluoride membrane. Then the membranes were blocked with 5% skimmed milk powder and incubated at 4 ℃ overnight with primary antibodies (Table 1). After recovery of the primary antibody, the membranes were incubated with horseradish peroxidase-labeled secondary antibody (HRP-labeled goat anti-mouse IgG, Sangon Biotech Cat# D110087, RRID: AB_2940948; HRP-labeled goat anti-rabbit IgG, Sangon Biotech Cat# D110058, RRID: AB_2940954) for 2 h at 37 ℃. The Enhanced Chemiluminescence (ECL) chemiluminescence reagent (Biosharp, China) was then used to develop imaging. The relative expression of the target protein was calculated using grey value analysis software.
Table 1
| Primary antibodies | Host | Dilution ratio | Brand | Catalogue number | RRID |
|---|---|---|---|---|---|
| GAPDH | Rabbit | 1:10,000 | Affinity, USA | AF7021 | AB_2839421 |
| AKT | Mouse | 1:2,000 | Proteintech, USA | 60203-2-Ig | AB_10912803 |
| P-AKT | Mouse | 1:2,000 | Proteintech, USA | 66444-1-Ig | AB_2782958 |
| PI3KP85 | Rabbit | 1:2,000 | Beyotime, China | AF7742 | AB_2844106 |
| ERK | Rabbit | 1:2,000 | Beyotime, China | AF1051 | AB_2923146 |
| P-ERK | Rabbit | 1:2,000 | Beyotime, China | AF1891 | AB_2923145 |
| P38 | Rabbit | 1:2,000 | Bioss, China | bs-0637R | AB_10856281 |
| P-P38 | Rabbit | 1:2,000 | Bioss, China | bs-0636R | AB_10856595 |
| STAT5b | Rabbit | 1:5,000 | Abcam, UK | ab178941 | AB_2885102 |
| MMP2 | Rabbit | 1:5,000 | Abcam, UK | ab92536 | AB_10561597 |
| MMP9 | Rabbit | 1:5,000 | Abcam, UK | ab76003 | AB_1310463 |
| EGFR | Rabbit | 1:5,000 | Abcam, UK | ab52894 | AB_869579 |
RRID, Research Resource Identifier.
Statistical analysis
Each experiment was repeated three times independently. Statistical analysis and drawing were performed using IBM SPSS Statistics 25 software (IBM Corp., Chicago, IL, USA), GraphPad Prism 8 (GraphPad Software, CA, USA). Variance analysis, independent t-tests and Chi-squared tests were performed to assess differences between groups. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001 were considered to indicate statistical significance.
Results
Upregulation of GGT5 expression in GC and its association with clinicopathological parameters
First, serum GGT expression levels of GC patients and healthy controls were analyzed, and the results showed that GGT levels were highly expressed in GC (P<0.05) (Figure 1A, Table 2). TCGA and GEO datasets were used to further confirm our results. We detected high expression of GGT5 in GC tissues and found that patients with high expression of GGT5 had a significantly poor prognosis (Figure 1B-1D). In addition, the association between GGT5 expression and clinicopathological parameters of GC was investigated. GGT5 expression was significantly increased in advanced tumor specimens, and was positively correlated with immune infiltration of GC and correlated with the T stage, grade and stage of GC (Figure 1E,1F). Meanwhile, the analysis of the datasets showed that the occurrence of GC was not related to sex and age (Figure 1G,1H).
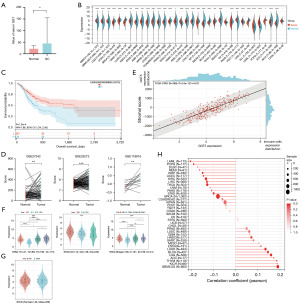
Table 2
| Variable | GC patients (n=124) | Healthy controls (n=287) | P value |
|---|---|---|---|
| GGT (U/L) | 44.48±111.48 | 21.62±14.9 | 0.02* |
| Age (years) | 60.17±11.21 | 57.80±10.08 | 0.50 |
| Gender (cases) | |||
| Male | 76 | 156 | |
| Female | 48 | 131 | 0.19 |
GGT and age were expressed as mean ± standard deviation, gender was expressed as number of cases. And n represented the number of cases in the GC patients or healthy controls. GGT and age were analyzed by using independent t-tests to assess differences between groups. Gender was analyzed by using Chi-squared tests to assess differences between groups. *, P<0.05. GC, gastric cancer; GGT, gamma-glutamyl transferase.
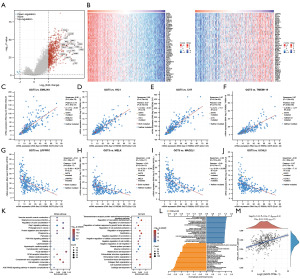
Through differential expression analysis, 806 differentially expressed genes were finally obtained, of which 802 were upregulated and 4 were downregulated in GC. Fold change and corrected P values were used for volcano plots, with red dots indicating significantly up-regulated genes, blue dots representing the genes with significant difference down, and gray dots indicating insignificant genes (Figure 2A). As the Figure 2B-2J shows, EMILIN1, HIC1, C1R and TMEM119 etc. were positively correlated genes with GGT5, LRPPRC, MELK, MAD2L1 and UCHL5 etc. were negatively correlated genes with GGT5 (Figure 2B-2J). Functional enrichment analysis of differential genes showed that the molecular functions of differential gene products were mainly related to vascular smooth muscle contraction, regulation of vascular development and angiogenesis, regulation of actin cytoskeleton, cell adhesion molecules, regulation of cell motility and migration, and extracellular matrix (ECM) organization (Figure 2K-2L). At the same time, the expression of GGT5 was found to be slightly positively correlated with the PI3K/AKT pathway (Figure 2M).
Low expression of GGT5 reduced GC cell proliferation
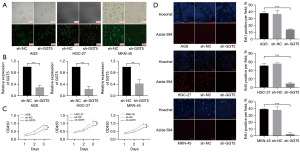
To identify the role of GGT5 on GC, first, we detected transfection efficiency after transfection of sh-NC and sh-GGT5 into GC cells. The fluorescence efficiency of transfection was observed by fluorescence microscopy to reach 75%, and then unstable cells were screened by puromycin. Subsequently, PCR revealed that GGT5 expression was significantly reduced by transfection with sh-GGT5 in GC cells (Figure 3A,3B). Next, to determine whether GGT5 could affect GC cell proliferation, EdU and CCK8 assays were performed. The results showed that the proliferation capacity was inhibited in sh-GGT5 GC cells (Figure 3C,3D).
Low expression of GGT5 inhibited GC cell proliferation by downregulating the PI3K/AKT-MAPK signaling pathway
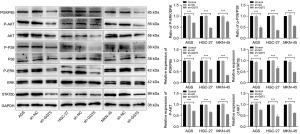
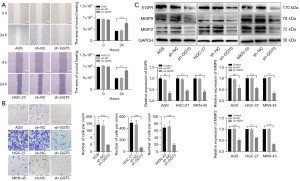
To further elucidate the detailed mechanisms of sh-GGT5 reduced proliferation in GC cells, PI3K/AKT-MAPK pathways were investigated. As expected, treatment with sh-GGT5 significantly decreased phosphorylation of P38 and ERK compared to the control group (P<0.001). At the same time, western blotting results showed that the expression of AKT, P-AKT and PI3KP85 protein was reduced (P<0.001) in GC cells when transfected with sh-GGT5. Furthermore, expression of the nuclear transcription factor STAT5b (P<0.001) was inhibited after GGT5 knockdown (Figure 4). The results evidently demonstrated low expression of GGT5 could affect the GC progression by regulating the PI3K/AKT‑MAPK signaling pathway.
GGT5 knockdown inhibited GC cell migration
Next, the functionalities of GGT5 on GC cell migration were investigated. As expected, scratch test results showed that GGT5 knockdown abrogated the migration capacity of GC cells (P<0.01). In addition, the Transwell assay was used to measure cell migration again. The results provided evidence that was consistent with the provided scratch test results (P<0.001) (Figure 5A,5B). To confirm the influence of GGT5 on MMPs signaling in GC cells, western blotting showed that expression of MMP2 (P<0.001), MMP9 (P<0.001) and EGFR (P<0.01) decreased when GGT5 was knocked down (Figure 5C). These results suggested that GGT5 promoted GC migration by activating the MMPs signaling pathway.
Discussion
GC is the fourth most common cancer in men and the seventh most common cancer in women. Early GC can be treated with endoscopic mucosal resection or endoscopic submucosal dissection for a good long-term prognosis (22,23). Improvements in hygiene and the elimination of H. Pylori have significantly lowered global statistics of GC, but there is still a long way to improve survival rates for advanced and metastatic GC (24). Carcinoembryonic antigen (CEA) and carbohydrate antigen 199 (CA199) are frequently used in GC diagnosis and prognosis. However, existing circulating biomarkers have showed low sensitivity and specificity (25). Therefore, it is important to find an effective therapeutic target and new biomarkers.
The GGT gene belongs to a multi-gene family and is involved in the occurrence and development of various malignant tumors (6,9,10). GGT1 and GGT5 are the only two extracellular enzymes in the GGT family that can cleave the γ-glutamyl bond. They produce precursor amino acids such as cysteine and cystine by hydrolyzing γ-glutamyl substrates, which are used to maintain the γ-glutamyl cycle in tumor cells and thereby promote the occurrence and development of tumors (26). Song et al. found that GGT5 may affect the trait of GGT (27). This study found that the preoperative serum GGT value of GC patients was higher than that of the healthy control group (Figure 1A). Analysis in the TCGA datasets revealed that GGT5 was highly expressed in GC tissues (Figure 1B). Therefore, this study focused on the functional study of GGT5 in GC.
GGT5 has been proved to be a catalytically active GGT enzyme, and is involved in regulating the body’s functions, including reoxidation, drug metabolism and immune functions (11,12). When there is unregulated uptake of amino acid metabolic enzymes and changes in metabolite-driven genes, it can affect the occurrence and development of tumors (28,29). GGT5 is mainly involved in the extracellular catabolism of GSH, hydrolyzing it to glutamic acid, cysteine and glycine (30). Tumor cells cannot directly absorb GSH, but rely on high concentrations of cysteine and cystine in serum and interstitial fluid to maintain intracellular GSH levels. Therefore, the cysteine generated by GGT5 decomposing GSH becomes an important raw material required for tumor cells to synthesize GSH (26). Meanwhile, GSH also plays a role in cell cycle regulation and cell signal transduction (31,32). Studies have shown that GSH has an inhibitory effect on PI3K/AKT pathway (33), and its expression level may decrease with the high expression of GGT5 in GC, while the decrease of GSH level will cause the activation of p38MAPK and AKT (34,35). Gene enrichment analysis showed that GGT5 overexpression may be closely related to MAPK pathway and PI3K/AKT pathway (36,37). In addition, GGT5 can convert LeukotrienceC4 into active leukotrienceD4, thus playing an important role in tumor immunity (38). Overexpression of GGT5 in tumor tissues is positively correlated with PD-L1 expression and CD8 T cell infiltration (39), and overexpression of GGT5 in follicular dendritic cells can impair the ability of P2RY8 to enhance B cell restriction of germinal center (40). Moreover, the balance between GGT5 and oxidative stress cascade regulation plays a key role in steroid production (11). In summary, GGT5 can regulate the occurrence and development of tumors through pathways such as the metabolism of GSH, immune infiltration, and signal pathways. GGT5, as an oncogene, was activated in breast cancer (41), lung cancer (13,42,43) and liver cancer (2,44), causing the enhancement of tumor cell proliferation and invasion. However, the mechanism of GGT5 in GC was still unclear. Therefore, this study mainly explored the mechanism of GGT5 in GC, and provided a basis for GGT5 to be used as a diagnostic target for GC.
In our work, we further found that GGT5 was indeed highly expressed in GC through GEO datasets. In addition, bioinformatics analysis showed that increased GGT5 expression levels would lead to poorer outcomes in GC patients and was positively correlated with immune infiltration of GC (Figure 1C-1E). The study also investigated the relationship between GGT5 expression and clinicopathological parameters of GC, and the results showed that GGT5 expression increased significantly in advanced GC tumor specimens, which was correlated with the T stage, grade and stage of GC, and was independent of age and sex (Figure 1F-1H). Our work explored the mechanism of GGT5 in GC cells. We found that low expression of GGT5 could inhibit the growth and migration of GC cells and alter the biological characteristics of GC cells by CCK-8, EdU, scratch and Transwell experiments (Figures 3C,3D,5A,5B). PI3K/AKT signaling may promote cell survival, which have been identified as drivers of oncogenesis and cancer progression, and MAPKs are also important pathways in regulating tumor development (45-47). The results were shown in Figure 4, GGT5 knockdown significantly decreased AKT, P-AKT, PI3KP85 levels as previously described (Figure 2E) and reduced phosphorylation of P38 and ERK compared to the control group.
The tumor microenvironment (TME) of GC is composed of ECM, fibroblasts, endothelial cells, mesenchymal stem cells, macrophages, lymphocytes, neutrophils and other cell components (48). The stomach has a strong acidic environment and a unique endocrine system, which also makes the TME of GC distinctive (49). In GC, the characteristics of stromal cells are associated with the deterioration of patient survival rate and treatment resistance, and promote tumor invasion through activating stromal remodeling, immune crosstalk, metabolic effects, and soluble secreted factors (50,51). ECM and matrix together constitute the first barrier in the process of tumor metastasis, and its degradation is a key link in tumor invasion and metastasis. Studies have shown that the matrix metalloproteinases (MMPs), as important enzymes that can degrade ECM, play an important role in mediating tumor angiogenesis, metastasis and invasion (52,53). Western blotting showed that MMP2, MMP9, EGFR were inhibited by knockdown of GGT5 (Figure 5C). Therefore, GGT5 knockdown could regulate MMPs protein expression and inhibit GC cell metastasis through the ECM of the TME.
By differential expression gene analysis, we concluded that the GGT5 gene might act as a prognostic biomarker in GC (Figure 2A). Co-expressed EMILIN1, HIC1, C1R, and TMEM119 genes were positively correlated with GGT5 and had been associated with tumor growth and poor cancer prognosis (54-57). Co-expressed genes LRPPRC, UCHL5, MAD2L1, and MELK negatively correlated with GGT5 might be novel therapeutic targets for GC in the future (58-62) (Figure 2B-2J). Angiogenesis is required for aggressive tumor growth and metastasis (63), impairment of PI3K and MAPK signaling pathways may affect hematological parameters (64), MMPs may also contribute to the formation of the premetastatic microenvironment by secreting proangiogenic factors (65). To investigate the probable biological processes of GGT5, we performed GO and KEGG analyses on the GGT5 gene. Enrichment analysis of the GGT5 gene showed that the regulation of angiogenesis, cell motility and migration, cell adhesion molecules, ECM, and other related processes were significantly enriched, indicating that GGT5 was associated with metastasis and invasion of GC (Figure 2C,2D).
The study found that high levels of GGT5 in CAFs of lung adenocarcinoma contribute to cancer cell survival and drug resistance, indicating that GGT5 may be a promising therapeutic target for lung adenocarcinoma (13). Nevertheless, the role of GGT5 in the treatment resistance of GC still needs to be further investigation, with the aim of providing a theoretical basis for the development of GGT5 as a therapeutic target for GC. It has been reported that cell surface receptors transduced signals from ECM into cells, which may govern a variety of cellular processes including survival, growth, migration, and differentiation (66). A recent review showed that the ECM may be involved in deregulation of pre-malignant lesions and GC (67). Therefore, the role of GGT5 in EMT needs further study. In addition, the competing endogenous RNA network has been extensively studied in recent years (68,69), so we will further investigate the mechanism of GGT5 in GC to further elucidate the role of GGT5 in GC.
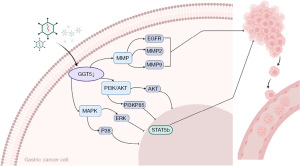
In summary, the potential function of GGT5 in GC was explored in our study. We concluded that GGT5 knockdown might inhibit GC progression by targeting PI3K/AKT-MAPK-MMPs pathways (Figure 6), and proposed that GGT5 might be a novel target for GC therapy.
Conclusions
In this study, we investigated the expression and function of GGT5 in GC and the underlying mechanism of GGT5 in tumor progression. Knockdown of GGT5 expression might inhibit GC cell proliferation and migration by inhibiting PI3K/AKT-MAPK-MMP pathways. GGT5 might be a potential therapeutic target for GC.
Acknowledgments
We acknowledge the Key Laboratory of Digestive Pathophysiology of Zhejiang Province and the Department of Cell and Immunology for providing GC cell lines.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2222/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2222/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2222/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2222/coif). W.C. reports receiving funding from Natural Science Foundation of Zhejiang Province (No. LQ19H160044). Z.Z. reports receiving funding from Zhejiang Province Traditional Chinese Medicine Science and Technology Project (No. 2023ZL056). Jian Xu reports receiving funding from Natural Science Foundation of Zhejiang Province (No. LGF21H160016). The other authors have no conflicts of interests to disclose.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Tian S, Li J, Guo Y, et al. Expression Status and Prognostic Significance of Gamma-Glutamyl Transpeptidase Family Genes in Hepatocellular Carcinoma. Front Oncol 2021;11:731144. [Crossref] [PubMed]
- Yu Y, Fan Y, Yang Z, et al. Elevated serum gamma-glutamyltransferase predicts advanced histological liver damage in chronic hepatitis B. Discov Med 2016;21:7-14. [PubMed]
- Mok Y, Son DK, Yun YD, et al. γ-Glutamyltransferase and cancer risk: The Korean cancer prevention study. Int J Cancer 2016;138:311-9. [Crossref] [PubMed]
- Xu XS, Miao RC, Zhang LQ, et al. Model Based on Alkaline Phosphatase and Gamma-Glutamyltransferase for Gallbladder Cancer Prognosis. Asian Pac J Cancer Prev 2015;16:6255-9. [Crossref] [PubMed]
- Xiao Y, Yang H, Lu J, et al. Serum gamma-glutamyltransferase and the overall survival of metastatic pancreatic cancer. BMC Cancer 2019;19:1020. [Crossref] [PubMed]
- Grimm C, Hofstetter G, Aust S, et al. Association of gamma-glutamyltransferase with severity of disease at diagnosis and prognosis of ovarian cancer. Br J Cancer 2013;109:610-4. [Crossref] [PubMed]
- Heisterkamp N, Groffen J, Warburton D, et al. The human gamma-glutamyltransferase gene family. Hum Genet 2008;123:321-32. [Crossref] [PubMed]
- Hanigan MH. gamma-Glutamyl transpeptidase, a glutathionase: its expression and function in carcinogenesis. Chem Biol Interact 1998;111-112:333-42. [Crossref] [PubMed]
- Wickham S, West MB, Cook PF, et al. Gamma-glutamyl compounds: substrate specificity of gamma-glutamyl transpeptidase enzymes. Anal Biochem 2011;414:208-14. [Crossref] [PubMed]
- Li W, Wu ZQ, Zhang S, et al. Augmented expression of gamma-glutamyl transferase 5 (GGT5) impairs testicular steroidogenesis by deregulating local oxidative stress. Cell Tissue Res 2016;366:467-81. [Crossref] [PubMed]
- Gallman AE, Wolfreys FD, Nguyen DN, et al. Abcc1 and Ggt5 support lymphocyte guidance through export and catabolism of S-geranylgeranyl-l-glutathione. Sci Immunol 2021;6:eabg1101. [Crossref] [PubMed]
- Wei JR, Dong J, Li L. Cancer-associated fibroblasts-derived gamma-glutamyltransferase 5 promotes tumor growth and drug resistance in lung adenocarcinoma. Aging (Albany NY) 2020;12:13220-33. [Crossref] [PubMed]
- Li X, Chen S, Zhu Y, et al. Comprehensive bioinformatics analyses identified Homeobox B9 as a potential prognostic biomarker and therapeutic target for gastric cancer. J Gastrointest Oncol 2021;12:2132-49. [Crossref] [PubMed]
- Wen F, Huang J, Lu X, et al. Identification and prognostic value of metabolism-related genes in gastric cancer. Aging (Albany NY) 2020;12:17647-61. [Crossref] [PubMed]
- Zhao Q, Zhao R, Song C, et al. Increased IGFBP7 Expression Correlates with Poor Prognosis and Immune Infiltration in Gastric Cancer. J Cancer 2021;12:1343-55. [Crossref] [PubMed]
- Shen W, Song Z, Zhong X, et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. Imeta 2022;1:e36. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Shahidi N, Bourke MJ. ESD, not EMR, should be the first-line therapy for early gastric neoplasia. Gut 2020;69:1-2. [Crossref] [PubMed]
- Ko WJ, Song GW, Kim WH, et al. Endoscopic resection of early gastric cancer: current status and new approaches. Transl Gastroenterol Hepatol 2016;1:24. [Crossref] [PubMed]
- Sexton RE, Al Hallak MN, Diab M, et al. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev 2020;39:1179-203. [Crossref] [PubMed]
- Necula L, Matei L, Dragu D, et al. Recent advances in gastric cancer early diagnosis. World J Gastroenterol 2019;25:2029-44. [Crossref] [PubMed]
- Lewerenz J, Hewett SJ, Huang Y, et al. The cystine/glutamate antiporter system x(c)(-) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal 2013;18:522-55. [Crossref] [PubMed]
- Song H, Li W, Li Y, et al. Genome-wide association study of 17 serum biochemical indicators in a chicken F(2) resource population. BMC Genomics 2023;24:98. [Crossref] [PubMed]
- Peeters R, Hubens A. The mesh skin graft--true expansion rate. Burns Incl Therm Inj 1988;14:239-40. [Crossref] [PubMed]
- Pavlova NN, Zhu J, Thompson CB. The hallmarks of cancer metabolism: Still emerging. Cell Metab 2022;34:355-77. [Crossref] [PubMed]
- Hanigan MH. Gamma-glutamyl transpeptidase: redox regulation and drug resistance. Adv Cancer Res 2014;122:103-41. [Crossref] [PubMed]
- Pallardó FV, Markovic J, García JL, et al. Role of nuclear glutathione as a key regulator of cell proliferation. Mol Aspects Med 2009;30:77-85. [Crossref] [PubMed]
- Dalle-Donne I, Rossi R, Giustarini D, et al. S-glutathionylation in protein redox regulation. Free Radic Biol Med 2007;43:883-98. [Crossref] [PubMed]
- Ting Hao W, Huang L, Pan W, et al. Antioxidant glutathione inhibits inflammation in synovial fibroblasts via PTEN/PI3K/AKT pathway: An in vitro study. Arch Rheumatol 2021;37:212-22. [Crossref] [PubMed]
- Chen X, Shi X, Gan F, et al. Glutamine starvation enhances PCV2 replication via the phosphorylation of p38 MAPK, as promoted by reducing glutathione levels. Vet Res 2015;46:32. [Crossref] [PubMed]
- Kim SM, Park JG, Baek WK, et al. Cadmium specifically induces MKP-1 expression via the glutathione depletion-mediated p38 MAPK activation in C6 glioma cells. Neurosci Lett 2008;440:289-93. [Crossref] [PubMed]
- Huang Y, Zhou H, Zou J, et al. GGT5 Is an Independent Prognostic Biomarker in Stomach Adenocarcinoma. Can J Gastroenterol Hepatol 2022;2022:9983351. [Crossref] [PubMed]
- Wang Y, Fang Y, Zhao F, et al. Identification of GGT5 as a Novel Prognostic Biomarker for Gastric Cancer and its Correlation With Immune Cell Infiltration. Front Genet 2022;13:810292. [Crossref] [PubMed]
- Hanigan MH, Gillies EM, Wickham S, et al. Immunolabeling of gamma-glutamyl transferase 5 in normal human tissues reveals that expression and localization differ from gamma-glutamyl transferase 1. Histochem Cell Biol 2015;143:505-15. [Crossref] [PubMed]
- Liu C, Tao Y, Lin H, et al. Classification of stomach adenocarcinoma based on fatty acid metabolism-related genes frofiling. Front Mol Biosci 2022;9:962435. [Crossref] [PubMed]
- Lu E, Wolfreys FD, Muppidi JR, et al. S-Geranylgeranyl-L-glutathione is a ligand for human B cell-confinement receptor P2RY8. Nature 2019;567:244-8. [Crossref] [PubMed]
- Passarelli MC, Pinzaru AM, Asgharian H, et al. Leucyl-tRNA synthetase is a tumour suppressor in breast cancer and regulates codon-dependent translation dynamics. Nat Cell Biol 2022;24:307-15. [Crossref] [PubMed]
- Serizawa M, Kusuhara M, Zangiacomi V, et al. Identification of metabolic signatures associated with erlotinib resistance of non-small cell lung cancer cells. Anticancer Res 2014;34:2779-87. [PubMed]
- Lukic A, Ji J, Idborg H, et al. Pulmonary epithelial cancer cells and their exosomes metabolize myeloid cell-derived leukotriene C4 to leukotriene D4. J Lipid Res 2016;57:1659-69. [Crossref] [PubMed]
- Xu YJ, He MK, Liu S, et al. Construction of a single nucleotide variant score-related gene-based prognostic model in hepatocellular carcinoma: analysis of multi-independent databases and validation in vitro. Cancer Cell Int 2021;21:610. [Crossref] [PubMed]
- Yap TA, Omlin A, de Bono JS. Development of therapeutic combinations targeting major cancer signaling pathways. J Clin Oncol 2013;31:1592-605. [Crossref] [PubMed]
- Braicu C, Buse M, Busuioc C, et al. A Comprehensive Review on MAPK: A Promising Therapeutic Target in Cancer. Cancers (Basel) 2019;11:1618. [Crossref] [PubMed]
- Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer 2020;20:74-88. [Crossref] [PubMed]
- Kang B, Camps J, Fan B, et al. Parallel single-cell and bulk transcriptome analyses reveal key features of the gastric tumor microenvironment. Genome Biol 2022;23:265. [Crossref] [PubMed]
- Liu Y, Li C, Lu Y, et al. Tumor microenvironment-mediated immune tolerance in development and treatment of gastric cancer. Front Immunol 2022;13:1016817. [Crossref] [PubMed]
- Shen J, Zhai J, You Q, et al. Cancer-associated fibroblasts-derived VCAM1 induced by H. pylori infection facilitates tumor invasion in gastric cancer. Oncogene 2020;39:2961-74. [Crossref] [PubMed]
- Sahai E, Astsaturov I, Cukierman E, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 2020;20:174-86. [Crossref] [PubMed]
- He L, Kang Q, Chan KI, et al. The immunomodulatory role of matrix metalloproteinases in colitis-associated cancer. Front Immunol 2023;13:1093990. [Crossref] [PubMed]
- Asgari R, Vaisi-Raygani A, Aleagha MSE, et al. CD147 and MMPs as key factors in physiological and pathological processes. Biomed Pharmacother 2023;157:113983. [Crossref] [PubMed]
- Qi Y, Lv J, Liu S, et al. TSPAN9 and EMILIN1 synergistically inhibit the migration and invasion of gastric cancer cells by increasing TSPAN9 expression. BMC Cancer 2019;19:630. [Crossref] [PubMed]
- Li Y, Yao M, Wu T, et al. Loss of hypermethylated in cancer 1 (HIC1) promotes lung cancer progression. Cell Signal 2019;53:162-9. [Crossref] [PubMed]
- Viiklepp K, Nissinen L, Ojalill M, et al. C1r Upregulates Production of Matrix Metalloproteinase-13 and Promotes Invasion of Cutaneous Squamous Cell Carcinoma. J Invest Dermatol 2022;142:1478-1488.e9. [Crossref] [PubMed]
- Zheng P, Wang W, Ji M, et al. TMEM119 promotes gastric cancer cell migration and invasion through STAT3 signaling pathway. Onco Targets Ther 2018;11:5835-44. [Crossref] [PubMed]
- Li X, Lv L, Zheng J, et al. The significance of LRPPRC overexpression in gastric cancer. Med Oncol 2014;31:818. [Crossref] [PubMed]
- Arpalahti L, Laitinen A, Hagström J, et al. Positive cytoplasmic UCHL5 tumor expression in gastric cancer is linked to improved prognosis. PLoS One 2018;13:e0193125. [Crossref] [PubMed]
- Zhang Z, Hu X, Kuang J, et al. LncRNA DRAIC inhibits proliferation and metastasis of gastric cancer cells through interfering with NFRKB deubiquitination mediated by UCHL5. Cell Mol Biol Lett 2020;25:29. [Crossref] [PubMed]
- Wang Y, Wang F, He J, et al. miR-30a-3p Targets MAD2L1 and Regulates Proliferation of Gastric Cancer Cells. Onco Targets Ther 2019;12:11313-24. [Crossref] [PubMed]
- Calcagno DQ, Takeno SS, Gigek CO, et al. Identification of IL11RA and MELK amplification in gastric cancer by comprehensive genomic profiling of gastric cancer cell lines. World J Gastroenterol 2016;22:9506-14. [Crossref] [PubMed]
- Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol 2002;29:15-8. [Crossref] [PubMed]
- Sun G, Liang X, Qin K, et al. Functional Analysis of KIT Gene Structural Mutations Causing the Porcine Dominant White Phenotype Using Genome Edited Mouse Models. Front Genet 2020;11:138.
- Peinado H, Lavotshkin S, Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin Cancer Biol 2011;21:139-46. [Crossref] [PubMed]
- Theocharis AD, Skandalis SS, Gialeli C, et al. Extracellular matrix structure. Adv Drug Deliv Rev 2016;97:4-27. [Crossref] [PubMed]
- Moreira AM, Pereira J, Melo S, et al. The Extracellular Matrix: An Accomplice in Gastric Cancer Development and Progression. Cells 2020;9:394. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]


