
Disulfidptosis-related gene SLC7A11 predicts prognosis and indicates tumor immune infiltration in lung adenocarcinoma
Highlight box
Key findings
• Three distinct clusters in lung adenocarcinoma (LUAD) were identified based on disulfidptosis-related genes.
• Elevated expression of SLC7A11 is associated with poor prognosis and immune evasion in LUAD.
• Disulfidptosis likely plays a significant role in LUAD progression and immune regulation.
What is known, and what is new?
• LUAD is heterogeneous and is influenced by factors such as smoking. Disulfidptosis, a novel form of cell death, is influenced by genes like SLC7A11, but little is known about its role in cancer immunity.
• This study identified LUAD clusters based on disulfidptosis genes and linked high SLC7A11 expression to a poor prognosis and immune evasion, offering insights into LUAD immunobiology.
What is the implication, and what should change now?
• An understanding of disulfidptosis-related genes in LUAD may lead to the development of novel therapies. Our findings suggest that SLC7A11 could serve as a potential target to enhance immunotherapy. Further research should focus on developing SLC7A11-targeted therapies and incorporating SLC7A11 status in patient stratification for personalized immunotherapy.
Introduction
Lung cancer is the most common cancer and leading cause of cancer-related death worldwide (1,2). Non-small cell lung cancer (NSCLC) represents approximately 85% of all lung cancer cases, and can be further classified into lung adenocarcinoma (LUAD), squamous cell carcinoma, and large cell carcinoma (3). Of these, LUAD is the most frequently occurring NSCLC subtype, and its development is linked to smoking, alcohol consumption, and metabolic imbalances (4,5). Despite advancements in LUAD treatments, which include surgery, chemotherapy, radiotherapy, targeted therapy, and immunotherapy, patient survival rates remain disappointingly low (6). Recent research has highlighted the insufficiency of traditional histological categorizations of LUAD, due to the inherent heterogeneity and complex nature of the disease (7). This has led to an increased focus on molecular subtype research to refine treatment approaches. Understanding how genetic alterations disrupt the tumor microenvironment (TME) and affect LUAD prognosis is crucial for identifying novel therapeutic targets (8,9).
Disulfidptosis is a novel form of cell death distinct from other known types such as apoptosis and ferroptosis (10,11). It occurs under certain conditions, such as glucose starvation in cells with high expression of the SLC7A11 gene (12,13). The other genes were FLNA, FLNB, MYH9, TLN1, ACTB, MYL6, MYH10, CAPZB, DSTN, IQGAP1, ACTN4, PDLIM1, CD2AP, and INF2, which are closely related to disulfidptosis. This process is characterized by the accumulation of intracellular disulfides, leading to aberrant disulfide bonds in cytoskeleton proteins, which in turn affect the actin cytoskeleton, causing cell death. That is caused by disulfide stress caused by the accumulation of excess cystine in cells. The study of genes related to disulfide death is of great significance for understanding the mechanism of cell death and exploring potential cancer therapies. Current research on the role of disulfidptosis in cancer is burgeoning, yet there are no reports on its effect on the immune microenvironment (13). Our study aimed to analyze the immunoregulatory role of disulfidptosis-related genes in LUAD to unveil and underscore their significance in the process of immune regulation.
In this study, we examined 15 disulfidptosis-related genes previously reported (14) and conducted a clustering analysis of the sequencing results of the LUAD cohort from The Cancer Genome Atlas (TCGA). Subsequently, we analyzed the immune infiltration scores of the clusters regulated by disulfidptosis. By integrating the results of bulk and single-cell RNA sequencing from immune therapy cohorts, our research findings shed light on the immunoregulatory role of disulfidptosis in cancers, exemplified by LUAD. We present this article in accordance with the STREGA reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1182/rc).
Methods
Preprocessing the RNA expression data
The RNA expression datasets for TCGA-LUAD were meticulously compiled using the UCSC Xena platform (https://xenabrowser.net/datapages/). A total of 511 cases were included in the study. Additionally, comprehensive somatic mutation data corresponding to these cases were acquired from the Genomic Data Commons (GDC) portal of the National Cancer Institute (NCI), which can be accessed at the NCI GDC Repository (https://portal.gdc.cancer.gov/). Single-cell sequencing data from patients treated with programmed cell death protein 1 (PD1) therapy were curated from a breast cancer study (15). Such datasets are invaluable in advancing understandings of the molecular intricacies in LUAD. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Principal component analysis (PCA) and consensus molecular clustering by non-negative matrix factorization (NMF)
A PCA, a critical technique in data dimensionality reduction, was conducted using highly variable genes. NMF, a method extensively employed for clustering in high-dimensional datasets, especially in the realm of computational biology, was then applied. Focusing on TCGA-LUAD cohort, we identified distinct disulfidptosis-related molecular clusters using a consensus clustering approach via the “NMF” function.
Data analysis for Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) screens
The CRISPR screen data were curated from lung cancer mouse in vitro and in vivo models (16). To process and analyze the CRISPR screen data, we employed MaGeCK software. Specifically, Fastq reads from the CRISPR screen underwent processing via the MaGeCK count module. Subsequently, using the default parameters, the robust rank aggregation (RRA) module was used to calculate the log2fold changes and P values associated with the genes. Additionally, custom scripts in R (version 4.1.3) were developed and used to visualize the data.
Immune analysis
We applied gene signatures of immune cells, as identified by Charoentong et al. (17), to compute the scores related to immune infiltration. This computation was performed using the single-sample gene set enrichment analysis (ssGSEA) technique.
Statistical analyses
All the statistical analyses were conducted using R software (version 4.1.2) and GraphPad Prism (version 9.50). The Wilcoxon test, log-rank test, and Kruskal-Wallis H test were employed to evaluate the data. For comprehensive details regarding the statistical tests used, please refer to the specific annotations provided in the figure legends.
Results
Genetic variation of disulfidptosis-related genes in LUAD
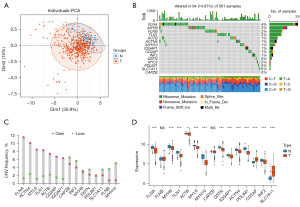
To study the regulatory role of disulfidptosis in lung cancer, we referenced a gene signature of disulfidptosis reported in previous article (18). Using TCGA-LUAD RNA expression cohort, we conducted a PCA of cancer and adjacent non-tumor tissues (Figure 1A). The results demonstrated that the genes related to disulfidptosis could be used to distinguish between the tumor and adjacent non-tumor tissues in LUAD. Figure 1B shows the frequency of gene mutations in this category; FLNA had the highest mutation frequency. The results of the copy number variation (CNV) analysis are presented in Figure 1C. Combining the expression levels of these genes in the tumor and adjacent non-tumor tissues in LUAD (Figure 1D), the copy number amplification of ACTB and CD2AP were consistent with their high expression levels in tumors, while the decreased copy number amplifications of MYH10, DSTN, and INF2 were consistent with their low expression levels in tumors. This suggests that these genes play a significant regulatory role in LUAD; thus, the regulatory mechanisms of disulfidptosis in the development of cancer urgently need to be explored.
Disulfidptosis-related clusters in LUAD
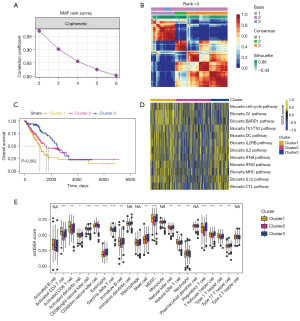
To better explore the regulatory role of the disulfidptosis in LUAD, we adopted the classical categorization algorithm (i.e., NMF) (19,20) to subgroup the disulfidptosis-related gene clusters based on gene expression levels in LUAD. The NMF rank algorithm (Figure 2A) suggested that dividing the LUAD into three groups was appropriate. Figure 2B shows the heatmap of the consensus matrix. Therefore, we concluded that under the regulation of disulfidptosis, LUAD can be divided into three significant groups; that is, clusters 1–3. A subsequent survival analysis (Figure 2C) showed that cluster 1 had the worst prognosis, while cluster 3 had the best prognosis. The pathway ssGSEA (21,22) revealed that the cell cycle activity in cluster 1 was more robust, which to some extent confirmed the reason for the poorer prognosis of cluster 1 (Figure 2D). Conversely, cluster 3 expressed higher levels of immune activation pathways, which suggests that patients in this LUAD subgroup might be more suitable for immunotherapy. To further corroborate this hypothesis, we used a common set of immune-related genes for a GSEA. The results showed that the activated cluster of differentiation 8 (CD8) T cell GSEA score (i.e., the CD8 activation-related score) was highly expressed in both clusters 1 and 3 (Figure 2E). This increase in cluster 3 confirmed that it was an immune-activated LUAD subgroup. However, the activation of CD8 T cells in the cluster1 subgroup, which had the poorest prognosis, might indicate the presence of factors leading to exhausted CD8 T cells, rendering it unresponsive to immunotherapy.
Analyses of TME infiltration of disulfidptosis-related clusters
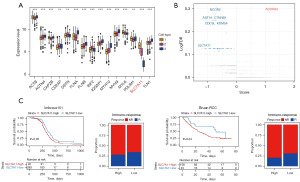
Given that clusters 1–3 demonstrated potential mechanisms of immune infiltration linked to prognosis, we proceeded to investigate the expression levels of disulfidptosis-related genes in these clusters. Notably, we found that SLC7A11 was significantly overexpressed in cluster 1 (Figure 3A). SLC7A11, a gene known for its strong association with disulfidptosis, has also been reported to be highly expressed in cancer (23). This finding aligned with our analysis, which suggested a poorer prognosis for cancer patients in cluster 1. High SLC7A11 expression was correlated with an immunosuppressed state in patients; however, it did not necessarily reflect the inherent immune-regulatory functions of the gene. To explore this further, we analyzed CRISPR screen single-guide RNA sequencing data (24) from co-cultures of OT1 mouse T cells and tumor cells (Figure 3B). Our analysis identified SLC7A11 as a significant contributor to immune evasion. Additionally, we used PD1 bulk tissue sequencing results for our analysis. The initial findings showed that patients with high SLC7A11 expression had a poorer prognosis than those with low SLC7A11 expression (Figure 3C). Further, we observed that patients with higher SLC7A11 expression demonstrated a greater tolerance to immunotherapy than those with lower SLC7A11 expression (Figure 3C). This indicates that SLC7A11, which is a critical gene regulating disulfidptosis, plays a key role in immune evasion.
Immune function analysis of disulfidptosis-related clusters
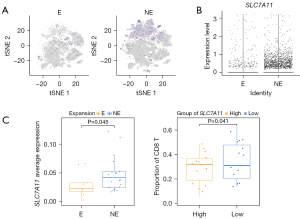
The bulk tissue sequencing levels indicated high SLC7A11 expression in the immunotherapy non-responsive group; however, we could not exclude the influence of the expression levels of immune microenvironment cells. Thus, we subsequently introduced single-cell sequencing data from PD1-treated patients (15). The data we referenced used T cell receptor (TCR) clonal expansion as an indicator of patient response to PD1 treatment. Thus, clonal expansion (“E”) indicated a response to immunotherapy, and no expansion (“NE”) indicated a tolerance to immunotherapy. The analysis revealed that SLC7A11 was highly expressed in the tumor cells in the NE group (Figure 4A). Additionally, we used the average expression level of epithelial cells to represent the SLC7A11 expression in each sample, and the results were consistent with the single-cell analysis; that is, the average expression of SLC7A11 in the tumor cells of the NE group was higher than that of the E group (Figure 4B). Following this, we categorized patients into high- and low-expression groups based on the median of the average expression level of SLC7A11 in tumor cells. We found that patients with low SLC7A11 expression had a lower proportion of CD8+ T cell infiltration. In summary, SLC7A11 plays a role in immune evasion (Figure 4C).
Discussion
In this study, we explored the transcriptomic landscapes shaped by disulfidptosis-related genes. We found that the LUAD cohort from TCGA could be stratified into three distinct clusters, each characterized by unique clinicopathological features. Notably, cluster 1 was characterized by the poorest survival outcomes and a high degree of immune cell infiltration. While cluster 3 was characterized by robust immune cell infiltration, a favorable prognosis, and the notable presence of immune-activating cells like activated CD8 T cells. This study effectively mapped the intratumoral transcriptomic diversity as influenced by disulfidptosis, shedding light on the tumor TME infiltration patterns. Disulfidptosis-driven molecular subtyping can offer fresh perspectives in LUAD pathogenesis.
By exploring the expression of disulfidptosis-related genes in our three distinct clusters, we observed that SLC7A11 was highly expressed in cluster 1. This study showed that the role of SLC7A11 in cancer is integral to the process of disulfidptosis, which is a unique form of cell death. When expressed at high levels, particularly in the context of glucose starvation, SLC7A11 leads to an excessive accumulation of intracellular disulfides (13). This accumulation causes aberrant disulfide bonding in actin cytoskeleton proteins, leading to cell death. A previous well-known study showed that disulfidptosis occurs in the SLC7A11-high condition of cancer (25). Further, it suggested that targeting disulfide stress, particularly in SLC7A11-high tumors, could be a promising therapeutic strategy in cancer treatment. In our research, we confirmed that SLC7A11 was highly expressed in LUAD patients with a poor prognosis, reinforcing its significant role in promoting the progression of LUAD. However, our study differed notably from previous research in that it focused more on the role of high SLC7A11 expression in regulating the immune microenvironment in LUAD tumors. We discovered that SLC7A11 is a gene associated with immune evasion and shows resistance to immunotherapy. Therefore, we believe that targeting and inhibiting SLC7A11 in LUAD patients could enhance the response to immunotherapy.
However, our study mainly focused on the bioinformatic analyses, which falls short of robust confirmation. Therefore, a more in-depth experimental validation of our findings is required.
Conclusions
In conclusion, the study provided new insights into the role of disulfidptosis in LUAD, particularly the immunoregulatory effects of SLC7A11. Our findings could lead to the development of novel therapeutic strategies targeting SLC7A11 to enhance immunotherapy responses in LUAD patients.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1182/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1182/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1182/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17-48. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 2008;83:584-94. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Hutchinson BD, Shroff GS, Truong MT, et al. Spectrum of Lung Adenocarcinoma. Semin Ultrasound CT MR 2019;40:255-64. [Crossref] [PubMed]
- de Groot P, Munden RF. Lung cancer epidemiology, risk factors, and prevention. Radiol Clin North Am 2012;50:863-76. [Crossref] [PubMed]
- Lambe G, Durand M, Buckley A, et al. Adenocarcinoma of the lung: from BAC to the future. Insights Imaging 2020;11:69. [Crossref] [PubMed]
- Bronte G, Rizzo S, La Paglia L, et al. Driver mutations and differential sensitivity to targeted therapies: a new approach to the treatment of lung adenocarcinoma. Cancer Treat Rev 2010;36:S21-9. [Crossref] [PubMed]
- Saito M, Suzuki H, Kono K, et al. Treatment of lung adenocarcinoma by molecular-targeted therapy and immunotherapy. Surg Today 2018;48:1-8. [Crossref] [PubMed]
- Zheng T, Liu Q, Xing F, et al. Disulfidptosis: a new form of programmed cell death. J Exp Clin Cancer Res 2023;42:137. [Crossref] [PubMed]
- Liu X, Zhuang L, Gan B. Disulfidptosis: disulfide stress-induced cell death. Trends Cell Biol 2024;34:327-37. [Crossref] [PubMed]
- Zheng P, Zhou C, Ding Y, et al. Disulfidptosis: a new target for metabolic cancer therapy. J Exp Clin Cancer Res 2023;42:103. [Crossref] [PubMed]
- Liu X, Nie L, Zhang Y, et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol 2023;25:404-14. [Crossref] [PubMed]
- Wang T, Guo K, Zhang D, et al. Disulfidptosis classification of hepatocellular carcinoma reveals correlation with clinical prognosis and immune profile. Int Immunopharmacol 2023;120:110368. [Crossref] [PubMed]
- Bassez A, Vos H, Van Dyck L, et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat Med 2021;27:820-32. [Crossref] [PubMed]
- Li F, Huang Q, Luster TA, et al. In Vivo Epigenetic CRISPR Screen Identifies Asf1a as an Immunotherapeutic Target in Kras-Mutant Lung Adenocarcinoma. Cancer Discov 2020;10:270-87. [Crossref] [PubMed]
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248-62. [Crossref] [PubMed]
- Liu H, Tang T. Pan-cancer genetic analysis of disulfidptosis-related gene set. Cancer Genet 2023;278-279:91-103. [Crossref] [PubMed]
- Hu G, Zhou T, Luo S, et al. Assessment of nonnegative matrix factorization algorithms for electroencephalography spectral analysis. Biomed Eng Online 2020;19:61. [Crossref] [PubMed]
- Frigyesi A, Höglund M. Non-negative matrix factorization for the analysis of complex gene expression data: identification of clinically relevant tumor subtypes. Cancer Inform 2008;6:275-92. [Crossref] [PubMed]
- Yi M, Nissley DV, McCormick F, et al. ssGSEA score-based Ras dependency indexes derived from gene expression data reveal potential Ras addiction mechanisms with possible clinical implications. Sci Rep 2020;10:10258. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021;12:599-620. [Crossref] [PubMed]
- Bock C, Datlinger P, Chardon F, et al. High-content CRISPR screening. Nat Rev Methods Primers 2022;2:9. [Crossref] [PubMed]
- Yan Y, Teng H, Hang Q, et al. SLC7A11 expression level dictates differential responses to oxidative stress in cancer cells. Nat Commun 2023;14:3673. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


