
PA-MSHA exerts potent activity against cetuximab-resistant colorectal cancer through the miR-7-5p/Akt3/Wnt-β-catenin pathway
Highlight box
Key findings
• Pseudomonas aeruginosa-mannose sensitive hemagglutinin (PA-MSHA) exerts potent activity against cetuximab-resistant colorectal cancer (CRC) in vitro and in vivo, indicating that PA-MSHA shows therapeutic promise for cetuximab-resistant CRC.
What is known and what is new?
• The prognosis and survival of individuals with cetuximab-resistant CRC remain severely impacted by therapy for this disease.
• In the present study, we revealed that PA-MSHA exerted great anticancer effects on cetuximab-resistant CRC by regulating the miR-7-5p/Akt3/Wnt-β-catenin pathway.
What is the implication, and what should change now?
• The results of our study will establish a theoretical framework for understanding the miRNA-mRNA regulatory gene network and treatment efficacy of PA-MSHA in cetuximab-resistant CRC. Other target genes of miR-7-5p possibly play a role in mediating the anticancer activities of PA-MSHA, and further research is necessary in areas that remain unresolved for future investigation.
Introduction
Colorectal cancer (CRC) is a prevalent malignancy that contributes significantly to global mortality rates (1,2). CRC’s high mortality rate is attributed in part to the fact that patients are often diagnosed at a late stage of the disease, which further leads to recurrence in a short time (2,3). The available information indicates that approximately 25% of patients exhibit metastasis at the time of diagnosis, whereas roughly 50% of patients will ultimately experience the development of metastatic CRC (4). Currently, notable advancements have been made in metastatic CRC treatment, such as targeted therapy (5). Cetuximab, a targeted medicine approved by Food and Drug Administration (FDA), is commonly employed as a conventional treatment for the majority of patients with advanced CRC (6,7). However, patients harboring primary or acquired kirsten rat sarcoma viral oncogene (KRAS) and B-Raf proto-oncogene (BRAF) mutations are resistant to cetuximab (8). In addition, the patients without common mutations who receive cetuximab monotherapy will develop resistance to the drug in 4 months (9). Thus, there is a high demand for developing effective therapeutic approaches to treat cetuximab-resistant CRC.
Bacterization has emerged as a potentially effective approach in the field of cancer therapy (10). Several bacterial toxins, such as denileukin diftitox (11), leukotoxin (12) and LukS-PV (the S component of Panton Valentineleukocidin secreted by Staphylococcus aureus) (13), have demonstrated the ability to specifically eliminate cancerous cells. A strain of Pseudomonas aeruginosa called Pseudomonas aeruginosa-mannose sensitive hemagglutinin (PA-MSHA) that carries MSHA fimbriae has been licensed for use as a medicinal biological product in China (14). To date, the utilization of PA-MSHA has been extensively documented in the realm of anticancer therapies, including bladder (15), non-small cell lung (16), breast (17) and cervical cancers (18). However, there is a lack of knowledge regarding the possible effects of PA-MSHA on cetuximab-resistant CRC. This research aims to evaluate the anticancer activities of PA-MSHA in cetuximab-resistant CRC and elucidate the relevant functional mechanisms. We present this article in accordance with the MDAR and ARRIVE reporting checklists (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2211/rc).
Methods
Cell lines, materials, and antibodies
The human CRC cell lines were acquired from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), including HCT116, HT-29 and SW480. Among them, the HT-29 cell line was BRAF mutated, and the HCT116 and SW480 cell lines carried mutations in the KRAS gene (19,20). The HCT116-R cell line, which exhibits resistance to cetuximab, was generated through the use of extracorporeal shock in our laboratory (21). HCT116 cells were grown in medium containing cetuximab at a dose of 20 µg/mL for 48 hours. Subsequently, the adherent cells were grown in cetuximab-free medium for a period of time, allowing for their progressive recovery, which took 24 hours. Following one generation, the cell state was reinstated, and the drug, specifically 20 µg/mL cetuximab, was subsequently delivered once again. The aforementioned technique was replicated in approximately eight instances. Subsequently, the HCT116-R cell line was successfully developed. The cells were cultivated in Roswell Park Memorial Institute (RPMI) 1640 media or Dulbecco’s modified eagle medium (DMEM), both of which were supplemented with 10% fetal bovine serum (FBS) from Gibco. The culture conditions were maintained at 37 ℃ with a 5% CO2.
PA-MSHA was donated, diluted, and kept at 4 ℃ thanks to Wanter Biopharma Company (Beijing, China). Primary rabbit or mouse antibodies, including anti-nonphosphorylated β-catenin (#19807), anti-E-cadherin (#14472), anti-survivin (#2808), anti-Akt3 (#4059) and anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (#2118), were purchased from Cell Signaling Technology (CST, Massachusetts, USA).
MiR-7-5p transfection
Lentiviral vectors, namely LV-miR-7-5p-mimic, LV-miR-7-5p-inhibitor and LV-miRNA-NC (negative control) (GenePharma, Shanghai, China) were transfected into HCT116-R cells. Briefly, one day before transfection, cells were seeded at a density of 2×105 cells/mL in six-well plates. Following the manufacturer’s protocol, the miR-7-5p mimic, miR-7-5p inhibitor and their respective NC vectors were transfected into cells. After transfection, the cells were subjected to PA-MSHA treatment.
Coculture assays
To examine the function of Akt3, HCT116-R cells were transfected with miR-7-5p inhibitor vector and subsequently grown in 24-well plates. Next, the cells were transfected with small interfering RNA (siRNA) specifically designed to target Akt3 utilizing Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, USA) according to the guidelines provided by the manufacturer. Within a 24-hour period, a concentration of 0.3×109/mL PA-MSHA was given to each of the experimental groups. Finally, the cells were harvested at 48 h and subjected to apoptosis, migration, invasion and western blot assays.
Cell proliferation analysis
In 96-well plates (100 µL per well), cells were spread out at a density of 8×104/mL. To investigate the sensitivity of cetuximab, cells were exposed to 0, 0.01, 0.5, 1, 5, 10 and 20 µg/mL cetuximab. To explore the effect of PA-MSHA, cells were treated with 0, 0.01, 0.1, 0.2, 0.3, 0.4 and 0.6×109/mL PA-MSHA. Then, the cells were incubated at 37 ℃ for 24 and 48 hours in a humidified 5% CO2 environment. Cell Counting Kit-8 (CCK-8) was used to measure cell proliferation. Each statistical analysis was performed three times.
Cell apoptosis analysis
Cells were placed into six-well plates at a density of 2×105/mL. After being treated with PA-MSHA for 48 hours, the cells were stained with Annexin V/Propidium Iodide (PI) to check for apoptosis (BB-4101-3, Beibo, Shanghai, China). Cells were collected by centrifugation at 1,500 rpm for 5 minutes and washed twice in cold phosphate buffered solution (PBS). Then, 5 µL Annexin V-Fluorescein Isothiocyanate (FITC) and 10 µL PI were applied to the cells, and they were stained for 15 minutes at room temperature in the absence of light. Finally, flow cytometry was implemented to assess cell apoptosis (Navious 10 COLORS/3 LASER, Beckman Coulter, Brea, USA). The experiments were conducted in triplicate. The data were analyzed with FlowJoV10 software.
Cell migration and invasion analyses
In six-well plates, cells were disseminated out at an average density of 2×105/mL and stimulated with PA-MSHA. The cells were gathered after 48 hours to perform migration and invasion assays. To test cell migration, wound-healing analysis was carried out. Artificial wounds were created in a continuous layer of cells that were cultivated in medium devoid of FBS. Photographs were captured 24 hours following the establishment of the wound.
Transwell analysis was conducted to assess cell invasion. The cells were suspended in 200 µL of medium lacking FBS and were then placed into the upper chamber together with 80 µL of Matrigel (Millipore, Billerica, USA) solution at a dose of 1 mg/mL. Subsequently, 600 µL of medium containing 10% FBS was placed in the lower chamber as the nutritional attractant. After 48 hours, the cells adhered to the lower surface of the chamber were fixed for 30 minutes with 20% methanol and dyed for 20 minutes with 0.1% crystal violet. We counted the cells in 3 different fields of vision, and the results were then used to determine the average number of cells. Every experiment was repeated three times.
MiRNA isolation and quantitative real time polymerase chain reaction (qRT-PCR) analysis
The manufacturer’s protocol was followed to isolate total RNA from cells or serum samples. The A260/A280 ratio was measured by Qubit 4.0 (Life Technologies, USA) to quantify the extracted RNA. MiRNA levels were measured using the miRNA qPCR Primer Set (RiboBio, T1027) according to the manufacturer’s instructions. The Roche LC480 system (Roche, Germany) was employed to perform qRT-PCR, with an initial incubation at 95 ℃ for 10 minutes, followed by 40 cycles of 95 ℃ for 15 seconds and 60 ℃ for 1 minute. The U6 RNA levels for cells and cel-miR-39 levels for serum samples (22,23) served as internal controls to ensure the accuracy of the data. The expression levels of miRNAs were compared using the 2−ΔΔCt method. There were three separate runs of each analysis. The specific primers of miRNAs are shown in Table 1.
Table 1
| miRNAs | Forward primers (5′-3′) | Reverse primers (5′-3′) |
|---|---|---|
| miR-7-5p | TGGAAGACTAGTGATTTT | GTGCAGGGTCCGAGGT |
| miR-3529-3p | GGCACCATTAGGTAGACTGG | CCCCATCTGGAAGACTAGTG |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| cel-miR-39 | GGCCTCACCGGGTGTAAATCAG | AGTGCAGGGTCCGAGGTAT |
qRT-PCR, quantitative real time polymerase chain reaction.
Western blot analysis
Following 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) separation, proteins were transferred to 0.45 µm polyvinylidene fluoride (PVDF) membranes (Amersham, UK). Next, the membranes were blocked with 5% milk-PBST (Phosphate Buffered Saline with Tween 20). They were then incubated overnight with primary antibodies against β-catenin, E-cadherin, Survivin, Akt3, and GAPDH at the right concentrations. After that, they were incubated for 1 hour with a secondary antibody attached to horseradish peroxidase (1:2,000). Images were taken using an LAS-4000 system (Olympus, Tokyo, Japan) following incubation with an enhanced chemiluminescence reagent (Life Technologies, NY, USA).
MiRNA sequencing and data analysis
HCT116-R cells treated with 0, 0.3 or 0.4×109/mL PA-MSHA for 48 h were collected for miRNA sequencing. A quantity of three micrograms of total RNA was used for library production, using a previously documented protocol with minor adjustments. The libraries that had undergone purification were subjected to sequencing using an Illumina HiSeq 2000 platform, which was performed by Annoroad Genome, a company headquartered in Beijing, China. In summary, the raw data were processed using Python scripts to assure data quality. Subsequently, the data were filtered based on Q30 statistics. We used Bowtie (v1.01) to map the clean data to the Ensemble library (GRCh37) (http://grch37.ensembl.org/index.html) and then compared it with miRBase to find mature miRNAs. The miRNA counts and RPM (reads per million total reads) values were gathered for each sample. DEGseq software version 1.18.0 was employed to perform differential expression analysis. The criteria utilized to identify differentially expressed miRNAs (DEMs) were a false discovery rate (FDR) <0.05 and |log2-fold change (FC)| ≥1.
MiR-7-5p target prediction and luciferase reporter analysis
The TargetScan and miRTarBase databases were used for the prediction of miR-7-5p targets. Luciferase analysis was performed by transfecting HEK 293T cells with miR-7-5p mimic, empty miR nonspecific control (NC), wild-type and mutant Akt3 3’-UTR (untranslated region) plasmids using Lipofectamine 2000 (Invitrogen, USA). A dual-luciferase analysis device (Promega, Madison, WI, USA) was used to measure luciferase activity 48 hours after transfection. Luminescence readings were obtained with the help of a Flexstation 3 Multiscan Spectrum (Molecular Devices, Sunnyvale, CA, USA).
In vivo efficacy of PA-MSHA
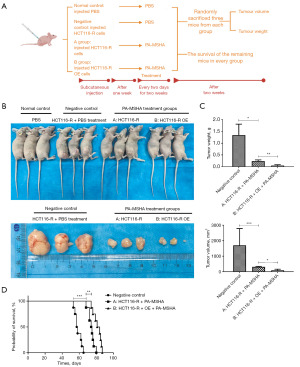
Female BALB/c nude mice (4–5 weeks old) were procured from the animal center of the University of Science and Technology of China and housed in a specific pathogen-free facility located at the Lab Animal Center of the First Affiliated Hospital of University of Science and Technology of China. A protocol was prepared before the study without registration. Animal experiments were performed under a project license (No. 2024-N (A)-158) granted by the Ethics Committee of the First Affiliated Hospital of the University of Science and Technology of China, in compliance with national guidelines for the care and use of animals. After a week of getting used to their fresh surroundings, 44 mice were randomly split into four groups, with 11 mice in each. These groups were the normal control, the negative control, the A group, and the B group. The mice in the normal control group received an injection of PBS and were administered PBS therapy. Tumors were initiated by subcutaneous injection (s.c.) of 2×106 HCT116-R cells (in 100 µL PBS) into the right lower flanks of the nude mice in the negative control and A groups. The mice in the B group were subcutaneously injected with 2×106 miR-7-5p-overexpressing HCT116-R cells. Seven days later, the mice were treated by peritumor injection every two days for two weeks according to the following protocols: (I) normal and negative groups: 0.1 mL PBS, (II) A and B groups: 0.1 mL 1.0×1010/mL PA-MSHA.
Two weeks after treatment, 3 randomly selected mice from each group were sacrificed by cervical dislocation without any anesthetics used, and their tumor samples were collected. Tumor volume was determined using the formula 1/2 × larger diameter × (smaller diameter)2. Tumor weight was also measured. The survival of each group of remaining mice was measured from the start of therapy to death. The death state is when the mouse cannot move, does not respond to gentle stimulation, or the body becomes rigid. The assessment of survival benefit was conducted using Kaplan-Meier analysis.
Clinical CRC patient samples
From January 2017 through July 2020, 96 patients with CRC were recruited at random from the Oncology Department of Ward 1 at the Western Branch of the First Affiliated Hospital of the University of Science and Technology of China (Hefei, Anhui, China) to provide serum samples. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All participants provided informed consent, and the study was authorized by the Ethics Committee of The First Affiliated Hospital of the University of Science and Technology of China (No. 2019-KY-28). The inclusion criteria were as follows: (I) pathology confirmed the patients’ diagnoses of CRC. (II) The patients had no serious underlying disease or other cancers. (III) The patients had data for the KRAS and BRAF genes (mutation or wild-type). The participating patients were divided into the cetuximab-sensitive and cetuximab-resistant groups. The cetuximab-sensitive group consisted of patients with both KRAS and BRAF wild-type. The cetuximab-resistant group consisted of patients with KRAS mutations only. In addition, serum samples were retrospectively selected from twenty-two patients with cetuximab therapy at the following stages: initial diagnosis, disease remission and progression. The initial diagnosis stage was defined as the point when the patient had not been given treatments. The response of every patient to cetuximab was defined as disease remission (complete or partial remission) and disease progression (stable or progressive disease). Finally, miR-7-5p levels were measured by qRT-PCR.
Statistical analysis
All data were presented as the mean ± standard deviation (SD). The statistical studies depicted in the figures were conducted utilizing t tests or one-way analysis. The statistical analyses were performed using SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). A significance level of P<0.05 was deemed to indicate statistical significance.
Results
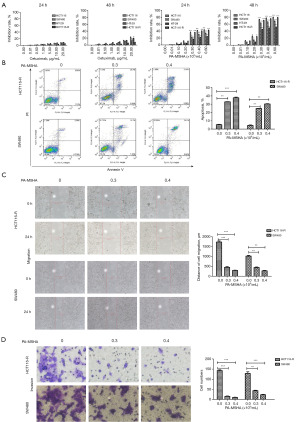
PA-MSHA exerts antitumor effects on cetuximab-resistant CRC in vitro
Cetuximab is ineffective in CRC patients harboring KRAS or BRAF mutations. We selected HT-29 cells with BRAF mutations and HCT116 and SW480 cell lines harboring KRAS mutations in the present study. Additionally, using extracorporeal shock, we were able to cultivate a panel of CRC HCT116 cells that were resistant to cetuximab (HCT116-R). The CCK-8 data showed that the highest inhibition rate of 20 µg/mL cetuximab was not more than 30% at 48 h (Figure 1A), confirming that these cell lines were not sensitive to cetuximab. Greater resistance to cetuximab was observed in HCT116-R cells, and the inhibition rate of 20 µg/mL cetuximab was only 7.6% at 48 h. Conversely, PA-MSHA markedly inhibited the growth of cetuximab-resistant CRC cells in a dose- and time-dependent manner. The inhibition rates of HCT116, HCT116-R, SW480 and HT-29 cells treated with 0.6×109/mL PA-MSHA for 48 h were 79.7%, 76.8%, 68.6% and 51.3%, respectively (Figure 1A). Moreover, PA-MSHA markedly induced apoptosis (Figure 1B) and restrained migration and invasion in cetuximab-resistant CRC cells (Figure 1C,1D). Taken together, these data indicated that PA-MSHA had potent anticancer activities against cetuximab-resistant CRC in vitro.
MiR-7-5p mediates the effects of PA-MSHA on cetuximab-resistant CRC
We examined the levels of 1483 miRNAs in HCT116-R cells with increasing doses of PA-MSHA through miRNA sequencing analysis. In the comparison of the 0 and 0.3×109/mL PA-MSHA groups, 22 miRNAs were upregulated, whereas 122 miRNAs were downregulated. In the comparison of the 0.3×109/mL and 0.4×109/mL PA-MSHA groups, 71 miRNAs were upregulated, whereas 8 miRNAs were downregulated. The cluster graph depicting the profile results is illustrated in Figure 2A. The miRNAs with the highest fold increase and decrease were miR-7-5p and miR-3529-3p, respectively. To validate the data, we quantified the levels of these miRNAs in HCT116-R and SW480 cells after PA-MSHA treatment. As shown in Figure 2B,2C, the expression of miR-7-5p increased in a dose-dependent manner (Figure 2B), whereas miR-3529-3p did not decrease with increasing doses of PA-MSHA (Figure 2C). Thus, we mainly selected miR-7-5p for the subsequent study.
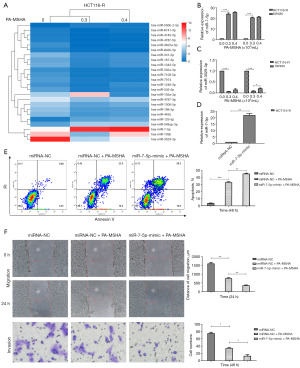
We next explored the association between miR-7-5p expression and PA-MSHA efficacy. The average expression of miR-7-5p in HCT116-R cells was observably upregulated after transfection with the miR-7-5p mimic vector (Figure 2D). MiR-7-5p overexpression significantly increased the apoptotic rate (Figure 2E) and markedly inhibited the migration and invasion of HCT116-R cells treated with PA-MSHA (Figure 2F). The data provided evidence that miR-7-5p overexpression dramatically enhanced the efficacy of PA-MSHA for cetuximab-resistant CRC.
Akt3 acts as the target of miR-7-5p to regulate the effect of PA-MSHA on cetuximab-resistant CRC
MiRNAs often exert their biological activity by inhibiting the expression of their target messenger RNAs (mRNAs). Based on the data from the literature review, TargetScan and miRTarBase databases, we speculate Akt3 might act as the target of miR-7-5p. As expected, the luciferase activity of the reporter plasmid carrying wild-type Akt3 3’-UTR sequences were markedly suppressed by miR-7-5p. However, this suppression was abrogated when the predicted sequences were subjected to mutation (Figure 3A). In addition, the level of Akt3 was markedly downregulated after miR-7-5p overexpression, whereas the protein showed the opposite trend by inhibiting miR-7-5p expression (Figure 3B).
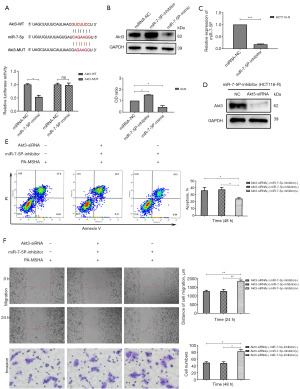
To further investigate the effects of Akt3 on the anticancer activities of PA-MSHA, we used the miR-7-5p inhibitor vector (Figure 3C) and Akt3 siRNA (Figure 3D) to reduce their expression in HCT116-R cells. MiR-7-5p downregulation significantly reduced the apoptosis rate (Figure 3E) and increased the migration and invasion of HCT116-R cells after PA-MSHA treatment (Figure 3F). Akt3 silencing obviously reversed these effects caused by miR-7-5p downregulation (Figure 3E,3F). These data suggested that Akt3 potentially acted as the target of miR-7-5p to regulate the anticancer activities of PA-MSHA in cetuximab-resistant CRC.
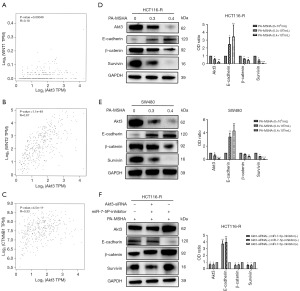
PA-MSHA inactivates the Wnt-β-catenin pathway in cetuximab-resistant CRC
Most cancers, particularly CRC, rely on aberrant Wnt-β-catenin signaling for their development and spread. By increasing downstream GSK3β phosphorylation and overall β-catenin expression, Akt activation can stimulate Wnt-β-catenin signaling. Hence, using the TCGA CRC databases, we calculated Spearman’s correlation coefficient between Akt3 expression and the expression of WNT and CTNNB1, two of the most important genes in the Wnt-β-catenin pathway. Akt3 expression was positively linked with WNT1 (Figure 4A, Spearman’s r=0.18, P=0.00049), WNT2 (Figure 4B, Spearman’s r=0.67, P=1.1e−49) and CTNNB1 (Figure 4C, Spearman’s r=0.33, P=4.5e−11) in TCGA CRC. The findings revealed that the Wnt-β-catenin signaling pathway was implicated in the efficacy of PA-MSHA in cetuximab-resistant CRC. As expected, the levels of β-catenin, the key protein of the Wnt-β-catenin pathway, were downregulated after PA-MSHA treatment. The levels of E-cadherin, a noteworthy indication of tumor migration and invasion, were upregulated. The expression of Survivin, a downstream protein of the Wnt-β-catenin pathway mediating cell apoptosis, was decreased (Figure 4D,4E). Moreover, we observed that miR-7-5p downregulation increased the levels of β-catenin and Survivin but decreased the levels of E-cadherin in HCT116-R cells treated with PA-MSHA. However, these proteins showed opposite trends after Akt3 silencing (Figure 4F). Collectively, the data suggest that PA-MSHA exerts potent activity against cetuximab-resistant CRC through miR-7-5p/Akt3/Wnt-β-catenin signaling pathway.
MiR-7-5p overexpression enhances the effects of PA-MSHA on cetuximab-resistant CRC in vivo
We further studied the function of PA-MSHA in cetuximab-resistant CRC in vivo. A tumorigenesis assay was conducted by subcutaneously administering cells into the right lower flanks of mice, divided into four distinct groups, including the normal control (injected PBS), the negative control (injected HCT116-R cells), A (injected HCT116-R cells) and B groups (injected miR-7-5p-overexpressing HCT116-R cells). The mice in the normal and negative control groups were given PBS treatment, but the mice in the A and B groups were given PA-MSHA treatment (Figure 5A). Compared with the negative control group, administration of PA-MSHA led to significant reductions in tumor weight and volume (Figure 5B,5C). Additionally, compared with the A group, miR-7-5p overexpression markedly slowed tumor growth in the B group (Figure 5B,5C). Moreover, the mice in the A group (74.3 days) had a significantly longer survival period than those in the negative control group (58.8 days), and miR-7-5p overexpression further prolonged the survival time of mice in the B group (81.5 days, Figure 5D). In the normal control group, all mice survived until the end of the experiment. These data indicated that PA-MSHA exerted potent activity against cetuximab-resistant CRC in vivo, and miR-7-5p overexpression further enhanced its efficacy.
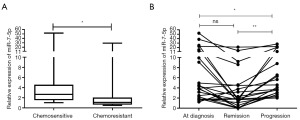
Serum miR-7-5p levels are associated with cetuximab resistance and therapeutic response in CRC
To better comprehend the clinical significance of miR-7-5p in CRC, we evaluated its level in serum samples. We found that serum miR-7-5p levels were significantly lower in cetuximab-resistant CRC than in cetuximab-sensitive CRC (Figure 6A). Next, we looked into the viability of miR-7-5p as a biomarker for tracking the effectiveness of cetuximab in CRC. The data revealed that the levels of miR-7-5p were significantly lower at the disease progression stage than at the remission or initial diagnosis stages in 22 CRC patients (Figure 6B). According to these findings, serum miR-7-5p was proven to be helpful for assessing cetuximab resistance and therapeutic response in CRC.
Discussion
For metastatic CRC, cetuximab—the first FDA-approved antibody for the treatment of CRC—has been used in conjunction with conventional chemotherapy. However, due to primary and acquired resistance, only a few patients could benefit from cetuximab treatment (24). Therefore, to combat cetuximab-resistant CRC, alternative treatments must be found. In the present study, we revealed that PA-MSHA, a medicinal biological product with Chinese regulatory approval, exerted great anticancer effects on cetuximab-resistant CRC by regulating the miR-7-5p/Akt3/Wnt-β-catenin pathway. These data have significant implications for gaining comprehension of the effectiveness and fundamental mechanics of PA-MSHA in cetuximab-resistant CRC.
PA-MSHA has been extensively documented as a chemotherapeutic agent in different cancers (18,25). However, the effects of PA-MSHA on cetuximab-resistant CRC remain unclear. A previous study reported that patients harboring primary and acquired KRAS and BRAF mutations were resistant to cetuximab (26). We selected KRAS-mutated CRC cell lines for this study (19,27). Our results showed that PA-MSHA significantly inhibited migration and invasion and induced apoptosis in cetuximab-resistant CRC cells. Similarly, PA-MSHA also displayed marked anticancer activities in animal studies.
We next explored the molecular mechanisms through which PA-MSHA exerted these functions. Cancer cells rely on miRNAs, a class of conserved short noncoding RNAs, to regulate many various aspects of their biology, including their response to treatment, differentiation, apoptosis, and migration (28,29). Generally, target mRNAs are silenced during translation or degraded after miRNA binds to the 3’-UTR (30). Most studies have demonstrated that chemicals or bacterization display obvious effects through the miRNA-mRNA gene network in various cancers. For instance, Sun et al. reported that LukS-PV, a component of PVL secreted by S. aureus, regulated miR-125a-3p to promote THP-1 macrophage differentiation and apoptosis by downregulating NF1 and Bcl-2 (13). However, the role of miRNAs in the anticancer activities of PA-MSHA is poorly understood. In our study, the analysis of the sequencing data identified that miR-7-5p and miR-3529-3p exhibited the most significant fold increase and decrease, respectively, in HCT116-R cells when treated with increasing doses of PA-MSHA. Given that only miR-7-5p expression was changed in a dose-dependent manner in the verified experiment, we speculated that it might be the key factor regulating PA-MSHA efficacy. As expected, miR-7-5p overexpression markedly enhanced the anticancer activities of PA-MSHA for cetuximab-resistant CRC in vitro and in vivo. Consistent with our data, several studies have reported that miR-7-5p functions as a suppressor in cancer tumorigenesis. Wang et al. showed that miR-7-5p mediated the antitumor activity of the N-terminal polypeptide in breast cancer (31). Wang et al. found that miR-7-5p stopped hepatocellular carcinoma cell lines from growing and migrating and sped up apoptosis by blocking SPC24 expression (32). Likewise, miR-7-5p plays a crucial role in CRC development (33,34).
To obtain an in-depth comprehension of the molecular pathways by which miR-7-5p affects the efficiency of PA-MSHA, we adopted bioinformatics techniques to anticipate its target genes. Based on the data from the literature review (35,36), TargetScan and miRTarBase databases, we speculated that Akt3 might act as the target gene of miR-7-5p to regulate the effects of PA-MSHA on cetuximab-resistant CRC. As expected, luciferase activity analysis confirmed that miR-7-5p directly targeted Akt3. Functional studies indicated that miR-7-5p suppression significantly reduced the anticancer activities of PA-MSHA, but the effects could be observably reversed by Akt3 silencing. As demonstrated in the report by Shiau et al., miR-7-5p was predicted to target AKT3 according to the miRDB database (37).
We further explored the downstream signaling of Akt3 involved in the effects of PA-MSHA on cetuximab-resistant CRC. Akt is a central node of many pathways. The activation of Akt is widely recognized for its ability to augment the phosphorylation of downstream GSK3β and the production of total β-catenin, hence leading to the activation of the Wnt-β-catenin signaling pathway (38,39). The Wnt-β-catenin pathway is commonly overactivated in many tumors (40,41), especially in CRC (42,43). Its activation is not only associated with tumor migration and invasion but also closely related to cell apoptosis in CRC (44,45). In addition, it is clear that the Akt/Wnt-β-catenin pathway plays a key role in most cancers, such as hepatocellular carcinoma (46) and CRC (47). Moreover, the miR-7/Akt pathway plays a momentous role in the development of lung cancer (48) and CRC (49). Based on the aforementioned results, our hypothesis posited that miR-7-5p may augment the efficacy of PA-MSHA in treating cetuximab-resistant CRC by impeding the activation of the Akt3/Wnt-β-catenin pathway. Consistent with our prediction, we demonstrated that Akt3 expression was positively correlated with key genes in the Wnt-β-catenin pathway, including WNT1, WNT2 and CTNNB1, by Spearman’s correlation coefficient from TCGA CRC databases. PA-MSHA treatment reduced the levels of β-catenin, which is a crucial protein involved in the Wnt-β-catenin pathway. In addition, the expression of proteins downstream of the Wnt-β-catenin pathway was significantly changed. For instance, the levels of the anti-metastatic factor E-cadherin were upregulated, and the expression of anti-apoptotic protein Survivin was decreased. Moreover, we found that Akt3 silencing could obviously reverse the variation tendency of these proteins caused by miR-7-5p downregulation. These data consumingly demonstrated that PA-MSHA exerted potent activity against cetuximab-resistant CRC through the miR-7-5p/Akt3/Wnt-β-catenin pathway.
Due to their amazing stability in plasma/serum, circulating miRNAs offer major potential as biomarkers for cancer screening and monitoring purposes (50-52). Most serum miRNAs, including miR-21, miR-210, miR-663 and miR-497, are related to the diagnosis, prognosis (53) and recurrence (54,55) of CRC. Our study showed that serum miR-7-5p levels were significantly higher in cetuximab-sensitive CRC than in cetuximab-resistant CRC. Besides, serum miR-7-5p levels were decreased in patients who achieved disease progression with cetuximab treatment. These results indicated that serum miR-7-5p levels could be useful in monitoring cetuximab resistance and response in CRC during treatment.
Conclusions
In summary, our study demonstrates that PA-MSHA exerts potent activity against cetuximab-resistant CRC in vitro and in vivo, indicating that PA-MSHA shows therapeutic promise for cetuximab-resistant CRC. In addition, we found that the miR-7-5p/Akt3/Wnt-β-catenin pathway plays a role in controlling PA-MSHA efficacy. Specifically, PA-MSHA treatment primarily upregulates miR-7-5p levels, inhibits the expression of Akt3, and finally inactivates the Wnt-β-catenin pathway (Figure 7). This study deeply enriches the regulatory network of PA-MSHA in cancer therapy. Moreover, we show that serum miR-7-5p might constitute a potential dynamic biomarker to monitor the resistance and therapeutic efficacy of cetuximab in CRC.
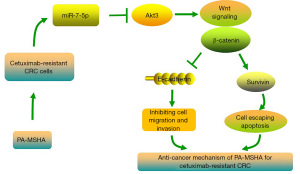
Indeed, our research exhibited certain constraints. First, although the obtained sequencing findings indicated that miR-3529-3p was observably decreased, its direct effect has not been verified. Shang et al. reported that miR-3529-3p was markedly increased in radioresistant CRC cells (56). These results indicated that miR-3529-3p might be a critical miRNA to regulate CRC radiotherapy. Second, other target genes of miR-7-5p possibly play a role in mediating the anticancer activities of PA-MSHA, and further research is necessary in areas that remain unresolved for future investigation. Notwithstanding these limitations, the results of our study will establish a theoretical framework for understanding the miRNA–mRNA regulatory gene network and treatment efficacy of PA-MSHA in cetuximab-resistant CRC.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2211/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2211/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2211/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2211/coif). W.S. reports that this work was supported by Natural Science Foundation of Anhui Province (No. 1908085QH375); New Medical Union Foundation of University of Science and Technology of China (No. WK9110000072); and the Foundation of the West Branch of the First Affiliated Hospital of University of Science and Technology of China (No. 2018YJQN018). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were approved by the Ethics Committee of the First Affiliated Hospital of University of Science and Technology of China (No. 2024-N (A)-158), in compliance with national guidelines for the care and use of animals. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of The First Affiliated Hospital of University of Science and Technology of China (No. 2019-KY-28). Informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tepeš B, Mlakar DN, Stefanovič M, et al. The impact of 6 years of the National Colorectal Cancer Screening Program on colorectal cancer incidence and 5-year survival. Eur J Cancer Prev 2021;30:304-10. [Crossref] [PubMed]
- Vetter LE, Merkel S, Bénard A, et al. Colorectal cancer in Crohn's colitis is associated with advanced tumor invasion and a poorer survival compared with ulcerative colitis: a retrospective dual-center study. Int J Colorectal Dis 2021;36:141-50. [Crossref] [PubMed]
- Islam MA, Hossen MB, Horaira MA, et al. Exploring Core Genes by Comparative Transcriptomics Analysis for Early Diagnosis, Prognosis, and Therapies of Colorectal Cancer. Cancers (Basel) 2023;15:1369. [Crossref] [PubMed]
- Jiang YL, Xun Y. Molecular Mechanism of Salvia miltiorrhiza in the Treatment of Colorectal Cancer Based on Network Pharmacology and Molecular Docking Technology. Drug Des Devel Ther 2024;18:425-41. [Crossref] [PubMed]
- Guren MG. The global challenge of colorectal cancer. Lancet Gastroenterol Hepatol 2019;4:894-5. [Crossref] [PubMed]
- Heinemann V, von Weikersthal LF, Decker T, et al. FOLFIRI plus cetuximab or bevacizumab for advanced colorectal cancer: final survival and per-protocol analysis of FIRE-3, a randomised clinical trial. Br J Cancer 2021;124:587-94. [Crossref] [PubMed]
- Masuishi T, Tsuji A, Kotaka M, et al. Phase 2 study of irinotecan plus cetuximab rechallenge as third-line treatment in KRAS wild-type metastatic colorectal cancer: JACCRO CC-08. Br J Cancer 2020;123:1490-5. [Crossref] [PubMed]
- Albadari N, Xie Y, Li W. Deciphering treatment resistance in metastatic colorectal cancer: roles of drug transports, EGFR mutations, and HGF/c-MET signaling. Front Pharmacol 2024;14:1340401. [Crossref] [PubMed]
- Mu M, Zhang Q, Zhao C, et al. 3-Bromopyruvate overcomes cetuximab resistance in human colorectal cancer cells by inducing autophagy-dependent ferroptosis. Cancer Gene Ther 2023;30:1414-25. [Crossref] [PubMed]
- Marzhoseyni Z, Shojaie L, Tabatabaei SA, et al. Streptococcal bacterial components in cancer therapy. Cancer Gene Ther 2022;29:141-55. [Crossref] [PubMed]
- Mohseni Z, Sedighian H, Halabian R, et al. Potent in vitro antitumor activity of B-subunit of Shiga toxin conjugated to the diphtheria toxin against breast cancer. Eur J Pharmacol 2021;899:174057. [Crossref] [PubMed]
- DiFranco KM, Johnson-Farley N, Bertino JR, et al. LFA-1-targeting Leukotoxin (LtxA; Leukothera®) causes lymphoma tumor regression in a humanized mouse model and requires caspase-8 and Fas to kill malignant lymphocytes. Leuk Res 2015;39:649-56. [Crossref] [PubMed]
- Sun XX, Zhang SS, Dai CY, et al. LukS-PV-Regulated MicroRNA-125a-3p Promotes THP-1 Macrophages Differentiation and Apoptosis by Down-Regulating NF1 and Bcl-2. Cell Physiol Biochem 2017;44:1093-105. [Crossref] [PubMed]
- Zhao Y, Zhang Y, Liu J. Regulatory effect of Pseudomonas aeruginosa mannose-sensitive hemagglutinin on inflammation and immune function in percutaneous nephrolithotomy patients with upper urinary tract calculi complicated with infection. Front Immunol 2023;14:1181688. [Crossref] [PubMed]
- Liu J, Duan X. PA-MSHA induces apoptosis and suppresses metastasis by tumor associated macrophages in bladder cancer cells. Cancer Cell Int 2017;17:76. [Crossref] [PubMed]
- Zhao X, Wu X, Yu W, et al. PA-MSHA inhibits proliferation and induces apoptosis in human non-small cell lung cancer cell lines with different genotypes. Mol Med Rep 2016;14:5369-76. [Crossref] [PubMed]
- Wei Y, Liu D, Jin X, et al. PA-MSHA inhibits the growth of doxorubicin-resistant MCF-7/ADR human breast cancer cells by downregulating Nrf2/p62. Cancer Med 2016;5:3520-31. [Crossref] [PubMed]
- Yin TQ, Ou-Yang X, Jiao FY, et al. Pseudomonas aeruginosa mannose-sensitive hemagglutinin inhibits proliferation and invasion via the PTEN/AKT pathway in HeLa cells. Oncotarget 2016;7:37121-31. [Crossref] [PubMed]
- Gong J, Chen Y, Yang L, et al. MEK162 Enhances Antitumor Activity of 5-Fluorouracil and Trifluridine in KRAS-mutated Human Colorectal Cancer Cell Lines. Anticancer Res 2017;37:2831-8. [PubMed]
- Forbes SA, Tang G, Bindal N, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res 2010;38:D652-7. [Crossref] [PubMed]
- Wen J, Zheng B, Hu Y, et al. Establishment and biological analysis of the EC109/CDDP multidrug-resistant esophageal squamous cell carcinoma cell line. Oncol Rep 2009;22:65-71. [PubMed]
- Franck M, Thon C, Schütte K, et al. Circulating miR-21-5p level has limited prognostic value in patients with hepatocellular carcinoma and is influenced by renal function. World J Hepatol 2020;12:1031-45. [Crossref] [PubMed]
- Murray MJ, Bell E, Raby KL, et al. A pipeline to quantify serum and cerebrospinal fluid microRNAs for diagnosis and detection of relapse in paediatric malignant germ-cell tumours. Br J Cancer 2016;114:151-62. [Crossref] [PubMed]
- Tapia-Galisteo A, Sánchez Rodríguez Í, Aguilar-Sopeña O, et al. Trispecific T-cell engagers for dual tumor-targeting of colorectal cancer. Oncoimmunology 2022;11:2034355. [Crossref] [PubMed]
- Wang C, Hu Z, Zhu Z, et al. The MSHA strain of Pseudomonas aeruginosa (PA-MSHA) inhibits gastric carcinoma progression by inducing M1 macrophage polarization. Tumour Biol 2016;37:6913-21. [Crossref] [PubMed]
- Su C, Olsen KA, Bond CE, et al. The Efficacy of Using Patient-Derived Organoids to Predict Treatment Response in Colorectal Cancer. Cancers (Basel) 2023;15:805. [Crossref] [PubMed]
- Huang F, Chang H, Greer A, et al. IRS2 copy number gain, KRAS and BRAF mutation status as predictive biomarkers for response to the IGF-1R/IR inhibitor BMS-754807 in colorectal cancer cell lines. Mol Cancer Ther 2015;14:620-30. [Crossref] [PubMed]
- Sun W, Li J, Zhou L, et al. The c-Myc/miR-27b-3p/ATG10 regulatory axis regulates chemoresistance in colorectal cancer. Theranostics 2020;10:1981-96. [Crossref] [PubMed]
- Charkiewicz R, Sulewska A, Charkiewicz A, et al. miRNA-Seq Tissue Diagnostic Signature: A Novel Model for NSCLC Subtyping. Int J Mol Sci 2023;24:13318. [Crossref] [PubMed]
- Su K, Wang N, Shao Q, et al. The role of a ceRNA regulatory network based on lncRNA MALAT1 site in cancer progression. Biomed Pharmacother 2021;137:111389. [Crossref] [PubMed]
- Wang HF, Dong ZY, Yan L, et al. The N-terminal polypeptide derived from vMIP-II exerts its antitumor activity in human breast cancer through CXCR4/miR-7-5p/Skp2 pathway. J Cell Physiol 2020;235:9474-86. [Crossref] [PubMed]
- Wang Y, Yang H, Zhang G, et al. hsa-miR-7-5p suppresses proliferation, migration and promotes apoptosis in hepatocellular carcinoma cell lines by inhibiting SPC24 expression. Biochem Biophys Res Commun 2021;561:80-7. [Crossref] [PubMed]
- Dong M, Xie Y, Xu Y. miR-7-5p regulates the proliferation and migration of colorectal cancer cells by negatively regulating the expression of Krüppel-like factor 4. Oncol Lett 2019;17:3241-6. [PubMed]
- Zheng Y, Nie P, Xu S. Long noncoding RNA CASC21 exerts an oncogenic role in colorectal cancer through regulating miR-7-5p/YAP1 axis. Biomed Pharmacother 2020;121:109628. [Crossref] [PubMed]
- Jia H, Ma T, Jia S, et al. AKT3 and related molecules as potential biomarkers responsible for cryptorchidism and cryptorchidism-induced azoospermia. Transl Pediatr 2021;10:1805-17. [Crossref] [PubMed]
- Zhang X, Niu W, Mu M, et al. Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop. J Exp Clin Cancer Res 2020;39:196. [Crossref] [PubMed]
- Shiau JP, Chuang YT, Yen CY, et al. Modulation of AKT Pathway-Targeting miRNAs for Cancer Cell Treatment with Natural Products. Int J Mol Sci 2023;24:3688. [Crossref] [PubMed]
- Honardoost M, Rad SMAH. Triangle of AKT2, miRNA, and Tumorigenesis in Different Cancers. Appl Biochem Biotechnol 2018;185:524-40. [Crossref] [PubMed]
- Ma J, Zhao J, Lu J, et al. Cadherin-12 enhances proliferation in colorectal cancer cells and increases progression by promoting EMT. Tumour Biol 2016;37:9077-88. [Crossref] [PubMed]
- Bugter JM, Fenderico N, Maurice MM. Mutations and mechanisms of WNT pathway tumour suppressors in cancer. Nat Rev Cancer 2021;21:5-21. [Crossref] [PubMed]
- Zhang Y, Wang X. Targeting the Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol 2020;13:165. [Crossref] [PubMed]
- Caspi M, Wittenstein A, Kazelnik M, et al. Therapeutic targeting of the oncogenic Wnt signaling pathway for treating colorectal cancer and other colonic disorders. Adv Drug Deliv Rev 2021;169:118-36. [Crossref] [PubMed]
- Predes D, Oliveira LFS, Ferreira LSS, et al. The Chalcone Lonchocarpin Inhibits Wnt/β-Catenin Signaling and Suppresses Colorectal Cancer Proliferation. Cancers (Basel) 2019;11:1968. [Crossref] [PubMed]
- Jearawuttanakul K, Khumkhrong P, Suksen K, et al. Cleistanthin A induces apoptosis and suppresses motility of colorectal cancer cells. Eur J Pharmacol 2020;889:173604. [Crossref] [PubMed]
- Liu Z, Gu Y, Cheng X, et al. Upregulation lnc-NEAT1 contributes to colorectal cancer progression through sponging miR-486-5p and activating NR4A1/Wnt/β-catenin pathway. Cancer Biomark 2021;30:309-19. [Crossref] [PubMed]
- Zhang N, Li H, Qin C, et al. Insufficient radiofrequency ablation promotes the metastasis of residual hepatocellular carcinoma cells via upregulating flotillin proteins. J Cancer Res Clin Oncol 2019;145:895-907. [Crossref] [PubMed]
- McCool EN, Xu T, Chen W, et al. Deep top-down proteomics revealed significant proteoform-level differences between metastatic and nonmetastatic colorectal cancer cells. Sci Adv 2022;8:eabq6348. [Crossref] [PubMed]
- Das K, Rao LVM. The Role of microRNAs in Inflammation. Int J Mol Sci 2022;23:15479. [Crossref] [PubMed]
- Moslehian MS, Shabkhizan R, Asadi MR, et al. Interaction of lncRNAs with mTOR in colorectal cancer: a systematic review. BMC Cancer 2023;23:512. [Crossref] [PubMed]
- Diao ZB, Sun TX, Zong Y, et al. Identification of plasma microRNA-22 as a marker for the diagnosis, prognosis, and chemosensitivity prediction of osteosarcoma. J Int Med Res 2020;48:300060520967818. [Crossref] [PubMed]
- Ghafouri-Fard S, Hussen BM, Badrlou E, et al. MicroRNAs as important contributors in the pathogenesis of colorectal cancer. Biomed Pharmacother 2021;140:111759. [Crossref] [PubMed]
- So JBY, Kapoor R, Zhu F, et al. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut 2021;70:829-37. [Crossref] [PubMed]
- Li G, Wang Q, Li Z, et al. Serum miR-21 and miR-210 as promising non-invasive biomarkers for the diagnosis and prognosis of colorectal cancer. Rev Esp Enferm Dig 2020;112:832-7. [Crossref] [PubMed]
- Wang N, Zeng L, Li Z, et al. Serum miR-663 expression and the diagnostic value in colorectal cancer. Artif Cells Nanomed Biotechnol 2019;47:2650-3. [Crossref] [PubMed]
- Zou G, Wang R, Wang M. Clinical response and prognostic significance of serum miR-497 expression in colorectal cancer. Cancer Biomark 2019;25:11-8. [Crossref] [PubMed]
- Shang Y, Wang L, Zhu Z, et al. Downregulation of miR-423-5p Contributes to the Radioresistance in Colorectal Cancer Cells. Front Oncol 2021;10:582239. [Crossref] [PubMed]


