
Analysis of the role of POC1A in the development and progression of hepatocellular carcinoma
Highlight box
Key findings
• This study was the first to identify a gene [i.e., POC1 centriolar protein A (POC1A)] associated with the progression of hepatocellular carcinoma (HCC).
• Through a multidimensional validation analysis and a correlation analysis of the clinical parameters, this study showed that this gene significantly affects the prognosis of patients with HCC through the regulation of cell-cycle pathways and chromosome segregation functions.
• In terms of immunity, the high expression of POC1A and its interacting genes may play an important role in B cells and macrophages.
What is known, and what is new?
• Gene signatures and biomarkers play a crucial role in HCC prognosis and personalized medicine.
• The identification of this new potential therapeutic target provides a theoretical basis for the development of new drugs.
What is the implication, and what should change now?
• Our findings provide a rationale for the treatment of HCC and the development of new drugs against target genes.
• Further validation studies need to be conducted to assess the specific molecular mechanisms by which genes influence the prognosis of HCC patients.
Introduction
There are various types of liver cancer, including hepatocellular carcinoma (HCC), cholangiocarcinoma, and mixed cell carcinoma, among which, HCC is the most common histological type (1). HCC is also the most common type of cancer (1). Numerous studies have shown that the occurrence and development of HCC is a complex multi-step process that involves sustained inflammatory damage, including liver cell necrosis and regeneration, and fibrotic deposition, and is not related to the absolute regulation of a single gene. After their activation in the immune system, inflammatory cells release a large amount of free radicals, such as reactive oxygen species and nitric oxide, which can also cause DNA damage and mutations in oncogenes (2,3).
Depending on the stage of HCC, the treatment methods differ; however, currently, there are insufficient biomarkers for the identification and diagnosis of HCC. Choosing the optimal treatment method can maximize the therapeutic effect and improve the survival rate. Presently, the widely used biomarkers for diagnosing HCC include serum alpha-fetoprotein (AFP), core-fused AFP, and deprotection-γ-carboxyprothrombin (4). These biomarkers play a certain role in the diagnosis of HCC, but their sensitivity is limited. Thus, there is an urgent need to identify novel biomarkers with greater specificity and sensitivity.
Bioinformatics is an emerging interdisciplinary area in the fields of biology, computer science, and informatics that can acquire, store, process, and interpret biological information. In recent years, the development of high-throughput sequencing has enabled this technology to more efficiently show the significance of a large and complex amount of biological data. Bioinformatics analysis can improve our understanding of the molecular mechanisms underlying cancer pathogenesis. Combining bioinformatics analysis with various omics approaches enables more detailed research in genomics, transcriptomics, proteomics, and other fields, and the identification of key genes through differential gene analysis. The identification of significant biomarkers provides new methods for tumor diagnosis, the prediction of clinical outcomes, and targeted therapy (5). Liang et al. conducted a bioinformatics analysis to examine the role of HELLS in promoting, inhibiting, and treating different cancers, and showed that HELLS had immune, targeted, and cytotoxic therapeutic value (6).
Bioinformatics analysis can also provide insights into drug treatment mechanisms. Yang et al. adopted a network pharmacology and multi-omics approach to investigate the active pharmaceutical ingredients and their mechanism of action in the treatment of sepsis (7). A fundamental goal of cancer research is to understand the mechanisms of cell transformation. The ability to identify all the genes that drive mutations in tumors plays an important role in cancer research. Carefully designed bioinformatics methods provide an effective means for identifying driver genes (8). In addition, bioinformatics technology can also help decipher the cancer epigenome to study cancer immune evasion (9).
POC1 centriolar protein A (POC1A), also known as WDR51, is a gene that encodes a protein that plays a critical role in the centrosome, and is one of the two subtypes of POC1. The POC1A protein has a WD40 protein domain at the N-terminus and a coiled-coil domain at the C-terminus. Mutations in this gene may lead to the occurrence of human fibrosis (10). POC1 is related to centromere replication, length control, and cilia formation. POC1A interacts with POC1B, and is also related to centromere integrity. The loss of one POC1 protein does not significantly disrupt mitosis, but the depletion of both proteins can lead to spindle tissue defects, resulting in inequality or unipolar spindle formation (11). POC1A plays an important role in the formation and maintenance of centrosomes, and the integrity and replication of centrosomes are crucial for the cell cycle. Thus, mutations in this gene may have adverse effects on the formation of mitotic spindles (12). In a study on the mechanism of gastric cancer, POC1A was identified as a significantly differentially expressed gene, and it was hypothesized that POC1A may act as a tumor suppressor in gastric cancer by regulating the cell cycle and cell growth (13). Kasavi et al. conducted a meta-analysis and differential analysis on a transcriptome dataset related to ovarian cancer, and identified POC1A as a novel candidate prognostic biomarker for ovarian cancer (14). Zhao et al. also found that POC1A was overexpressed in almost 33 types of tumors, and a gene set enrichment analysis (GSEA) showed that POC1A was associated with the cell cycle and immune-related pathways (15). However, there is currently no research on the role of POC1A in HCC.
In this study, we investigated the role of POC1A in the occurrence and development of HCC. First, we analyzed the expression of POC1A in various types of HCC patients and its effect on survival using HCC patient information from the Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA), the Human Protein Atlas (HPA), and Hepatocellular Carcinoma Cell DataBase (HCCDB). We then explored the major enrichment pathways and gene functions of POC1A in HCC using the GSEA method and examined its protein-protein interactions (PPIs). Finally, we predicted the potential transcription factors (TFs) and target microRNAs (miRNAs) of POC1A and analyzed the single nucleotide variation (SNV) and copy number variation (CNV) mutations of POC1A and related genes in HCC, as well as their effects on immune cells. We present this article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2398/rc).
Methods
Analysis of POC1A pan-cancer expression
The differential expression of the POC1A gene between pan-cancerous tumor tissues and adjacent normal tissues was analyzed using the DiffExp module in the Tumor Immune Estimation Resource (TIMER; https://cistrome.shinyapps.io/timer/) database. The statistical significance of the differential expression was assessed using the Wilcoxon test.
Expression and prognostic analysis of POC1A in HCC
GEO2R was used to analyze the expression of POC1A in the liver cancer datasets GSE159220 and GSE55758. GSE159220 comprised three HCC samples and three normal samples, while GSE55758 comprised eight HCC samples and eight adjacent samples. A log fold change ≥|0.5|, and a P value <0.05 were set as the analysis criteria. The expression and prognosis of the POC1A gene in liver hepatocellular carcinoma (LIHC) samples from TCGA database were analyzed and validated using the Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) database. The normal samples were matched with LIHC sample data and Genotype-Tissue Expression (GTEx) sample data. At the prognostic level, the overall survival (OS) and disease-free survival (DFS) rates were analyzed using the quartile cut-off method.
Analysis of POC1A expression and prognosis in the HCCDB
The HCCDB (http://lifeome.net/database/hccdb/about.html), which is a liver cancer–specific analysis database, was used to analyze the expression of POC1A in liver cancer patient samples. The HCCDB contains 15 common liver cancer expression datasets, with up to 4,000 clinical samples, and was developed as a one-stop online resource to explore the expression of liver cancer genes through a user-friendly interface. It can also show the specific role of genes in liver cancer patients.
Expression characterization of POC1A in LIHC patient samples
The University of ALabama at Birmingham CANcer data analysis Portal (UALCAN; http://ualcan.path.uab.edu/) database was used to analyze the expression of POC1A in liver cancer patients of different ages, genders, races, and cancer stages. It enabled us to collect potentially valuable information and data for further targeted analysis and problem solving. We used the Sangerbox database (http://vip.sangerbox.com/home.html) to download the unified and standardized LIHC dataset from the University of California, Santa Cruz (UCSC; https://xenabrowser.net/) database, and further extracted the expression data of the ENSG00000164087 (POC1A) gene in each sample. We screened the samples from the Primary Blood Derived Cancer – Peripheral Blood and Primary Tumor database, and performed log2(x+0.001) transformation for each expression value to investigate the expression of POC1A at different tumor stages.
Expression confirmation of POC1A at the protein level
To analyze the expression of POC1A in normal liver samples and liver cancer samples, we used the HPA (https://www.proteinatlas.org/) database to analyze the staining degree of POC1A in tissue sections.
Cell culture, RNA extraction, and qRT-PCR
The human normal hepatocyte cell line LO2 (Servicebio Technology Co., Ltd., Wuhan, China) was cultured in Roswell Park Memorial Institute 1640 medium, and the hepatoma cell line HepG2 (Procell Life Science & Technology Co., Ltd., Wuhan, China) was cultured in Dulbecco’s Modified Eagle Medium (Hyclone, Thermo Fisher Scientific Inc., Waltham, MA), supplemented with 10% fetal bovine serum and antibiotics (10,000 U/mL penicillin and 10 mg/mL streptomycin) (Procell Life Science & Technology Co., Ltd.). The cells were incubated in a humidified atmosphere containing 5% carbon dioxide at 37 ℃. The total RNA was isolated from the intestinal tissues using the TRIzol Reagent Kit (ComWin Biotech Co., Ltd., Beijing, China) and then subjected to quantitative real-time polymerase chain reaction (qRT-PCR). RNA (1 µg) was reverse transcribed using the PrimeScript™ RT reagent Kit with genomic DNA Eraser (Takara Bio Inc., Ltd., Japan). For the messenger RNA (mRNA) analyses, the resulting complementary DNAs were analyzed in triplicate using SYBR® Premix Ex Taq™ II (Takara Bio Inc., Ltd.) on the Applied Biosystems 7500 qRT-PCR system. The relative mRNA expression level was calculated by 2−△△Ct, and the expression values were normalized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primers. The following qRT-PCR primers were used: GAPDH 5'-CAGGAGGCATTGCTGATGAT-3', 3'-GAAGGCTGGGGCTCATTT-5'; and POC1A 5'-GCATATCAACTGGGTCCGCT-3', 3'-CACATAGGTGACAAAGCCGC-5'.
HCC-specific GO and KEGG analyses of POC1A
The LinkedOmics (http://www.linkedomics.org/login.php) database was used for the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses and the related GSEA of POC1A. The patient sample was taken using the LIHC RNA sequencing sample from the University of North Carolina at Chapel Hill (UNC) institution. The LinkFinder module and a Pearson correlation test were used to analyze the genes that were significantly correlated with POC1A. The LinkInterpreter module and the GSEA algorithm was used for the GO and KEGG analyses. A false discovery rate (FDR) P value ≤0.05 was considered statistically significant.
POC1A major enrichment pathway-related genes for the PPI, miRNA, and TF prediction analyses
The analysis of POC1A-related PPIs, miRNAs targets and TFs were performed using the Network Analyst 3.0 (https://www.networkAnalyst.ca/) database. The significantly correlated POC1A pathway genes obtained from the KEGG analysis were included as predictive genes in the PPI analysis. The PPIs of POC1A were analyzed using liver samples from the DifferentialNet database, and the POC1A-associated target miRNAs were analyzed using the miRTarBase8.0 database. The POC1A-associated TFs were analyzed using the JASPER database. The website defaults were selected as the parameters.
Characterization of POC1A-interacting gene alterations
The SNV and CNV mutations of the POC1A gene in LIHC were analyzed using the Gene Set Cancer Analysis (GSCA; http://bioinfo.life.hust.edu.cn/GSCA/#/) database.
Prognostic analysis of the POC1A reciprocal genes in patients with HCC
The GEPIA2 (http://gepia2.cancer-pku.cn/#index) database was used to draw a prognostic heatmap of the POC1A interaction genes in the liver cancer patient samples. The median was chosen as the grouping threshold, and a P value <0.05 indicated that the gene had a significant effect on patient prognosis.
mRNA correlation analysis of the POC1A interaction genes in patients with HCC
The GEPIA2 database was used to analyze the correlation between the mRNA and the POC1A interaction genes in the LIHC samples. The Spearman algorithm was used to perform the paired-gene expression correlation analysis of a given TCGA and GTEx expression dataset. A P value <0.05 indicated a significant correlation between two genes.
Analysis of POC1A and the interaction genes in various immune cells
The uniformly standardized LIHC dataset was downloaded from the UCSC (https://xenabrowser.net/) database, and the samples were screened from the Primary Blood Derived Cancer-TCGA-Acute Myeloid Leukemia, Primary Tumor, Metastatic-TCGA-Skin Cutaneous Melanoma, Primary Blood Derived Cancer-Bone Marrow, Primary Solid Tumor, and Recurrent Blood Derived Cancer-Bone Marrow datasets. We performed a log2(x+0.001) transformation for each expression value, and we extracted the gene expression profile of each tumor separately from these datasets, mapped each expression profile to GeneSymbol, and used the R package ESTIMATE to calculate the stromal, immune, and ESTIMATE scores for each patient with LIHC tumors based on gene expression (16). The relationship between the expression of POC1A, the major interacting genes, and the grade of immune infiltration of various immune cell types in the HCC samples was analyzed by the TIMER database. The results showed the Rho values and statistical significance of some Spearman correlations corrected for purity. A P value <0.05 indicated a significant correlation between two genes. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
The t-test was used for the intergroup analysis. Survival curves were generated using the Kaplan-Meier method. A P value <0.05 was considered statistically significant.
Results
POC1A expression was significantly upregulated in multiple cancers
First, we examined the expression of POC1A in the pan-cancer samples. The TIMER database was used to perform the differential gene analysis of the POC1A gene in pan-cancer samples (Figure 1A). The results revealed that the POC1A gene was highly expressed in various cancer tissues compared to normal tissues.
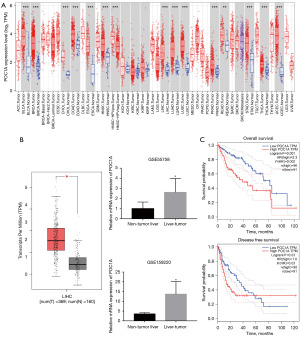
POC1A was highly expressed in HCC and significantly correlated with a poor OS rate
We selected HCC for further study and explored the expression of POC1A using the GEO dataset. The results showed that the expression of POC1A was significantly higher in TCGA-LIHC tumor samples than the normal samples (Figure 1B). Next, we analyzed the OS and DFS rates of LIHC patients based on POC1A expression. The Kaplan-Meier analysis showed that the LIHC patients with high POC1A expression had a significantly lower OS rate (log-rank P=0.001) and DFS rate (log-rank P=0.03) than those with low POC1A expression. The hazard ratio (HR) results showed that the risk of the POC1A high-expression group was significantly higher than that of the POC1A low-expression group in terms of both OS (HR =2.3, P=0.002) and DFS (HR =1.6, P=0.03). Thus, POC1A could serve as an independent prognostic risk factor for HCC (Figure 1C). Table 1 sets out the results for the analysis of the relationship between POC1A expression in the LIHC tissues and the clinicopathologic characteristics of the patients. The high expression of POC1A significantly affected the AFP level, histologic grade, and pathologic tumor (T) stage of the patients.
Table 1
| Characteristics | Low expression of POC1A (n=187) | High expression of POC1A (n=187) | P |
|---|---|---|---|
| Gender, n (%) | 0.22 | ||
| Female | 66 (17.6) | 55 (14.7) | |
| Male | 121 (32.4) | 132 (35.3) | |
| Age (years), n (%) | 0.38 | ||
| ≤60 | 84 (22.5) | 93 (24.9) | |
| >60 | 102 (27.3) | 94 (25.1) | |
| Alpha-fetoprotein, n (%) | 0.009 | ||
| ≤400 ng/mL | 116 (41.4) | 99 (35.4) | |
| >400 ng/mL | 23 (8.2) | 42 (15) | |
| Grade, n (%) | 0.009 | ||
| G1 | 29 (7.9) | 26 (7) | |
| G2 | 98 (26.6) | 80 (21.7) | |
| G3 | 56 (15.2) | 68 (18.4) | |
| G4 | 1 (0.3) | 11 (3) | |
| Pathologic T stage, n (%) | 0.03 | ||
| T1 | 101 (27.2) | 82 (22.1) | |
| T2–T4 | 83 (22.4) | 105 (28.3) | |
| Overall survival event, n (%) | 0.009 | ||
| Alive | 134 (35.8) | 110 (29.4) | |
| Dead | 53 (14.2) | 77 (20.6) |
The statistical analyses were performed using the Chi-square test. POC1A, POC1 centriolar protein A.
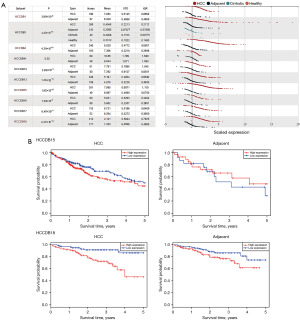
Cross-validation in the HCC-specific HCCDB
To cross-validate the accuracy of the conclusion that POC1A is highly expressed in HCC and significantly correlated with a poor OS rate, the HCCDB, a HCC-specific database, was used to further analyze the differential expression levels of the POC1A gene. The results of 10 different HCC datasets showed that in all datasets, the expression levels of the POC1A gene in the HCC tissue was significantly higher compared to that in the adjacent liver tissue (Figure 2A). In HCCDB18, there was a significant difference between the high and low expression of POC1A in the LIHC and adjacent tissues. The observation graph (Figure 2B) clearly showed that the survival rate of patients with low POC1A expression was higher than that of patients with high expression for the follow-up period, and the difference in the survival rate between the groups gradually increased as the survival years increased. However, in the HCCDB15 dataset, the effects of the POC1A expression on the prognosis of the diseased patients were the same, but the difference was not statistically significant (P=0.17). Even in the adjacent tissues with a long-term follow-up period of 2 years, the survival rate of patients with high POC1A expression was higher than that of patients with low POC1A expression (Figure 2B).
Characterization of POC1A expression in HCC
To investigate the expression of POC1A in different subtypes of LIHC patients, we compared the expression of POC1A in the LIHC tissue samples of patients with different clinical and pathological parameters. As Figure 3A shows, compared with the normal group, the expression level of POC1A was significantly increased in the HCC group. The expression levels of POC1A in both the male and female HCC patients were higher than those of the control patients, and the expression levels were higher in males than females (Figure 3B). The expression level of POC1A in the HCC patients aged ≥21 years was generally higher than that of the control patients, and the expression level of POC1A in the HCC patients aged 41–60 years was also significantly higher than that in the patients aged 61–80 years (Figure 3C). The expression levels of POC1A were all higher in the Caucasian, African-American, and Asian HCC patients than the normal patients. The expression of POC1A was also significantly higher in the tissue of Asian LIHC patients than the tissue of Caucasian LIHC patients (Figure 3D).
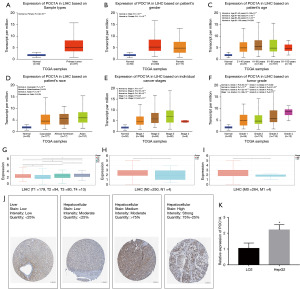
In terms of tumor staging, the expression level of POC1A in the HCC patients was positively correlated with the first three stages of the tumor, but not with the fourth stage (Figure 3E,3F). In the tumor, node, metastasis (TNM) staging criteria, T refers to the condition of the primary tumor, which is sequentially represented by T1–T4 as the volume of the tumor increases and the extent of adjacent tissue involvement increases. The results showed that POC1A was significantly correlated with the progression of staging, but there was no such significant correlation in terms of the N and M staging (Figure 3G-3I).
Immunohistochemical (IHC) staining data obtained from the HPA database showed the protein expression of POC1A (Figure 3J). Notably, the expression level of the POC1A protein was higher in the HCC tissues than the normal liver tissues. The mRNA expression levels of POC1A were examined using the normal hepatocyte cell line LO2 and the hepatoma cell line HepG2. As Figure 3K shows, the mRNA expression of POC1A in the HepG2 cell line was significantly higher than that in the LO2 cell line, which was consistent with the results of our analysis.
GO and KEGG analyses of POC1A in HCC
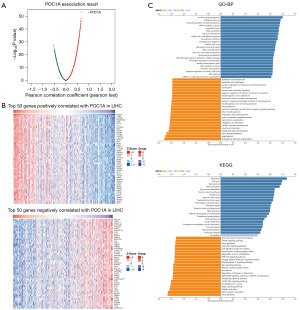
To explore the functions and possible regulatory role of POC1A in HCC, the LinkedOmics database was used to explore the co-expressed genes of POC1A. The results showed that there were 19,922 co-expressed genes with FDR values ≤0.05 and 500 times statistical analysis simulations were conducted. The co-expressed genes of POC1A is presented in the form of a volcanic map (Figure 4A). The top 50 genes that were positively or negatively correlated with POC1A were selected and presented in a heat map (Figure 4B). The heat map used the Z-score to standardize the gene expression values and displays the gene expression patterns clustered by sample grouping. GO and KEGG enrichment analyses were conducted of the co-expressed genes. The GO analysis showed that the related genes were mainly involved in chromosome separation, non-coding RNA processing, ribonucleoprotein complex biogenesis, ribosomal RNA metabolism, organelle division, DNA replication, and cell-cycle checkpoints. The KEGG analysis showed that the related genes were mainly enriched in ribosome, the cell cycle, splicing bodies, DNA replication, pyrimidine metabolism, and other processes (Figure 4C). These results suggest that POC1A may act as a part of the centrosome to affect DNA replication, regulate the cell cycle, and thus affect HCC cell proliferation, promoting tumor occurrence and development.
Prediction of interacting proteins of POC1A and potential miRNAs and TFs
We selected the cell-cycle pathway for further analysis. Figure 5A shows the GSEA enrichment map of POC1A in the cell-cycle pathway. We then conducted a PPI analysis of the enriched genes of this pathway. The results are shown in Figure 5B-5D. In this pathway, POC1A interacted with the NUDC protein. Next, we further predicted the potential miRNAs and TFs of POC1A, and the results showed that POC1A may be regulated by miRNAs such as hsa-mir-148a-3p, hsa-mir-10a-5p, and hsa-mir-10b-5p. Genes such as FOXC1, YY1, and NFIC may be the TFs of POC1A. The specific results are provided in the Figure S1.
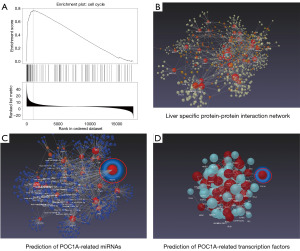
Expression and prognostic mapping of POC1A and interacting genes in HCC
To preliminarily explore the possible role of the POC1A interaction genes in HCC, we first used TCGA-LIHC patient samples from the GEPIA database to examine the expression of the POC1A interaction genes and to predict the POC1A TFs. We then plotted a prognosis map. The results are shown in Figure 6A,6B. Notably, the expression of the PPARG, NUDC, YY1, and HNF4A genes was found to be increased in the LIHC samples, but there was no significant difference between normal samples and tumor samples. The prognosis map showed that the high expression of PPARG and NUDC significantly affected the OS rate of the LICH patients. We then analyzed the correlation between the POC1A interacting genes and patient prognosis in the LIHC patient samples, and the results are shown in Figure 6C. Notably, POC1A was significantly correlated with NUDC (P=5.8×10−13) and PPARG (P=8.7×10−9). Figure 6D shows the expression of different genotypes of POC1A, NUDC, and PPARG in the LIHC samples.
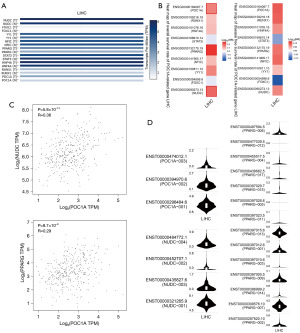
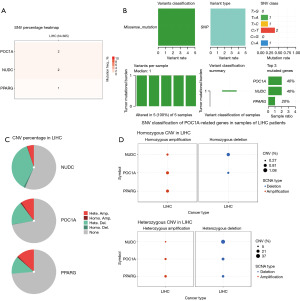
Analysis of genetic changes in POC1A and interacting genes, and analysis of multiple immune cell types in LIHC samples
To analyze the SNV and CNV changes in POC1A and the genes interacting with POC1A in the LIHC samples, we used the GSCA database. In the LIHC patient samples, POC1A, NUDC, and PPARG each had SNV mutations of varying degrees. Among the SNVs, missense mutations and single nucleotide polymorphisms, and Cytosine > Thymine were the main types of variants (Figure 7A,7B). Among the CNVs, homozygous CNVs and heterozygous CNVs were also present in POC1A, NUDC, and PPARG. Among the heterozygous CNVs, POC1A, NUDC, and PPARG were found to have different levels of deletion and amplification (Figure 7C,7D).
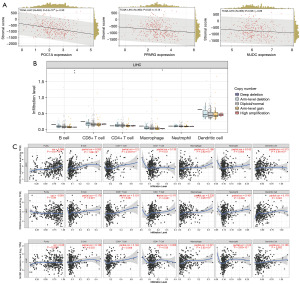
To explore POC1A and the related genes in the immune cells of the LIHC patients, we first calculated the Pearson correlation coefficients between the genes and immune infiltration scores in individual tumors using the “corr.test” function of the R package psyche (version 2.1.6) to identify the significantly correlated immune infiltration scores. We found that both POC1A (P=3.2×10−9) and PPARG (P=0.02) were significantly correlated with the immune infiltration of the LIHC samples (Figure 8A). We then used the TIMER database to analyze the possible roles of core interacting genes with POC1A in immune cells. Figure 8B shows the relationship between the infiltration levels of POC1A and six types of immune cells under different somatic copy number alterations levels in the LIHC samples. There was a significant difference in the infiltration level of high amplification between the B cells and macrophages. Figure 8C shows that the high expression of POC1A and its interacting genes play a significant role in various immune cell types. These genes and the way they interacted with POC1A warrant further analysis. Taken together, we can conclude that POC1A is significantly overexpressed in HCC, and promotes the occurrence and development of HCC by affecting the cell cycle. The use of POC1A as a potential molecular marker may play an important role in the treatment of HCC.
Discussion
Causes of HCC include chronic liver disease caused by hepatitis B virus and hepatitis C virus, long-term alcohol consumption, and non-alcoholic fatty liver disease (17). HCC has an extremely high mortality rate. This study aimed to explore whether POC1A could serve as a new therapeutic target for HCC. The analyses in this study were mainly conducted using a number of public databases and bioinformatics methods. First, the TIMER database was used to show that the expression level of POC1A was significantly upregulated in pan-cancer. Next, the effect of POC1A on LIHC tissue was closely explored. The results showed that the expression level was significantly elevated in patients with LIHC, and high gene expression was significantly negatively correlated with survival prognosis. High levels of POC1A were associated with lower OS rates in HCC patients, and a further correlation was found between POC1A and some clinical and pathological features, including gender, age, race, and tumor stage. GO and KEGG enrichment analyses were then conducted. The genes associated with POC1A were presented in volcanic maps, and the top 50 genes positively or negatively correlated with POC1A were presented in heat maps. The GO enrichment analysis revealed that POC1A is involved in the biological process of chromosome separation, which may result in aneuploidy and micronuclei, leading to chromosomal instability (CIN).
CIN is a significant marker of cancer. It is caused by persistent errors in chromosome separation during mitosis, and is the main factor promoting tumor development. Micronuclei have been found in the regenerative and developmental nodules of the liver, which suggests that CIN can occur in the early stages of liver cancer (18). The number of abnormal chromosomes is a prominent feature of cancer cells, and polyploidy, where the DNA content increases to an integer multiple of the entire chromosome set, is also considered a possible mechanism leading to the occurrence of liver cancer. Chromosome aberrations can endow cancer cells with drug resistance, ultimately leading to treatment failure. The degree of drug resistance is related to the degree of CIN (19,20).
POC1A is also enriched in the process of non-coding RNA, which has been continuously studied and was previously considered useless in mediating the regulatory factors involved in many functions but has since been identified as a carcinogenic driving factor and tumor suppressor (21). The KEGG enrichment analysis showed that POC1A is involved in ribosome related pathways, and genetic mutations in ribosomal biogenesis are associated with an increased risk of cancer. Dysregulated ribosomal biogenesis has a broader role in the development and progression of most spontaneous cancers (22). The protein encoded by the POC1A gene is an important protein that regulates the assembly of centrioles. As a protein located on the centrosome, it regulates tumor cells by participating in the cell cycle and DNA replication.
The use of PPIs and small molecule antagonisms in therapeutic approaches has become increasingly feasible. POC1A-encoded proteins contain WD40 repeat (WDR) domains, one of the most abundant PPI domains in human proteomics, consisting of WDR5 and EED proteins. The propeller domain has been successfully targeted by effective, specific, and cellular active drug-like chemical probes (23). Thus, POC1A is likely to be a new target for the early diagnosis and treatment of HCC. In this study, we focused on the pathways by which POC1A interacts to affect cell-cycle progression. We found that POC1A interacts with the NUDC protein, and genes such as FOXC1, YY1, and NFIC may be the TFs of POC1A.
Moreover, the high expression of PPARG and NUDC significantly affected the OS rate of the LIHC patients. Research has shown that NUDC may serve as a new natural killer cell-related marker to predict the prognosis of and efficacy of immunotherapy in liver cancer patients (24). PPARG may play an important role in cell proliferation, the complement system, and immune cell invasion of HCC, and may be independently associated with susceptibility to progressive liver fibrosis (25,26). In addition, many studies have found that the occurrence and development of tumors are closely related to the tumor microenvironment. Tumor tissue does not simply comprise tumor cells; rather, various cells form the tumor microenvironment. The difference in the infiltration degree of different immune cells is highly correlated with tumor progression and prognosis. The high expression of POC1A and its interaction genes could play a significant role in various immune cell types. These genes and their interaction with POC1A are worthy of further research.
There have been some advances in science and technology in recent years, and there have been improvements in the treatment of HCC, of which liver transplantation is still one of the most effective methods. However, due to many limiting factors such as a shortage of donor organs and long waiting times, patients may not be able to undergo transplantation due to tumor progression and may ultimately die. In addition to studying effective treatment methods after diagnosis, more accurate early screening and diagnosis methods are needed to develop a more targeted and effective method to improve the survival rate, especially in patients in the middle and late stages. Further validation studies are needed to assess the specific molecular mechanisms by which the POC1A gene affects the prognosis of HCC patients.
Conclusions
This study investigated the mode and mechanism of POC1A in HCC patients by integrating and analyzing a large number of HCC data from multiple databases. We found that POC1A was significantly highly expressed in HCC patients, and the OS and DFS rates of these patients were significantly reduced. The overexpression of POC1A may have an important effect on the development of HCC via the modulation of the cell-cycle pathway. This study provides a theoretical basis for the development of HCC, and suggests that POC1A could be used as a new potential biomarker for the treatment and prevention of HCC, which is of great significance in the study of liver cancer.
Acknowledgments
Funding: The work was financially supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2398/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2398/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2398/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2398/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Claasen MPAW, Ivanics T, Beumer BR, et al. An international multicentre evaluation of treatment strategies for combined hepatocellular-cholangiocarcinoma☆. JHEP Rep 2023;5:100745. [Crossref] [PubMed]
- Kelley RK, Greten TF. Hepatocellular Carcinoma - Origins and Outcomes. N Engl J Med 2021;385:280-2. [Crossref] [PubMed]
- Pinato DJ, Fulgenzi CAM, D'Alessio A. Immunotherapy at all stages of hepatocellular carcinoma. Nat Med 2024;30:640-1. [Crossref] [PubMed]
- Tayob N, Kanwal F, Alsarraj A, et al. The Performance of AFP, AFP-3, DCP as Biomarkers for Detection of Hepatocellular Carcinoma (HCC): A Phase 3 Biomarker Study in the United States. Clin Gastroenterol Hepatol 2023;21:415-423.e4. [Crossref] [PubMed]
- Chen W, Xu Z, Jiang J, et al. Identification of LPCAT1 as a key biomarker for Crohn's disease based on bioinformatics and machine learnings and experimental verification. Gene 2024;920:148519. [Crossref] [PubMed]
- Liang X, Li L, Fan Y. Diagnostic, Prognostic, and Immunological Roles of HELLS in Pan-Cancer: A Bioinformatics Analysis. Front Immunol 2022;13:870726. [Crossref] [PubMed]
- Gu Y, Li Z, Li H, et al. Exploring the efficacious constituents and underlying mechanisms of sini decoction for sepsis treatment through network pharmacology and multi-omics. Phytomedicine 2024;123:155212. [Crossref] [PubMed]
- Shen Y, Huang J, Jia L, et al. Bioinformatics and machine learning driven key genes screening for hepatocellular carcinoma. Biochem Biophys Rep 2023;37:101587. [Crossref] [PubMed]
- Chen E, Zou Z, Wang R, et al. Predictive value of a stemness-based classifier for prognosis and immunotherapy response of hepatocellular carcinoma based on bioinformatics and machine-learning strategies. Front Immunol 2024;15:1244392. [Crossref] [PubMed]
- Shaheen R, Faqeih E, Shamseldin HE, et al. POC1A truncation mutation causes a ciliopathy in humans characterized by primordial dwarfism. Am J Hum Genet 2012;91:330-6. [Crossref] [PubMed]
- Venoux M, Tait X, Hames RS, et al. Poc1A and Poc1B act together in human cells to ensure centriole integrity. J Cell Sci 2013;126:163-75. [Crossref] [PubMed]
- Koparir A, Karatas OF, Yuceturk B, et al. Novel POC1A mutation in primordial dwarfism reveals new insights for centriole biogenesis. Hum Mol Genet 2015;24:5378-87. [Crossref] [PubMed]
- Lu J, Huang XY, Wang YH, et al. POC1A acts as a promising prognostic biomarker associated with high tumor immune cell infiltration in gastric cancer. Aging (Albany NY) 2020;12:18982-9011. [Crossref] [PubMed]
- Kasavi C. Gene co-expression network analysis revealed novel biomarkers for ovarian cancer. Front Genet 2022;13:971845. [Crossref] [PubMed]
- Zhao Q, Gao S, Chen X, et al. POC1A, prognostic biomarker of immunosuppressive microenvironment in cancer. Aging (Albany NY) 2022;14:5195-210. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Llovet JM, Willoughby CE, Singal AG, et al. Nonalcoholic steatohepatitis-related hepatocellular carcinoma: pathogenesis and treatment. Nat Rev Gastroenterol Hepatol 2023;20:487-503. [Crossref] [PubMed]
- Bakhoum SF, Ngo B, Laughney AM, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018;553:467-72. [Crossref] [PubMed]
- Tahmasebi-Birgani M, Ansari H, Carloni V. Defective mitosis-linked DNA damage response and chromosomal instability in liver cancer. Biochim Biophys Acta Rev Cancer 2019;1872:60-5. [Crossref] [PubMed]
- Du G, Yang R, Qiu J, et al. Multifaceted Influence of Histone Deacetylases on DNA Damage Repair: Implications for Hepatocellular Carcinoma. J Clin Transl Hepatol 2023;11:231-43. [PubMed]
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer 2018;18:5-18. [Crossref] [PubMed]
- Xue M, Dong L, Zhang H, et al. METTL16 promotes liver cancer stem cell self-renewal via controlling ribosome biogenesis and mRNA translation. J Hematol Oncol 2024;17:7. [Crossref] [PubMed]
- Schapira M, Tyers M, Torrent M, et al. WD40 repeat domain proteins: a novel target class? Nat Rev Drug Discov 2017;16:773-86. [Crossref] [PubMed]
- Guo C, Tang Y, Li Q, et al. Deciphering the immune heterogeneity dominated by natural killer cells with prognostic and therapeutic implications in hepatocellular carcinoma. Comput Biol Med 2023;158:106872. [Crossref] [PubMed]
- Wang C, Yu S, Qian R, et al. Prognostic and immunological significance of peroxisome proliferator-activated receptor gamma in hepatocellular carcinoma based on multiple databases. Transl Cancer Res 2022;11:1938-53. [Crossref] [PubMed]
- Rybicka M, Verrier ER, Baumert TF, et al. Polymorphisms within DIO2 and GADD45A genes increase the risk of liver disease progression in chronic hepatitis b carriers. Sci Rep 2023;13:6124. [Crossref] [PubMed]


