
Integrative analysis of the role of the DPH gene family in hepatocellular carcinoma and expression validation
Highlight box
Key findings
• Diphthamide genes (DPHs) are significantly differentially expressed in cancer, particularly in hepatocellular carcinoma (HCC).
• DPH2,3 demonstrated significant diagnostic and prognostic potential in HCC, with strong correlations to various immune-related genes and tumor-infiltrating immune cells, suggesting a role in immune modulation.
What is known and what is new?
• It is known that the diphthamide (DPH) gene family modifies eukaryotic elongation factor 2 (eEF2) and is linked to carcinogenesis.
• This study provides new insights into the roles of DPH2,3 in HCC, highlighting their expression patterns and diagnostic and prognostic relevance, as well as their potential mechanisms related to immune-related genes and pathways.
What is the implication, and what should change now?
• The findings suggest that DPH2,3 could serve as valuable biomarkers for the diagnosis and prognosis of HCC.
• The expression of DPH2,3 should be further explored as potential therapeutic targets, particularly in the context of immunotherapy. Given their association with immune-related genes and pathways, DPH2,3 hold promise for enhancing the efficacy of immunotherapies in HCC, potentially leading to more effective treatment strategies.
Introduction
Hepatocellular carcinoma (HCC) accounts for a large majority of liver cancer cases, and it is one of the leading causes of cancer-related deaths worldwide (1,2). Sadly, only 18% of HCC patients survive for at least 5 years, which indicates a poor prognosis (3). There are several treatment options available for individuals with HCC, such as surgical resection and liver transplantation, as well as transarterial, radiotherapy, and percutaneous ablation. However, systemic therapy has become the preferred treatment for those with intermediate to advanced disease or individuals who are not eligible for local treatment (4). Long-term survival of more than a year is still unusual, although systemic medication therapy has lately given hope for the treatment of HCC patients a new lease of life (5). Immunotherapy is crucial in treating HCC and can synergize with other treatments (6-8). Biomarkers play a vital role in precision medicine by helping to identify the most effective treatment options (4,9). Therefore, novel biomarker studies are essential for early detection, treatment, and prognosis follow-up of HCC.
The diphthamide (DPH) gene family members, including DPH1, DPH2, DPH3, DPH5, DPH6, and DPH7, produce enzymes that play a crucial role in creating diphthamide. Diphthamide is a modified form of histidine that is only found in eukaryotic elongation factor 2 (eEF2). These post-translational modifications are critical in ensuring the accuracy and efficiency of the translation process (10). However, diphthamide is the target of pseudomonas exotoxin and diphtheria toxin ADP-ribosylation, halting protein synthesis and causing cell death (11-13). The “active state” of diphthamide directly affects the immunotoxicity of tumor cells, in which the DPH genes (DPHs) play an indispensable role (14). Studies have shown that eEF2 overexpression promotes the proliferation, migration, and invasion of a wide range of tumors (15) and may be a potential target for drug therapy (16). Recent studies have shown that DPH1 deletion or deletion of diphthamide inhibits the development of N-diethylnitrosamine (DEN)-induced periportal HCC, but leads to an increase in K19-positive periportal stem, progenitor cells, which nevertheless promotes Pten, Trp53-deficient HCC development (17). In addition, colorectal cancer cells affect their proliferation and invasion through the MiR218-5p, DPH1 axis (18). Mutations in the DPH3 gene have been associated with basal cell carcinoma (19,20). And fusions of the DPH7 and PTP4A3 genes may be associated with tumor grading and recurrence in endometrial cancer (21). However, the specific role of DPHs in HCC is unclear.
Our primary objective was to analyze the expression patterns of DPHs in HCC. Moreover, we investigated their diagnostic and prognostic value in HCC patients and their associated clinical features. Our research revealed that DPH2 and DPH3 (DPH2,3) are independent prognostic factors for HCC patients. We also delved into the methylation and mutation of DPH2,3 in HCC to determine the reasons for its increased expression. After analyzing the reasons for the increased expression of DPH2,3, we conducted a gene sets enrichment analysis (GSEA) and discovered a potential connection between DPH2,3 and the tumor microenvironment (TME). We further examined the role of DPH2,3 in TEM by studying its relationship with immune-related genes and immune-infiltrating cells. The research suggests a strong link between HCC and DPHs, with DPH2,3 being a possible target for treating HCC. The theoretical basis of our current work contributes to the development of a new effective molecular pathway for the treatment of HCC. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-147/rc).
Methods
Ethical statement
The research conducted has adhered to the moral guidelines set in the Declaration of Helsinki (as revised in 2013) by the World Medical Association while acquiring and classifying all human tissue samples. The study was approved by The Medical Ethics Committee of the Affiliated Hospital of North Sichuan Medical College (protocol No. 2023ER153-1). All patients who participated in the study provided written informed consent before the collection of tissue samples.
Clinical samples
All tissue samples were collected in November 2019 and June 2020. Patients over 18 who had undergone surgical resection of the primary tumor and were confirmed to have HCC through pathology assessment were included. All tissue samples were obtained from the Affiliated Hospital of North Sichuan Medical College’s Department of Hepatobiliary Surgery. None of the study participants had received any radiation, chemotherapy, or immunotherapy therapies before surgery. Two independent pathologists independently verified the identification of each material to guarantee correct post-surgical diagnosis. The tissue samples were immediately submerged in liquid nitrogen following collection and were stored at −80 ℃. All tissue samples are stored at the Hepato-Biliary-Pancreatic-Intestinal Disease Department of the North Sichuan Medical College.
Cell culture
HCC cell lines, including Huh7, Hep3B, HepG2, SMMC7721, HCCLM3, MHCC97L, and MHCC97H, were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The Huh7, Hep3B, HepG2, HCCLM3, MHCC97L, and MHCC97H cell lines were cultured in DMEM (VivaCell, Shanghai, China) supplemented with 10% fetal bovine serum (VivaCell). While the SMMC7721 cell line was cultured in RPMI-1640 medium (VivaCell) supplemented with 10% fetal bovine serum. They were cultured at 37 ℃ and 5% CO2 under recommended conditions. Only the cells in the logarithmic growth phase were collected for subsequent experiments.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNAs were isolated from the samples using Trizol reagent (Invitrogen, Waltham, MA, USA). The RNA was then extracted with chloroform, isopropanol, and alcohol. The extracted RNA was reverse transcribed to complementary DNA (cDNA) using a HiScript III RT SupperMix for Qpcr (+gDNA wiper) kit (Vazyme Biotech, Nanjing, China). RT-qPCR was performed using Taq Pro Universal SYBR qPCR Master Mix kit (Vazyme Biotech). The relative expression of DPHs mRNA was calculated using the delta-delta Cq (2−∆∆Ct) method with GAPDH as the internal control. The specific primers (Sangon Biotech, Chengdu, China) used for amplifying each gene are listed in Table 1.
Table 1
| Primer name | Sequence |
|---|---|
| DPH1 | |
| Forward | CTGAAAGCCGAGTATCGTGTG |
| Reverse | TGTTCTCTGGATAGGACTTTGCT |
| DPH2 | |
| Forward | CCTGGACGGAGTGTACGAG |
| Reverse | AGCATCTCCCAATAGCTGGTC |
| DPH3 | |
| Forward | GCAGTGTTTCATGACGAGGTG |
| Reverse | TTGCCACGTCTTCCCCATTC |
| DPH5 | |
| Forward | ATGCTTTATCTCATCGGGTTGG |
| Reverse | GCAGCGTCTAACAACTTCCAG |
| DPH6 | |
| Forward | TGCTGGGCATCAGATCGTTG |
| Reverse | GAGGGGAAGAGCCATTGCTT |
| DPH7 | |
| Forward | AGCCTCAGGTCCGTTTAGG |
| Reverse | CCTCGACCAGAGGGTGAATAG |
| GAPDH | |
| Forward | GGAGCGAGATCCCTCCAAAAT |
| Reverse | GGCTGTTGTCATACTTCTCATGG |
RT-qPCR, real-time quantitative polymerase chain reaction; DPH, diphthamide.
Data sources and processing
We obtained RNA-seq data in transcripts per-million (TPM) format for The Cancer Genome Atlas (TCGA) and Genotype-Tissue Txpression (GTEx) from the UCSC XENA (https://xenabrowser.net/) using the Toil process (22). We extracted (pan-cancer) data corresponding to TCGA and the corresponding normal tissue data in GTEx. The full names and abbreviations of the pan-cancers are shown in Table 2. We downloaded RNA-seq data from the TCGA database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga) for the STAR process of the TCGA-LIHC project and extracted data in TPM format and clinical data. Depending on the characteristics of the data format, we selected appropriate statistical methods for analysis using R packages “stats” (version 4.2.1) and “car” (version 3.1.0), and visualized the data using the ggplot2 (version 3.3.6) package. To ensure accuracy and consistency, we transformed these values by log2(TPM + 1) (22). We retrieved immunohistochemistry (IHC) and immunofluorescence images from the Human Protein Atlas (HPA) database (https://www.proteinatlas.org/) to assess expression levels and subcellular localization of DPHs proteins in HCC (23). Additionally, Information about DPHs protein expression was obtained from the University of Alabama Cancer database (UALCAN) (https://ualcan.path.uab.edu/index.html) (24).
Table 2
| Abbreviations | Full names |
|---|---|
| ACC | Adrenocortical carcinoma |
| BLCA | Bladder urothelial carcinoma |
| BRCA | Breast invasive carcinoma |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma |
| CHOL | Cholangiocarcinoma |
| COAD | Colon adenocarcinoma |
| DLBC | Lymphoid neoplasm diffuse large B-cell lymphoma |
| ESCA | Esophageal carcinoma |
| GBM | Glioblastoma multiforme |
| HNSC | Head and neck squamous cell carcinoma |
| KICH | Kidney chromophobe |
| KIRC | Kidney renal clear cell carcinoma |
| KIRP | Kidney renal papillary cell carcinoma |
| LAML | Acute myeloid leukemia |
| LGG | Lower grade glioma |
| LIHC | Liver hepatocellular carcinoma |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| MESO | Mesothelioma |
| OV | Ovarian serous cystadenocarcinoma |
| PAAD | Pancreatic adenocarcinoma |
| PCPG | Pheochromocytoma and paraganglioma |
| PRAD | Prostate adenocarcinoma |
| READ | Rectum adenocarcinoma |
| SARC | Sarcoma |
| SKCM | Skin cutaneous melanoma |
| STAD | Stomach adenocarcinoma |
| TGCT | Testicular germ cell tumor |
| THCA | Thyroid carcinoma |
| THYM | Thymoma |
| UCEC | Uterine corpus endometrial carcinoma |
| UCS | Uterine carcinosarcoma |
| UVM | Uveal melanoma |
TCGA, The Cancer Genome Atlas.
The relationship between clinicopathological characteristics and DPHs mRNA
We analyzed the clinical data from 374 HCC patients in the TCGA database, to investigate the correlation between DPHs mRNA expression levels and clinicopathological characteristics, such as T, N, M stage, pathologic stage, gender, age, height, body mass index (BMI), prothrombin time, and vascular invasion. We used R and appropriate statistical methods to analyze the relationship between the expression of DPH genes and relevant clinical data.
The diagnostic and prognostic value of DPHs in HCC
We carried out an in-depth study to assess the diagnostic and prognostic significance of DPHs in HCC using data obtained from the TCGA. We used receiver operating characteristic (ROC) analysis with the R package “pROC” (version 1.18.0) and calculated the area under the curve (AUC). Next, we divided the cases into two groups (high-expression and low-expression groups) based on the median expression values of DPHs mRNA. We used the “survival” (version 3.3.1) R package to perform proportional risk hypothesis testing and fit survival regressions. This allowed us to make a comparison between the overall survival (OS) of the two groups. We then executed proportional risk hypothesis testing using the R package “survival” (version 3.3.1) and “rms” (version 6.3.0). We used TNM stage, pathologic stage, gender, age, and DPHs as statistical variables for Cox regression analysis (when the univariate analysis met P<0.1, it was entered into a multivariate COX to construct the model) (25).
Mutation and DNA methylation analysis of DPH2,3 in HCC
The analysis of mutations and gene amplifications in 1348 HCC samples was conducted using the cBioPortal database (https://www.cbioportal.org/). The samples were sourced from reputable institutions such as “Peking University, Cancer Cell 2019”, “INSERM, Nat Genet 2015”, “AMC, Hepatology 2014”, “RIKEN, Nat Genet 2012”, “MERiC, Basel, Nat Commun, 2022”, “MSK, Clin Cancer Res 2018”, and “TCGA Firehose Legacy”, were utilized for the analysis of mutations and gene amplifications (26). We retrieved mutations and copy number alterations in the DPH2,3 genes in HCC from the cBioPortal database. Furthermore, we used the UALCAN database to examine the promoter methylation status of DPH2,3 in HCC (24). Additionally, MethSurv (https://biit.cs.ut.ee/methsurv/) was utilized for prognostic analysis related to the methylation probes of DPH2,3 (27).
GSEA of DPH2,3 in HCC
We used GSEA (28) to explore the mechanism behind the high expression of DPH2,3 in HCC (29). First, line ID conversion was performed using the R package “rg.Hs.eg.db”, followed by GSEA analysis using the “clusterProfiler” package (version 4.4.4) (30). The significance thresholds were set at P<0.05 and q<0.25. From the MSigDB collection (https://www.gsea-msigdb.org/gsea/msigdb), we selected “c2.cp.all.v2022.1.Hs.symbols.gmt” as the reference gene set for pathway analysis. Finally, we chose the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway from the MSigDB collection for presentation.
Relationship analysis between immune-related genes and immune cell infiltration in DPH2,3
We performed correlation analyses of the variables in the data and the results were visualised in heatmaps and scatterplot to investigate the correlation between DPH2,3 and immune-related genes. We analyzed the immune infiltration of DPH2,3 based on the “ssGSEA” algorithm provided in the R package “GSVA” (version 1.46.0) in combination with 24 markers of immune cells provided in the immunity article (31,32).
Statistical analysis
The data was analyzed and visualized using R (version 4.2.1) and GraphPad (version 8.0.1). We conducted an analysis of the expression of DPHs in various types of cancer, including HCC. To do this, we used the Wilcoxon rank sum test to compare gene expression levels. For two-sample data, we utilized paired t-tests, and for multiple-sample data, we used one-way ANOVA. In order to assess the correlation between the gene expression levels of DPHs and clinical characteristics, we chose the appropriate statistical methods based on the characteristics of the data. To predict the prospects of DPHs as diagnostic biomarkers, we performed ROC curve analysis with the pROC R package. We also analyzed the OS of DPHs in HCC using the Log-rank test. To study the correlation of DPH2,3 with immune-related genes and immune cells, we used Spearman. Additionally, we compared the difference in immune infiltrating cells between high and low DPH2,3 expression groups using the Wilcoxon rank sum test. We defined P<0.05 as statistically significant.
Results
Patterns of DPHs expression in pan-cancer and HCC
We conducted a thorough pan-cancer analysis of the DPHs using the TCGA-GTEx pan-cancer dataset, looking at the mRNA expression levels in 33 distinct cancer types. Our findings demonstrated that DPH1,7 was considerably downregulated in 20 tumors, including adrenocortical carcinoma (ACC), breast invasive carcinoma (BRCA), and liver hepatocellular carcinoma (LIHC). However, DPH1,7 was overexpressed in head and neck squamous cell carcinoma (HNSC), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), and cholangiocarcinoma (CHOL) (Figure 1A,1B). DPH2,3,5,6 showed significant overexpression in a broad variety of cancer types, including 17 malignancies such as BRCA, CHOL, and LIHC, except acute myeloid leukemia (LAML) and kidney chromophobe (KICH) (Figure 1C-1F). These data implied that DPH2,3,5,6, which consistently overexpressed across a broad range of malignancies, may be crucial in the formation of carcinogenesis and tumors. On the other hand, DPH1,7 was downregulated in a variety of cancers.

TCGA database RNA-seq data paired analysis revealed that DPHs were considerably increased in HCC (Figure 2A). DPH2,3,5,6 were significantly overexpressed in HCC, according to an unpaired analysis, while DPH1,7 showed significantly downregulated expression (Figure 2B). DPH2,3,5,6 protein were significantly increased in HCC, while DPH1,7 protein were expressed at lower levels, according to IHC data from the HPA database. The UALCAN database displays the total protein expression of the DPHs, with DPH1,2,3,5 expression being up-regulated in HCC and DPH7 expression being down-regulated (Figure 2C). Overall, between normal and HCC tissues, there were considerable differences in the mRNA and protein expression of DPHs.

The relationship between DPHs mRNA and clinicopathological features
Based on clinical data from TCGA, such as T stage (Figure 3A), N stage (Figure 3B), histological grade (G1, G2, G3, G4) (Figure 3C), pathological stage (stage I, II and stage III, IV) (Figure 3D), and OS (Figure 3E), the expression of DPHs were evaluated in HCC and normal liver tissue. Our research showed that HCC tissues exhibited significantly higher expression levels of DPHs compared to normal liver tissue. Analysis of the clinicopathological correlations between DPHs found interesting correlations. Specifically, DPH2 exhibited correlations with T stage (P=0.005), race (P=0.03), gender (P=0.001), histological grade (P=0.008), and OS (P=0.01). DPH3 demonstrated correlations with weight (P=0.004), histological grade (P=0.001), alpha-fetoprotein (AFP) levels (P<0.001), and OS (P=0.01) (Table 3 and Table S1). Furthermore, DPH5 displayed correlations with T stage (P=0.005), race (P=0.02), histological grade (P<0.001), and vascular invasion (P=0.02). DPH6 showed correlations with histological grade (P<0.001), race (P=0.001), weight (P<0.001), and prothrombin time (P=0.006). Lastly, DPH7 exhibited correlations with weight (P<0.001), age (P=0.02), height (P=0.008), BMI (P=0.03), histological grade (P<0.001), AFP (P<0.001) and vascular infiltration (P=0.02) (Table 4 and Table S1).

Table 3
| Characteristics | DPH1 expression, n (%) | DPH2 expression, n (%) | DPH3 expression, n (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Low (N=185) | High (N=186) | P value | Low (N=185) | High (N=186) | P value | Low (N=185) | High (N=186) | P value | |||
| Pathologic T stage (N=368) | 0.91 | 0.005 | 0.92 | ||||||||
| T1 | 90 (24.5) | 91 (24.7) | 103 (28) | 78 (21.2) | 90 (24.5) | 91 (24.7) | |||||
| T2 & T3 & T4 | 94 (25.5) | 93 (25.3) | 79 (21.5) | 108 (29.3) | 92 (25) | 95 (25.8) | |||||
| Histologic grade (N=366) | 0.82 | 0.008 | 0.001 | ||||||||
| G1 & G2 | 115 (31.4) | 117 (32) | 127 (34.7) | 105 (28.7) | 131 (35.8) | 101 (27.6) | |||||
| G3 & G4 | 68 (18.6) | 66 (18) | 54 (14.8) | 80 (21.9) | 52 (14.2) | 82 (22.4) | |||||
| Race (N=359) | 0.20 | 0.03 | 0.58 | ||||||||
| Asian | 83 (23.1) | 75 (20.9) | 67 (18.7) | 91 (25.3) | 74 (20.6) | 84 (23.4) | |||||
| Black or AA & White | 92 (25.6) | 109 (30.4) | 109 (30.4) | 92 (25.6) | 100 (27.9) | 101 (28.1) | |||||
| Weight (N=344) | 0.73 | 0.08 | 0.004 | ||||||||
| ≤70 kg | 91 (26.5) | 91 (26.5) | 84 (24.4) | 98 (28.5) | 78 (22.7) | 104 (30.2) | |||||
| >70 kg | 78 (22.7) | 84 (24.4) | 90 (26.2) | 72 (20.9) | 95 (27.6) | 67 (19.5) | |||||
| AFP (N=278) | 0.62 | 0.50 | <0.001 | ||||||||
| ≤400 ng/mL | 109 (39.2) | 104 (37.4) | 108 (38.8) | 105 (37.8) | 128 (46) | 85 (30.6) | |||||
| >400 ng/mL | 31 (11.2) | 34 (12.2) | 36 (12.9) | 29 (10.4) | 20 (7.2) | 45 (16.2) | |||||
| OS event | 0.26 | 0.01 | 0.01 | ||||||||
| Alive | 115 (31) | 126 (34) | 132 (35.6) | 109 (29.4) | 132 (35.6) | 109 (29.4) | |||||
| Dead | 70 (18.9) | 60 (16.2) | 53 (14.3) | 77 (20.8) | 53 (14.3) | 77 (20.8) | |||||
HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; AA, African American; AFP, alpha-fetoprotein; OS, overall survival; DPH, diphthamide.
Table 4
| Characteristics | DPH5 expression, n (%) | DPH6 expression, n (%) | DPH7 expression, n (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Low (N=185) | High (N=186) | P value | Low (N=185) | High (N=186) | P value | Low (N=185) | High (N=186) | P value | |||
| Pathologic T stage (N=368) | 0.005 | 0.07 | 0.07 | ||||||||
| T1 | 104 (28.3) | 77 (20.9) | 99 (26.9) | 82 (22.3) | 98 (26.6) | 83 (22.6) | |||||
| T2 & T3 & T4 | 80 (21.7) | 107 (29.1) | 85 (23.1) | 102 (27.7) | 84 (22.8) | 103 (28) | |||||
| Histologic grade (N=366) | <0.001 | <0.001 | <0.001 | ||||||||
| G1 & G2 | 134 (36.6) | 98 (26.8) | 136 (37.2) | 96 (26.2) | 136 (37.2) | 96 (26.2) | |||||
| G3 & G4 | 49 (13.4) | 85 (23.2) | 48 (13.1) | 86 (23.5) | 45 (12.3) | 89 (24.3) | |||||
| Race (N=359) | 0.02 | 0.001 | 0.07 | ||||||||
| Asian | 68 (18.9) | 90 (25.1) | 62 (17.3) | 96 (26.7) | 70 (19.5) | 88 (24.5) | |||||
| Black or AA & White | 112 (31.2) | 89 (24.8) | 113 (31.5) | 88 (24.5) | 108 (30.1) | 93 (25.9) | |||||
| Weight (N=344) | 0.23 | <0.001 | <0.001 | ||||||||
| ≤70 kg | 86 (25) | 96 (27.9) | 76 (22.1) | 106 (30.8) | 74 (21.5) | 108 (31.4) | |||||
| >70 kg | 87 (25.3) | 75 (21.8) | 97 (28.2) | 65 (18.9) | 98 (28.5) | 64 (18.6) | |||||
| AFP (N=278) | 0.67 | 0.054 | <0.001 | ||||||||
| ≤400 ng/mL | 108 (38.8) | 105 (37.8) | 111 (39.9) | 102 (36.7) | 127 (45.7) | 86 (30.9) | |||||
| >400 ng/mL | 31 (11.2) | 34 (12.2) | 25 (9) | 40 (14.4) | 14 (5) | 51 (18.3) | |||||
| OS event (N=371) | 0.97 | 0.26 | 0.20 | ||||||||
| Alive | 120 (32.3) | 121 (32.6) | 115 (31) | 126 (34) | 126 (34) | 115 (31) | |||||
| Dead | 65 (17.5) | 65 (17.5) | 70 (18.9) | 60 (16.2) | 59 (15.9) | 71 (19.1) | |||||
HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; AA, African American; AFP, alpha-fetoprotein; OS, overall survival; DPH, diphthamide.
Diagnostic and predictive value of DPHs in HCC
We evaluated the ability of DPHs as diagnostic indicators of HCC using ROC analysis. The results showed that DPHs had considerable diagnostic value for HCC, such as DPH1 (AUC =0.623, 95% CI: 0.562–0.685; Figure 4A); DPH2 (AUC =0.950, 95% CI: 0.923–0.977; Figure 4B); DPH3 (AUC =0.893, 95% CI: 0.855–0.932; Figure 4C); and DPH5 (AUC =0.924, 95% CI: 0.895–0.953, Figure 4D); DPH6 (AUC =0.883, 95% CI: 0.846–0.919; Figure 4E); DPH7 (AUC =0.962, 95% CI: 0.941–0.983; Figure 4F). We separated the HCC patient data from TCGA into high and low-expression groups based on median expression levels to further investigate the predictive significance of DPHs in HCC. Kaplan-Meier survival curves showed that patients had worse OS with high expression levels of DPH2 (HR =1.84, 95% CI: 1.30–2.61, P<0.001; Figure 4G) and DPH3 (HR =1.81, 95% CI: 1.27–2.57, P<0.001; Figure 4H). However, the OS of HCC patients was higher in those with high DPH7 expression levels (HR =1.46, 95% CI: 1.03–2.07, P=0.03; Figure 4I), DPH1,5,6 did not affect the prognosis of HCC patients (Figure 4J-4L).

To further explore the diagnostic value of DPH2,3 in HCC, we performed univariate and multivariate Cox regression analyses using RNA-seq data and clinical information from the TCGA database.
We conducted univariate and multivariate Cox regression analyses using RNA-seq data and clinical information from the TCGA database to better understand the predictive importance of DPHs in HCC. The variables considered in our analysis were tumor node metastasis (TNM) stage, histological grade, gender, age, and DPH family gene expression levels. Our univariate Cox regression analysis showed a strong correlation between the expression of DPH2 (HR =1.947, 95% CI: 1.371–2.766, P<0.001; Figure 5) and DPH3 (HR =1.828, 95% CI: 1.287–2.597, P<0.001; Figure 5) with OS. Furthermore, our multivariate Cox regression analysis indicated that DPH2 (HR =1.738, 95% CI: 1.093–2.763, P=0.02; Figure 5) and DPH3 (HR = 1.833, 95% CI: 1.164–2.887, P=0.009; Figure 5) are independent predictors in HCC patients.

Methylation, mutation analysis, and GSEA of DPH2,3 in HCC
To understand the reasons for the higher expression of DPH2,3 in HCC, we examined the mutations and methylation patterns associated with DPH2,3 in HCC. Only a very small proportion of the 1,348 HCC samples in the cBioPortal database had genetic alterations, including 0.4% for DPH2 and 0.3% for DPH3, and most of them were in the form of mutations and amplifications (Figure 6A,6B). Therefore, we propose that copy number amplification or mutation may not be the main cause of DPH2,3 overexpression in HCC.

According to data from the TCGA database, levels of DPH2,3 methylation in HCC were lower than those found in normal tissue (Figure 6C,6D). The MethSurv database revealed that the DPH2 probes Body-Island-cg05050882 (HR =0.638, P=0.01; Figure 6E) and TSS1500-Island-cg20577479 (HR =0.556, P<0.001; Figure 6F) showed a poor prognosis in the hypomethylated group. Similarly, the DPH3 probe 3’UTR-Open Sea-cg18650518 (HR =0.664, P=0.03; Figure 6G) demonstrated that the hypomethylation group had a poor prognosis. Based on these findings, we suggest that elevated DPH2,3 expression in HCC patients may be linked to hypomethylation levels, which could adversely impact their prognosis.
To investigate the mechanism of DPH2,3 in HCC, we performed a GSEA analysis. The results showed that DPH2 was positively correlated with 12 pathways, including cell cycle, DNA replication, homologous recombination, mismatch repair, pathogenic Escherichia coli infection, Fc gamma R mediated phagocytosis, P53 signaling pathway, gap junction, small cell lung cancer, pathways in cancer, neuroactive ligand receptor interaction and calcium signaling pathway (Figure 7A,7B), but with complement and coagulation cascades, peroxisome, peroxisome proliferators-activated recepto (PPAR) signaling pathway, drug metabolism cytochrome P450, oxidative phosphorylation and metabolism of xenobiotics by cytochrome P450 (Figure 7C) were negatively correlated. DPH3 was positively correlated with MAPK signaling pathway, focal adhesion, small cell lung cancer, cytokine cytokine receptor interaction, pathways in cancer, Wnt signaling pathway, ECM receptor interaction, pathogenic Escherichia coli infection, gap junction, primary immunodeficiency, Fc Gamma R mediated phagocytosis, and cell cycle (Figure 7D,7E). and negatively correlated with drug metabolism cytochrome P450, metabolism of xenobiotics by cytochrome P450, peroxisome, oxidative phosphorylation, drug metabolism other enzymes and PPAR signaling pathway (Figure 7F) were negatively correlated. The above results suggest that DPH2,3 may be involved in the progression of HCC by influencing immune regulation, cell cycle, cancer pathways, and substance metabolism.

DPH2,3 is correlated to immune-related genes and immune cell infiltration
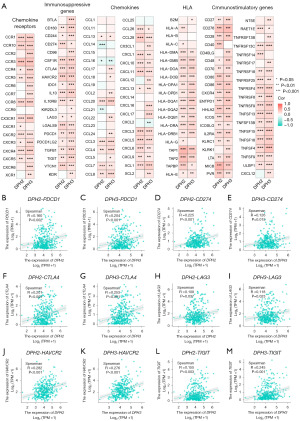
To investigate the immune action mechanism of DPH2,3, we analyzed its association with various types of immunity, including immunosuppressive, immunostimulatory, human leukocyte antigen (HLA), chemokine, and chemokine receptor genes. Our analysis showed a positive correlation between DPH2,3 and most of the immune-related genes (Figure 8A). Additionally, we studied the correlation between DPH2,3 and immune checkpoint molecules, such as PDCD1 (DPH2: R=0.16, P=0.002; DPH3: R=0.204, P<0.001; Figure 8B,8C), CD274 (DPH2: R=0.225, P<0.001; DPH3: R=0.126, P=0.02; Figure 8D,8E), CTLA-4 (DPH2: R=0.201, P<0.001; DPH3: R=0.253, P<0.001; Figure 8F,8G), LAG3 (DPH2: R=0.108, P=0.04; DPH3: R=0.118, P=0.02; Figure 8H,8I), HAVCR (DPH2: R=0.282, P<0.001; DPH3: R=0.276, P<0.001; Figure 8J,8K), and TIGIT (DPH2: R=0.155, P=0.003; DPH3: R=0.245, P<0.001; Figure 8L,8M), which are widely studied in HCC. Our analysis revealed a positive correlation between DPH2,3 and these immune checkpoint molecules.
We investigated the correlation between DPH2,3 expression and 24 different types of immune cells in HCC. Our findings revealed that DPH2 was positively correlated only with Th2 cells (R=−0.345, P<0.001). On the other hand, it was negatively associated with Th17 cells (R=−0.309, P<0.001), CD8 T cells (R=−0.288, P<0.001), natural killer (NK) cells (R=−0.253, P<0.001), cytotoxic cells (R=−0.251, P<0.001), plasmacytoid dendritic cells (pDC) (R=−0.243, P<0.001), eosinophils (R=−0.233, P<0.001), dendritic cells (DC) (R=−0.232, P<0.001), mast cells (R=−0.197, P<0.001), neutrophils (R=−0.186, P<0.001), NK CD56dim cells (R=−0.176, P<0.001), and B cells (R=−0.171, P<0.001) (Figure 9A). On the other hand, DPH3 was positively correlated with T helper cells (R=0.242, P<0.001), Th2 cells (R=0.202, P<0.001), macrophages (R=0.182, P<0.001), and activated dendritic cells (aDC) (R=0.104, P=0.02). However, it was negatively associated with Th17 cells (R=−0.339, P<0.001), NK cells (R=−0.163, P=0.002), and neutrophils (R=−0.116, P=0.02) (Figure 9B).

Next, we wanted to understand how DPH2,3 expression affects immune cell infiltration in the TME. To this end, we divided HCC samples into two groups based on the expression level of DPH2,3-low-expression and high-expression groups. Further, we calculated the enrichment scores of immune cells in both groups. The results showed that the higher scores in the DPH2 high-expression group were for Th2 cells, while the higher scores in the low-expression group were for 14 immune cell types including Th17 cells, CD8 T cells, and NK cells. These differences were statistically significant (all P<0.05; Figure 9C). Similarly, the higher scores in the DPH3 high-expression group were for T helper cells and Th2 cells, whereas the higher scores in the DPH3 low-expression group were for 8 immune cell types including Th17 cells (all P<0.05; Figure 9D). Thus, our findings suggest that increased expression of DPH2,3 in HCC affects the infiltration of immune cells in the TME, which may contribute to the survival of tumor cells.
Validation of DPHs mRNA expression in HCC cell lines and tissue samples using RT-qPCR
To verify the expression of DPHs, we first analyzed the mRNA levels in tissues using RT-qPCR. We found that the expression of DPH1 (P<0.001; Figure 10A), DPH2 (P<0.001; Figure 10B), DPH3 (P<0.001; Figure 10C), DPH5 (P<0.001; Figure 10D), and DPH6 (P<0.001; Figure 10E), DPH7 (P<0.001; Figure 10F) was elevated in HCC. Next, we analyzed the mRNA levels in tissues using RT-qPCR and found that DPH1 expression was highest in MHCC97-H cells and relatively low in HCC-LM3 cells (Figure 10G), DPH2 expression was highest in Hep-G2 cells and relatively low in Hep3B cells (Figure 10H), DPH3 expression was highest in Hep3B cells and relatively low in SMMC-7721 cells (Figure 10I), DPH5 had the highest expression in Hep-G2 cells and relatively low expression in HCC-LM3 cells (Figure 10J), DPH6 had the highest expression in Hep-G2 cells and relatively low expression in SMMC-7721 cells (Figure 10K), and DPH7 had the highest expression in Hep-G2 cells and relatively low expression in HCC-LM3 cells (Figure 10L). Overall, these results suggest that the DPH gene family is differentially expressed in HCC cells.

Discussion
Tumor cells promote their genesis and progression by reprogramming the translational process, in which high levels of protein synthesis play a key role in maintaining the metabolism of cancer cells (33). In this process, eEF2 plays an important role (34). It has been found that Huanglian Jiedu Tang (HLJDD) may promote the phosphorylation of eEF2 by activating the AMPK, mTOR pathway, which inhibits the activity of eEF2 and thus prevents the progression of HCC (35). Notably, overexpression and enhanced activity of eEF2 in cancer are closely associated with cancer cell progression as well as early tumor recurrence (36,37). DPHs encode a key group of enzymes involved in the synthesis of diphthamide, a specifically modified histidine presents only in eEF2. the status of diphthamide influences the active state of eEF2 (11). Nevertheless, there is a lack of detailed studies reported on the expression pattern and the possible functions of DPHs in HCC.
A recent study of HCC based on a mouse model found that reduced DPH1 expression was strongly associated with advanced disease and shorter survival time in patients with HCC. Nevertheless, DPH1 deletion alone does not seem to be sufficient to trigger hepatocarcinogenesis. Strikingly, while the downregulation of DPH1 limited translation prolongation rates, it increased total eEF2 levels (17). On the other hand, a study noted that the inactivation of either allele of DPH1 or DPH2 resulted in the accumulation of unmodified eEF2 and suggested that DPH1 and DPH2 were critical base for the synthesis of diphtheria (38). Meanwhile, DPH2,3 was found to maintain DPH1[4Fe-4S] cluster activity and reducibility, thus ensuring diphthamide function (39). In addition, a study revealed that the development and recurrence of basal cell carcinoma are associated with a DPH3 promoter mutation, a mutation that also promotes migration and invasion of malignant cells (20). In our study, we observed up-regulation DPH1 and DPH2,3 expression, which may be a key mechanism adopted by tumor cells to maintain a high level of protein synthesis. DPH5 serves as a key enzyme required for the second step in the process of diphthamide synthesis. Even in cases where diphthamide synthesis is hindered or incomplete, the presence of DPH5 ensures the accuracy of eEF2 during translation (12). Another study revealed that diphthamide deficiency syndrome triggered by DPH5 deficiency is a novel autosomal recessive Mendelian disease (40). In our study, the expression of DPH5 was significantly increased, which may be a key strategy adopted by tumor cells to ensure the accuracy of protein translation. In the third step of diphthamide synthesis, DPH6 and DPH7 are jointly involved in completing the process. Specifically, DPH7 is a methyl esterase that hydrolyzes the diphthyl ether generated by the methylation of DPH5, thus paving the way for the amidation of DPH6 to complete the final step of diphthamide synthesis (41). Strikingly, our study showed an increase in DPH6 expression. However, the expression of DPH7 was unstable, which may be caused by tissue variability. This needs to be verified with more experiments. Unexpectedly, the expression of DPH7 was inconsistent with the expected results. This phenomenon may be related to its role as a potential negative regulator of RNA polymerase I (12). and it has been shown that the tumor’s protein requirements for growth and cell division are dependent on RNA polymerase I (42). It was found that DPH1, DPH2, and DPH5 play a crucial role in diphthamide synthesis. Although the deletion of diphthamide itself is not fatal, the deletion of DPH3, DPH6, and DPH7 cannot be compensated by other genes (38,43). By analyzing the expression pattern of DPHs in pan-carcinomas, especially HCC, we found that the expression of DPH1,7 was unstable, while the expression of DPH2,3,5,6 increased. In this study, we used a sample of 18 cases to validate this expression pattern. We plan to expand the sample size in future studies to further validate and enhance our findings, ensuring their reliability and applicability on a larger scale. Based on this expression pattern, we hypothesized that HCC cells can ensure high levels of expression of their proteins through this expression pattern, thus driving tumorigenesis and progression.
To gain insight into the mechanism of DPHs in HCC, we further investigated their association with clinicopathologic features. The results showed that DPHs were strongly correlated with histologic grade, especially DPH2,3 was significantly correlated with OS. We also evaluated the efficacy of DPHs in the diagnosis and prognosis of HCC. The results of ROC analysis confirmed the high diagnostic value of DPHs. By Kaplan-Meier survival curve analysis, we found that patients with high expression of DPH2, DPH3, and DPH7 had a poor prognosis. Further univariate and multivariate Cox regression analyses also showed that the expression of DPH2,3 was significantly associated with OS time. Thus, DPH2,3 is not only valuable in the diagnosis of HCC but also independent factors affecting the prognosis of HCC patients. Next, we first used the cBioportal database to analyze the mutation status of these two genes, and then we performed an analysis of the promoter methylation status of DPH2,3 genes and their prognosis with the help of the UALCAN database and MethSurv to explore the reasons for the increased expression of DPH2,3 in HCC. The results suggested that in HCC, the overexpression of DPH2,3 was likely to be caused by the hypomethylation status of their promoters rather than gene mutations. Furthermore, this hypomethylation status may be a key factor contributing to the poor prognosis of HCC patients. To gain more insight into the regulatory mechanisms of DPH2,3 in HCC, we performed a GSEA. The results showed that DPH2,3 were positively correlated with cell cycle, Fc Gamma R mediated phagocytosis, and pathways in cancer, whereas they were positively correlated with peroxisome, PPAR signaling pathway, drug metabolism cytochrome P450, oxidative phosphorylation and oxidative phosphorylation were negatively correlated. Therefore, we hypothesized that HCC cells may promote their growth and development by regulating the cell cycle, cancer-related pathways, and certain immune and metabolic pathways.
TME in HCC is highly immunosuppressive, comprising various cell types, cytokines, and other components that impair antitumor immunity (44). The DPH gene family, particularly DPH2 and DPH3, may play a crucial role in regulating the immune microenvironment of HCC by influencing immune-related gene expression and immune cell distribution. To gain insight into the immune mechanisms of DPH2,3 within HCC, we first investigated the relationship between mRNA expression of DPH2,3 and the five major immune modalities, including chemokines receptor genes, immunosuppressive genes, chemokines genes, HLA genes, and immunostimulatory genes (43). in particular, we presented the association between DPH2,3 and common immune checkpoints (PD-1, PD-L1, CTLA-4, LAG3, TIM3, and TIGIT) (45). A previous study has shown that therapeutic strategies for HCC targeting immune checkpoints are highly promising (46). Our results showed that DPH2,3 was significantly and positively associated with most immune-related genes and common immune checkpoints. Despite the success of immune checkpoint inhibitors in treating HCC, drug resistance remains a significant issue, resulting in poor therapeutic outcomes (47). Our study indicates that high DPH2/3 expression may inhibit anti-tumor cell activity by regulating immunosuppressive pathways (e.g., PD-1/PD-L1 and CTLA-4), enabling tumor cells to evade immune surveillance and attack. Therefore, silencing DPH2/3 expression may enhance the efficacy of immune checkpoint inhibitors against tumor cells. The presence of immune-associated cells in the TME has a major impact on the progression of HCC. Tumor cells employ a variety of strategies to evade immune detection and clearance (48). Effective antitumor immunotherapy relies on the synergistic action of the components of the immune system (49) and utilizes immune cells inside and outside the TME to localize and destroy cancer cells (50-52), where combinations of immune checkpoint inhibitors and immune cell therapy are effective (53). We hypothesize that HCC cells disturb the immune mechanism by overexpressing DPH2/3, enabling tumor cells to evade immune detection and clearance. By altering the distribution of immune cells within the immune microenvironment, tumor cells evade immune attack. Therefore, we conducted an in-depth study of the association of 24 immune-infiltrating cells with DPH2,3 expression. The results showed that high DPH2/3 expression was associated with an increase in various pro-tumorigenic immune cells, including Th2 cells, helper T cells, and macrophages. These cells promote tumor growth and metastasis by secreting pro-inflammatory and growth factors (e.g., IL-4, IL-10, and TGF-β). Conversely, high DPH2/3 expression suppressed immune cells with anti-tumor effects, such as Th17 cells, CD8 T cells, and NK cells. These cells exert their effects by secreting cytotoxic factors (e.g., IFN-γ and TNF-α) and directly killing tumor cells (48,49). These results suggest that DPH2/3 modifies the immune status of the TME by adjusting the ratio and activity of immune cells, thereby influencing HCC progression. High DPH2/3 expression predicts poor prognosis for HCC patients. Targeting DPH2/3 and its pathways may restore anti-tumor immune responses and enhance immunotherapy, thus improving patient survival. Therefore, DPH2/3 is a potential immunotherapy target in HCC patients. Next, we will investigate the mechanisms of DPHs in tumors and conduct further experiments to verify their feasibility and effectiveness as therapeutic targets. These studies will aid in developing new therapeutic strategies and providing more effective treatment options for HCC patients.
Another concern is that when diphthamide is intact, the toxin causes ADP-ribosylation of the diphthamide, leading to blocked protein synthesis and thus cell death. Based on this, we hypothesized whether it would be possible to treat tumor cells by restoring their diphthamide integrity and then targeting the toxin to the tumor cells. Therefore, we will further investigate the mechanism of action of DPHs in tumors to explore their potential in the treatment of tumors.
Conclusions
Overall, HCC maintains its genesis and progression by finely regulating the expression patterns of DPHs. Particularly noteworthy, DPH2,3 showed significant clinical value in the diagnosis and prognosis of HCC. Further studies revealed that DPH2,3 plays crucial roles in the immune microenvironment and immunoregulation, which may make them potential therapeutic targets for immunotherapy.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding: This project was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-147/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-147/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-147/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at: https://tcr.amegroups.com/article/view/10.21037/tcr-24-147/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by The Medical Ethics Committee of the Affiliated Hospital of North Sichuan Medical College (protocol No. 2023ER153-1). All patients who participated in the study provided written informed consent before the collection of tissue samples.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ganesan P, Kulik LM. Hepatocellular Carcinoma: New Developments. Clin Liver Dis 2023;27:85-102. [Crossref] [PubMed]
- Toh MR, Wong EYT, Wong SH, et al. Global Epidemiology and Genetics of Hepatocellular Carcinoma. Gastroenterology 2023;164:766-82. [Crossref] [PubMed]
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin 2024;74:12-49. Erratum in: CA Cancer J Clin 2024;74:203. [Crossref] [PubMed]
- Vogel A, Meyer T, Sapisochin G, et al. Hepatocellular carcinoma. Lancet 2022;400:1345-62. [Crossref] [PubMed]
- Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019;156:477-491.e1. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Llovet JM, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol 2022;19:151-72. [Crossref] [PubMed]
- Llovet JM, De Baere T, Kulik L, et al. Locoregional therapies in the era of molecular and immune treatments for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2021;18:293-313. [Crossref] [PubMed]
- Wang H, Shi F, Zheng S, et al. Feasibility of hepatocellular carcinoma treatment based on the tumor microenvironment. Front Oncol 2022;12:896662. [Crossref] [PubMed]
- Zhang H, Quintana J, Ütkür K, et al. Translational fidelity and growth of Arabidopsis require stress-sensitive diphthamide biosynthesis. Nat Commun 2022;13:4009. [Crossref] [PubMed]
- Mateus-Seidl R, Stahl S, Dengl S, et al. Interplay between reversible phosphorylation and irreversible ADP-ribosylation of eukaryotic translation elongation factor 2. Biol Chem 2019;400:501-12. [Crossref] [PubMed]
- Uthman S, Bär C, Scheidt V, et al. The amidation step of diphthamide biosynthesis in yeast requires DPH6, a gene identified through mining the DPH1-DPH5 interaction network. PLoS Genet 2013;9:e1003334. [Crossref] [PubMed]
- Liu S, Milne GT, Kuremsky JG, et al. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol Cell Biol 2004;24:9487-97. [Crossref] [PubMed]
- Mayer K, Schröder A, Schnitger J, et al. Influence of DPH1 and DPH5 Protein Variants on the Synthesis of Diphthamide, the Target of ADPRibosylating Toxins. Toxins (Basel) 2017;9:78. [Crossref] [PubMed]
- Erdogan MA, Ashour A, Yuca E, et al. Targeting eukaryotic elongation factor-2 kinase suppresses the growth and peritoneal metastasis of ovarian cancer. Cell Signal 2021;81:109938. [Crossref] [PubMed]
- Zhu S, Liao M, Tan H, et al. Inhibiting Eukaryotic Elongation Factor 2 Kinase: An Update on Pharmacological Small-Molecule Compounds in Cancer. J Med Chem 2021;64:8870-83. [Crossref] [PubMed]
- Tu WL, Chih YC, Shih YT, et al. Context-specific roles of diphthamide deficiency in hepatocellular carcinogenesis. J Pathol 2022;258:149-63. [Crossref] [PubMed]
- Liu M, Yin K, Guo X, et al. Diphthamide Biosynthesis 1 is a Novel Oncogene in Colorectal Cancer Cells and is Regulated by MiR-218-5p. Cell Physiol Biochem 2017;44:505-14. [Crossref] [PubMed]
- Maturo MG, Rachakonda S, Heidenreich B, et al. Coding and noncoding somatic mutations in candidate genes in basal cell carcinoma. Sci Rep 2020;10:8005. [Crossref] [PubMed]
- Nawrocka PM, Galka-Marciniak P, Urbanek-Trzeciak MO, et al. Profile of Basal Cell Carcinoma Mutations and Copy Number Alterations - Focus on Gene-Associated Noncoding Variants. Front Oncol 2021;11:752579. [Crossref] [PubMed]
- Yao T, Liu JJ, Zhao LJ, et al. Identification of new fusion genes and their clinical significance in endometrial cancer. Chin Med J (Engl) 2019;132:1314-21. [Crossref] [PubMed]
- Vivian J, Rao AA, Nothaft FA, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol 2017;35:314-6. [Crossref] [PubMed]
- Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Chandrashekar DS, Karthikeyan SK, Korla PK, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022;25:18-27. [Crossref] [PubMed]
- Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400-416.e11. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018;10:277-88. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Bilanges B, Stokoe D. Mechanisms of translational deregulation in human tumors and therapeutic intervention strategies. Oncogene 2007;26:5973-90. [Crossref] [PubMed]
- Kovalski JR, Kuzuoglu-Ozturk D, Ruggero D. Protein synthesis control in cancer: selectivity and therapeutic targeting. EMBO J 2022;41:e109823. [Crossref] [PubMed]
- Wang N, Feng Y, Tan HY, et al. Inhibition of eukaryotic elongation factor-2 confers to tumor suppression by a herbal formulation Huanglian-Jiedu decoction in human hepatocellular carcinoma. J Ethnopharmacol 2015;164:309-18. [Crossref] [PubMed]
- Vasamsetti BMK, Liu Z, Park YS, et al. Muscarinic acetylcholine receptors regulate the dephosphorylation of eukaryotic translation elongation factor 2 in SNU-407 colon cancer cells. Biochem Biophys Res Commun 2019;516:424-9. [Crossref] [PubMed]
- Zhang X, Hu L, Du M, et al. Eukaryotic Elongation Factor 2 (eEF2) is a Potential Biomarker of Prostate Cancer. Pathol Oncol Res 2018;24:885-90. [Crossref] [PubMed]
- Stahl S, da Silva Mateus Seidl AR, Ducret A, et al. Loss of diphthamide pre-activates NF-κB and death receptor pathways and renders MCF7 cells hypersensitive to tumor necrosis factor. Proc Natl Acad Sci U S A 2015;112:10732-7. [Crossref] [PubMed]
- Zhang Y, Su D, Dzikovski B, et al. Dph3 Enables Aerobic Diphthamide Biosynthesis by Donating One Iron Atom to Transform a [3Fe-4S] to a [4Fe-4S] Cluster in Dph1-Dph2. J Am Chem Soc 2021;143:9314-9. [Crossref] [PubMed]
- Shankar SP, Grimsrud K, Lanoue L, et al. A novel DPH5-related diphthamide-deficiency syndrome causing embryonic lethality or profound neurodevelopmental disorder. Genet Med 2022;24:1567-82. [Crossref] [PubMed]
- Lin Z, Su X, Chen W, et al. Dph7 catalyzes a previously unknown demethylation step in diphthamide biosynthesis. J Am Chem Soc 2014;136:6179-82. [Crossref] [PubMed]
- Ferreira R, Schneekloth JS Jr, Panov KI, et al. Targeting the RNA Polymerase I Transcription for Cancer Therapy Comes of Age. Cells 2020;9:266. [Crossref] [PubMed]
- Wu B, Fu L, Guo X, et al. Multi-omics profiling and digital image analysis reveal the potential prognostic and immunotherapeutic properties of CD93 in stomach adenocarcinoma. Front Immunol 2023;14:984816. [Crossref] [PubMed]
- Shen KY, Zhu Y, Xie SZ, et al. Immunosuppressive tumor microenvironment and immunotherapy of hepatocellular carcinoma: current status and prospectives. J Hematol Oncol 2024;17:25. [Crossref] [PubMed]
- Liu Z, Liu X, Liang J, et al. Immunotherapy for Hepatocellular Carcinoma: Current Status and Future Prospects. Front Immunol 2021;12:765101. [Crossref] [PubMed]
- Oura K, Morishita A, Tani J, et al. Tumor Immune Microenvironment and Immunosuppressive Therapy in Hepatocellular Carcinoma: A Review. Int J Mol Sci 2021;22:5801. [Crossref] [PubMed]
- Ladd AD, Duarte S, Sahin I, et al. Mechanisms of drug resistance in HCC. Hepatology 2024;79:926-40. [Crossref] [PubMed]
- Hao X, Sun G, Zhang Y, et al. Targeting Immune Cells in the Tumor Microenvironment of HCC: New Opportunities and Challenges. Front Cell Dev Biol 2021;9:775462. [Crossref] [PubMed]
- Paijens ST, Vledder A, de Bruyn M, et al. Tumor-infiltrating lymphocytes in the immunotherapy era. Cell Mol Immunol 2021;18:842-59. [Crossref] [PubMed]
- Chakraborty E, Sarkar D. Emerging Therapies for Hepatocellular Carcinoma (HCC). Cancers (Basel) 2022;14:2798. [Crossref] [PubMed]
- Ma S, Li X, Wang X, et al. Current Progress in CAR-T Cell Therapy for Solid Tumors. Int J Biol Sci 2019;15:2548-60. [Crossref] [PubMed]
- Xie G, Dong H, Liang Y, et al. CAR-NK cells: A promising cellular immunotherapy for cancer. EBioMedicine 2020;59:102975. [Crossref] [PubMed]
- Mizukoshi E, Kaneko S. Immune cell therapy for hepatocellular carcinoma. J Hematol Oncol 2019;12:52. [Crossref] [PubMed]


