
Prognostic biomarkers based on GUF1, EFTUD2 and GSPT1 targets affecting migration of gastric cancer cells
Highlight box
Key findings
• Genes (GUF1, EFTUD2 and GSPT1) was selected to dissect the proliferation and migration in gastric cancer (GC) cell lines.
What is known and what is new?
• GUF1, EFTUD2, and GSPT1 regulate protein translation and play an important role in tumor
• This study identified that high levels of GUF1, EFTUD2 and GSPT1 expression are predictive biomarkers for a poor prognosis in GC.
What is the implication, and what should change now?
• GUF1, EFTUD2 and GSPT1 have diverse functions and complex regulatory mechanisms in cancer. The mechanism in tumors has not been studied to a great degree yet. However, there is no doubt that GUF1, EFTUD2 and GSPT1 play an important role in GC tumorigenesis and development and may be potential targets for clinical diagnosis and treatment of GC.
Introduction
The translational process of protein synthesis is considered to be an important clue for understanding oncogenesis (1). Alternatively, eukaryotic elongation factor 1 alpha (eEF1A) proteins may enhance tumorigenesis independent of the protein translation network. eEF1A proteins are known to associate with actin and tubulin, and ectopic eEF1A expression can decrease the length of actin filaments and tubulin microtubules (2,3). Perhaps, the ability of eEF1A to alter cell structure somehow contributes to a neoplastic phenotype. While it is unclear how eEF1A2 might control cell growth, the process of protein synthesis in eukaryotic cells is controlled by various translation factors, such as eukaryotic initiation factor (eIF), eukaryotic elongation factor (eEF), and eukaryotic release factor (eRF) that, respectively, regulate initiation, elongation, and termination, eEF1A2 is a protein coding gene. eEF1A2 is associated with conditions such as mental retardation, autosomal dominant, and developmental and epileptic encephalopathy. The pathways associated with eEF1A2 include protein metabolism and viral mRNA translation. Results of eEF1A2 cDNA hybridization in the cancer profiling array reveals significant overexpression of the transcript for eEF1A2 among several different cancer samples, including breast (50 pairs), uterus (42 pairs), colon (34 pairs), stomach (28 pairs) (4). Further investigation is necessary to determine the precise role for eEF1A2 in controlling the development of gastric cancer (GC). To date, 18 eEF1A2 genes have been identified in mammalian cells, which are numbered in the order of their discovery (eEF1A1, HBS1L, GSPT1, GSPT2, EEF2, TUFM, EIF5B, EFL1, GFM1, EIF2S3, MTIF2, GTPBP1, GFM2, EIF2S3B, GTPBP2, EFTUD2, EEFSEC, GUF1). In a comprehensive analysis, only GUF1, EFTUD2, and GSPT1 are shown to be associated with GC. Therefore, we undertook the investigation of the involvement of these genes in GC progression and prognosis. GUF1, EFTUD2, GSPT1 regulate protein translation and play an important role in tumor (5-7). Minor sites of normal eEF1A2 expression have been found in several specialized human body parts (8), including breast acini (9), glucagon-producing islet cells in the pancreas, Purkinje cells of the cerebellum (10), and lung alveoli (11). Nevertheless, the roles of GUF1, EFTUD2 and GSPT1 have been less studied in GC.
GC is one of the most common malignancies, ranking fifth and fourth in incidence and mortality among all malignancies worldwide respectively (12). In most countries, early GC screening is not carried out effectively, often resulting in a diagnosis at an advanced stage (13,14). It is important to examine the molecular mechanism underlying GC development and explore specific and efficient diagnostic markers and therapeutic targets. GC is a multifactorial disease caused by genetic and epigenetic changes, including alteration in DNA methylation and aberrant expression of noncoding RNAs. The treatment of GC has advanced recently; however, its prognosis has not been significantly enhanced and the prognosis of different individuals varies greatly. In recent years, to better guide the clinical treatment of GC, researchers have been working hard to identify biological markers of GC that could enable effective screening of people for susceptibility to GC and predict the survival patient with GC. Such biological markers of tumors can be of immense benefit in early diagnosis, clinical staging, prognosis and treatment efficacy assessment of GC.
While there is research (15) established that the eEF1A2 is genetically amplified and overexpressed in ovarian tumors and has oncogenic properties, it is yet to be shown whether eEF1A2 expression can directly cause other cancers such as GC in animals. Furthermore, we are yet to understand the mechanism by which eEF1A2 promotes tumorigenesis. In the absence of mechanistic insight, there are still important issues that need to be addressed with regard to eEF1A2 and GC. Firstly, is eEF1A2 copy number or eEF1A2 protein expression an GC prognostic factor? Secondly, is eEF1A2 a suitable target for anticancer therapy? In this study, we examined the expression of GUF1, EFTUD2, GSPT1 and GSPT2 from eEF1A2 in normal tissues and GC tissues, and various cell lines. Meanwhile, we also evaluated the prognostic value of GUF1, EFTUD2 and GSPT1 in GC based on The Cancer Genome Atlas (TCGA) dataset. Subsequently, we analyzed the association between the GUF1, EFTUD2 and GSPT1 expression levels and individual pathologic stage, clinical T stage and individual primary therapy outcome, and diagnostic capabilities. The Gene Expression Omnibus (GEO) databases were used to verify the expressions of GUF1, EFTUD2 and GSPT1 in GC. We also evaluated their role in the development of GC through bioinformatics analysis and experimental verification. Our study provides information to help in the early diagnosis of GC. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-125/rc).
Methods
Databases
We investigated the expression level of GUF1, EFTUD2 and GSPT1 in various types of normal tissues and tumor in the integrated datasets combining TCGA (https://portal.gdc.cancer.gov) with the GTEx (Genotype-Tissue Expression) database (https://www.gtexportal.org/home/-index.html). TCGA are open-ended and public and do not need the approval of a local ethics committee. We obtained the profiles of RNA expression (RNA-Seq2 level 3 data; format: TPM; platform: Illumina HiSeq 2000) and clinical samples of GUF1, EFTUD2 and GSPT1 patients from the TCGA database. TCGA included 375 GC samples and 32 normal gastric tissue samples, which contain general information, clinicopathological details, and prognostic information. The GEO databases were filtered to remove missing and duplicated results by stringr and dplyr, the difference analysis by package limma, and whole data transformed by log2 (TPM +1) using R package of “ggstatsplot” in an R environment (R version: 3.6.1).
For analysis, we chose the following detailed databases: Gene Cards, Oncomine (https://www.oncomine.org/resource/login.html, an online cancer microarray database), TCGA (https://portal.gdc.cancer.gov) with the GTEx database (https://www.gtexportal.org/home/-index.html). Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia2.cancer-pku.cn/#index), and Kaplan-Meier Plotter (www.kmplot.com). The Human Protein Atlas (https://www.proteinatlas.org/humanproteome/pathology), and the GEO databases (GSE66229, GSE62254). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Expression differential analysis
Oncomine gene expression array dataset was used to analyze the transcription levels of eEF1A2 in different cancers. The mRNA level of eEF1A2 in clinical cancer specimens were compared with those in controls, using Student’s t-test to generate a P value. The cutoffs of P value and fold change were 0.01 and 0.5.
Kaplan-Meier plot analysis
The prognostic value of the signal transducer and the activator of transcription (STAT) mRNA expression was evaluated using an online database, Kaplan-Meier Plotter contained gene expression data and survival information of GC patients (http://www.kmplot.com/analysis/index.php?p=service & cancer=gastric). To analyze the overall survival (OS), progression-free survival (FP), and post-progression survival (PPS) of patients with GC, the samples were split into two groups by median expression (high versus low expression) and assessed by a Kaplan-Meier survival plot, with a hazard ratio (HR) of 95% confidence intervals (CIs) and log rank P value. Only the JetSet best probe set of eEF1A2s was chosen to obtain Kaplan-Meier plots, where the number-at-risk is indicated below the main plot.
Protein immunohistochemical staining analysis
The data related to each protein are arranged in the database (The Human Protein Atlas:https://www.proteinatlas.org/humanproteome/pathology). The starting page is a summary interface, where the basic information of the protein is displayed. The different modules can be visited thereafter to collect the sets of data. The modules included in this database are: tissue expression profile, cellular localization profile, pathological expression profile and RNA expression profile.
GEPIA dataset
GEPIA (http://gepia2.cancer-pku.cn/) is a newly developed interactive web server for analyzing the RNA sequencing expression data of 9,736 tumors and 8,587 normal samples from TCGA and the Genotype Tissue Expression (GTEx) projects, using a standard processing pipeline. GEPIA can profile the tissue-wise expression of one gene in different cancer types, using a dot plot.
Cell lines and plasmid
The human GC cell lines AGS (CL-0022) were kindly provided by Procell LifeScience & Technology Co., Ltd. (Wuhan, China), and the normal gastric cell line GES-1 were kindly gifted by the Ms. Juan An’s research group (Department of Basic Medicine Science, Qinghai University Medical College). Mycoplasma testing has been carried out for the cell lines used; and the cell lines used have been authenticated. AGS and GES cells were maintained with 5% CO2 at 37 °C in Dulbecco’s modified Eagle Medium (DMEM) (Biosharp, Beijing, China). The knockdown sense sequence of GUF1, EFTUD2 and GSPT1 are provided in Table 1.
Table 1
| Gene | Direction | Primer sequences |
|---|---|---|
| GUF1 | PF | ACATAAGCAACCAGTGGAGCCC |
| GUF1 | PR | TCCAGCCAGCACCCAGAGC |
| EFTUD2 | PF | CAGCATCGTTCAAGGTTTC |
| EFTUD2 | PR | CATCAGACGAGGAGTAGCC |
| GSPT1 | PF | CGTAGCCCCAGGTGAAAA |
| GSPT1 | PR | AAATGTGCGTCCAGAATGA |
| GSPT2 | PF | TTGGCTGTGCTGGTCATC |
| GSPT2 | PR | GTTTTACCCCTGCCGTTT |
| GAPDH | PF | GAAGGTGAAGGTCGGAGT |
| GAPDH | PR | CATGGGTGGAATCATATTGGAA |
Cell transfection
Specific small interfering RNAs (siRNAs) targeting GUF1, EFTUD2 and GSPT1 (si-GUF1, si-EFTUD2 and si-GSPT1, respectively) and negative control siRNA (si-NC) were acquired from Beijing Xianghong Biotechnology Co., Ltd. (Beijing, China). For the siRNA experiments, 100 pmol siRNA and sample control (NC) were mixed with 125 µL serum-free medium and 4 µL Lipo8000 Transfection Reagent (Beyotime, Shanghai, China) and transfection was carried out. All transfected cells were cultured for 48 h before they were used for the various assays, unless indicated otherwise.
RNA extraction and quantitative real-time polymerase chain
Reaction
The total RNA from the cell lines was isolated using Total RNA Isolation Reagent (Biosharp), according to the manufacturer’s instructions. The cDNA was reverse transcribed with 1 µg of total RNA, using the SPARKscriptIIRT Plus Kit (with gDNA Eraser, AG0304). The quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the 2X SYBR Green qPCR Mix (with ROX, AH0104) with Roche LightCycler 96 instrument. Glyceraldehyde-3-phosphate dehydrogenase was used for normalization of the mRNA expression. All results were calculated using the 2−ΔΔCq method. Each experiment was performed three times. The primer sequences are listed in Table 2.
Table 2
| si-RNA | Direction | Primer sequences |
|---|---|---|
| hGUF1 | PF | CCUCCUAAAGUGCAUCGCAAATT |
| hGUF1 | PR | UUUGCGAUGCACUUUAGGAGGTT |
| hEFTUD2 | PF | GCCUCUCACAGAACCCAUUAUTT |
| hEFTUD2 | PR | AUAAUGGGUUCUGUGAGAGGCTT |
| hGSPT1 | PF | CCCGAUGAUGUAGAGACUGAUATT |
| hGSPT1 | PR | UAUCAGUCUCUACAUCAUCGGTT |
Cell proliferation assay
Cell proliferation was examined through 5-ethynyl-2'-deoxyuridine assay. EdU staining was performed using BeyoClickTM EdU Cell Proliferation Kit with Alexa Fluor 555, in accordance with the manufacturer’s instruction (C0075S, Beyotime).
Cell migration assays
To the chamber, 100 µL serum-free medium and 200 µL cell suspension (2.5×105/mL in serum-free medium) were added. Add 700 µL of culture medium (with serum) and it was incubated at 37 °C for 12–16 h. Subsequently, the medium was removed from the chamber and the chamber was washed twice in phosphate buffered saline (PBS). Following that, cells were fixed for 10 min with 4% paraformaldehyde at room temperature (15–20 °C). Crystal Violet staining solution was used to stain cells for 10 min at room temperature, formaldehyde was removed, and the cells were washed with PBS twice. Finally, migrated cells were counted under a light microscope, and the number of migrated cells was calculated for each group.
Statistical analysis
Statistical analyses were performed using SPSS 26.0 (IBM, Armonk, NY, USA). A fold change >2 and P value <0.05 were used as the screening criteria to filter the differentially expressed genes. Two-tailed unpaired Student’s t-test was used to compare the differences between the two groups. The Kaplan-Meier method and log-rank test were used for the survival rate analysis. For all analyses, a P value <0.05 was considered statistically significant. All measurement data were expressed as the mean ± standard deviation (SD) obtained from three independent experiments.
Results
Expression levels of GUF1, EFTUD2, GSPT1 and GSPT2 in patients with cancers
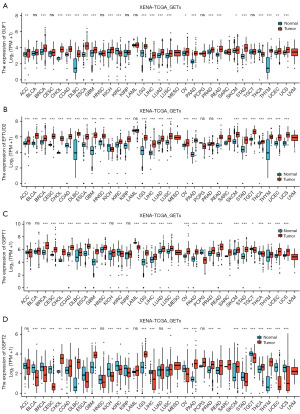
We compared the expression levels of GUF1, EFTUD2, GSPT1 and GSPT2 in cancer samples against those in normal samples using data from Oncomine databases. The analysis revealed that the expression of GSPT1 was mainly higher in bladder cancer, breast cancer, colorectal cancer (CRC) and myeloma samples as compared to that in the corresponding normal tissues, whereas the expression of GSPT2 was higher in CRC and leukemia samples, but lower or not detected in other cancer samples. GUF1 and EFTUD2 were expressed higher in samples of CRC, lung cancer, GC, leukemia and lymphoma (Figure 1A-1D).
Relationship between the GUF1, EFTUD2, GSPT1 and GSPT2 mRNA levels and the clinical parameters of patients with GC
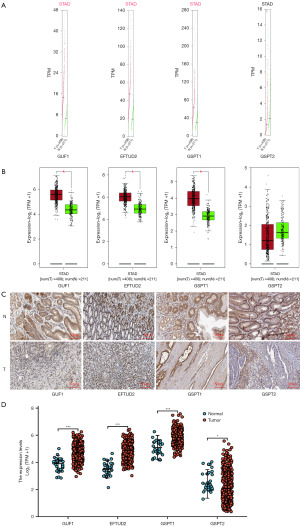
We used the GEPIA and TCGA datasets for the analysis and using log2 (TPM + 1) we derived the log-scale. Using the matched normal data, TCGA normal and GTEx data were matched, we compared the mRNA expression of GUF1, EFTUD2, GSPT1 and GSPT2 between GC and gastric tissues (Figure 2). The analysis showed higher expression levels of GUF1, EFTUD2 and GSPT1 in GC tissues than in normal tissues (Figure 2A). Furthermore, data from immunohistochemistry analysis (The Human Protein Atlas dataset) showed that the expression of GUF1, EFTUD2 and GSPT1 proteins was higher in the GC tissues than in the normal tissues (Figure 2C,2D). Furthermore, we compared the mRNA expression of GUF1, EFTUD2, GSPT1 and GSPT2 factors between GC and gastric normal tissues. The results indicated that the expression levels of GUF1, EFTUD2 and GSPT1 were higher in GC tissues than in normal tissues, and the expression level of GSPT2 had no statistically significant difference between GC and normal tissues. At the same time, GC tissues with high expression of GUF1, EFTUD2 and GSPT1 were disordered and lost its normal morphology (Figure 2B). Therefore, based on GC data in TCGA, we analyzed the expression level of GUF1, EFTUD2 and GSPT1 in AGS cell lines and GES control cell. The results revealed that mRNA levels of GUF1, EFTUD2 and GSPT1 were significantly higher in AGS, (P<0.001, Figure 2D). The mRNA levels of GUF1, EFTUD2 and GSPT1 were determined to be highly expressed in the AGS cell line.
Relationship between expressions of GUF1, EFTUD2 and GSPT1 with clinical profile
The correlation between GUF1, EFTUD2 and GSPT1 expression and clinical baseline data was analyzed. As the result of the analysis, statistically significant (P<0.05) differences were found between the GUF1 expression and the tumor T-stage and patient’s age and between GSPT1 expression and tumor T-stage (Table 3).
Table 3
| Parameter | Low expression | High expression | P |
|---|---|---|---|
| GUF1, n | 187 | 188 | |
| T stage | 50.6% | 49.3% | 0.02* |
| N stage | 49.8% | 50.2% | 0.87 |
| M stage | 48.7% | 52.3% | 0.59 |
| Gender | 49.9% | 50.1% | 0.72 |
| Age, years, median [IQR] | 64 [57, 71] | 69 [61, 75] | <0.001* |
| EFTUD2, n | 187 | 188 | |
| T stage | 50.1% | 49.9% | 0.10 |
| N stage | 49.6% | 50.4% | >0.99 |
| M stage | 50.2% | 49.8% | 0.09 |
| Gender | 49.9% | 50.1% | 0.47 |
| Age, years, median [IQR] | 66 [57, 72] | 68 [59, 74] | 0.09 |
| GSPT1, n | 187 | 188 | |
| T stage | 50.1% | 49.9% | 0.004* |
| N stage | 49.6% | 50.4% | 0.43 |
| M stage | 50.4% | 49.6% | 0.38 |
| Gender | 49.8% | 50.2% | 0.35 |
| Age, years, median [IQR] | 67 [58, 73] | 68 [58, 73] | 0.49 |
*, P<0.05 was considered statistically significant. GC, gastric cancer; IQR, interquartile range.
Association of GUF1, EFTUD2 and GSPT1 mRNA Levels with the prognosis of patients with GC
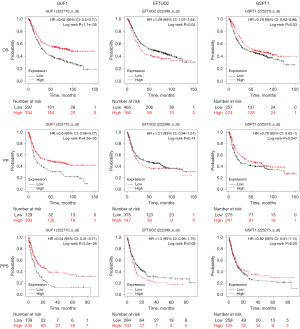
The Kaplan-Meier method was used to assess the difference between “high” and “low” risk groups based on the best separation of GUF1, EFTUD2 and GSPT1 expression, and the correlation between the GUF1, EFTUD2 and GSPT1 mRNA levels and the survival of patients with GC was analyzed, the publicly available Kaplan-Meier dataset was used. The Kaplan-Meier curve and log rank test analysis revealed that the increased GUF1 mRNA level was significantly associated with patients’ OS, FP and PPS (P<0.05) (Figure 3). The increased EFTUD2 mRNA level was significantly associated with OS (P<0.05) (Figure 3); however, it did not show the same association with FP and PPS (P>0.05) of all the patients with GC. The increased GSPT1 mRNA level showed significant association with OS and FP (P<0.05) (Figure 3). The GC patients with high mRNA levels of GUF1, EFTUD2 and GSPT1 were predicted to have high OS, FP and PPS.
Relationship between the mRNA expression level of GUF1, EFTUD2 and GSPT1 clinicopathological parameters in GC patients
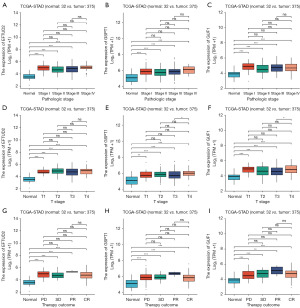
We analyzed the relationship between the mRNA expression of GUF1, EFTUD2, GSPT1 and clinicopathological parameters of GC patients (individual pathologic stage, clinical T stage and individual primary therapy outcome) through TCGA. The mRNA expression levels of GUF1, EFTUD2 and GSPT1 were correlated with individual cancer stage: patients with a more advanced cancer stage tended to express higher mRNA levels of GUF1, EFTUD2 and GSPT1 (Figure 4A-4I). Similarly, the expression levels of GUF1, EFTUD2 and GSPT1 were significantly related to primary therapy outcome. The mRNA levels of GUF1, EFTUD2 and GSPT1 tended to be related with individual pathologic stage, clinical T stage and individual primary therapy outcome.
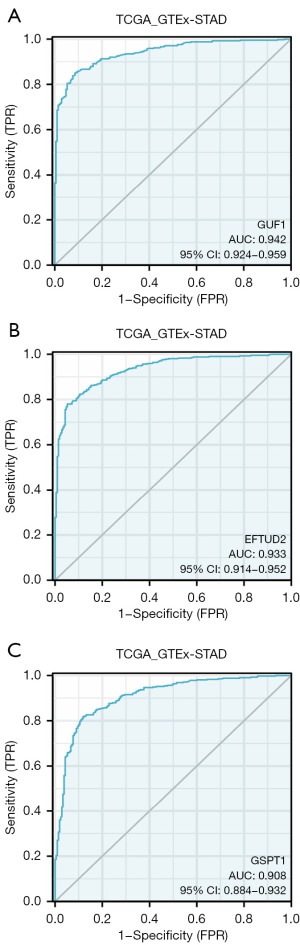
Diagnostic capability of GUF1, EFTUD2 and GSPT1 for GC
We analyzed the disease diagnostic capabilities of GUF1, EFTUD2 and GSPT1 expression in GC. Receiver operating characteristic (ROC) curves for each gene are shown in Figure 5. Please note that the sensitivity range had been limited to show only values ≥90% and (1 − specificity) limited to show only values ≤20% to focus on high sensitivity and specificity operational conditions that would be suitable for the ROC triage application. The result showed that the expression of GUF1, EFTUD2 and GSPT1 genes could be called as early diagnostic markers for GC.
Predicted functions of GUF1, EFTUD2 and GSPT1 in GC cells
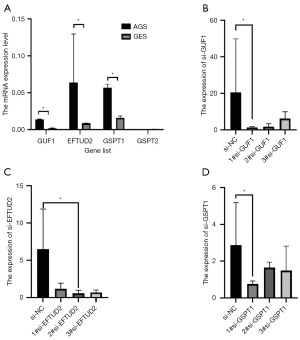
GUF1, EFTUD2 and GSPT1 expression level in cell lines and cell transfection
Figure 6A shows that GUF1, EFTUD2 and GSPT1 were highly expressed in AGS than in GES cell line, and the difference was statistically significant (P<0.05). For the RNAi experiments, according to the instructions of siRNA manufacturer, we tested the 100, 50, 30, 20, and 10 nM siRNA concentrations. As shown in Figure 6A, we performed siRNA-mediated silencing of GUF1, EFTUD2 and GSPT1. Compared to control si-NC (nonsense siRNA), si-GUF1, si-EFTUD2 and si-GSPT1 (transfected with specific siRNA) reduced the mRNA levels of GUF1, EFTUD2 and GSPT1, respectively over 60% gene silencing efficiency: indicative of efficient silence of the corresponding genes in AGS cells (Figure 6B-6D).
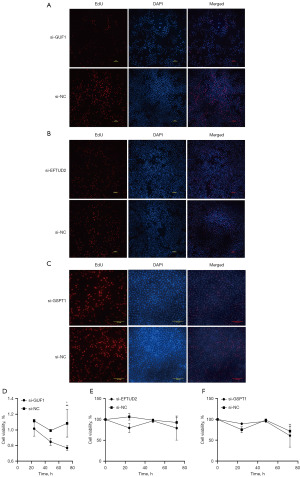
GUF1, EFTUD2 and GSPT1 knock-down suppressed cellular proliferation of cancer cell
We evaluated the changes of DNA synthesis in AGS cells with EdU immunofluorescence assay after GUF1, GSPT1 and EFTUD2 silencing, and cell proliferation was evaluated by CCK-8 assay. The results showed that the proportion of the cells in the S phase in the si-GUF1, si-EFTUD2, and si-GSPT1 groups was distinctly lower than that in the si-NC group (Figure 7A-7C). At the same time, cell proliferation was inhibited (Figure 7D-7F). These results revealed that GUF1, EFTUD2 and GSPT1 had proliferation-promoting properties in GC cells.
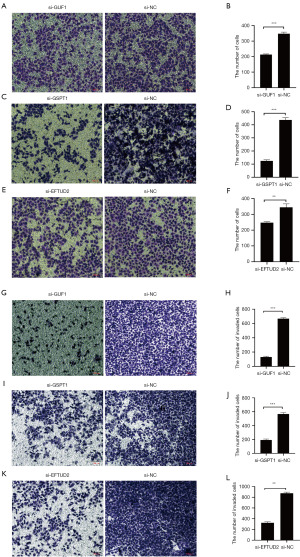
Silencing of GUF1, EFTUD2 and GSPT1 inhibited AGS cell migration and invasion ability in vitro
Metastasis in GC is characterized by increased cell motility, angiogenesis, and epithelial-mesenchymal transition. We investigated the roles of GUF1, EFTUD2, and GSPT1 in advanced GC. The tumor cell migration and invasion abilities were examined with Transwell assay. GUF1, EFTUD2 and GSPT1 knockdown by si-RNA dramatically reduced the invasion capacity of AGS cells compared to that of the si-NC group, suggesting that the knockdowns of GUF1, EFTUD2 and GSPT1 could inhibit the migration (Figure 8A-8F) and invasion (Figure 8G-8L) ability of GC cells.
Discussion
GC is one of the most fatal cancers worldwide (16,17), which manifests as highly prevalent malignant tumors in China. GC ranks fifth in cancer incidence and fourth in mortality worldwide (18). The 5-year survival rate for GC patients increased from 15.3% between 1995 and 1999 to 31.3% between 2005 and 2009, as screening became more common and treatment strategies improved (19). Despite the advancements in comprehensive treatment, including surgery, chemotherapy and radiation therapy, the prognosis of GC patients has not significantly improved. Currently, studies on the pathogenesis, occurrence and development of GC are facilitated by the modern molecular biology techniques.
The role of eEF1A2 in tumors remains debatable. Increased eEF1A2 expression may lead to an overall increase in protein translation. An increase in bulk protein synthesis may enhance cell replication because cell division requires sufficient protein production to fulfill the metabolic and size requirements of two new daughter cells (20). Increasing bulk protein abundance may decrease the time required to translate the overall mass of proteins necessary for cell division. If this is the case, then it would be expected that anything increasing protein translation rates would be predicted to be oncogenic. The reverse is certainly true, and inhibitors of protein translation are universally and highly toxic to cells and organisms. The molecular mechanism underlying its role in tumorigenesis and progression is not clearly understood.
Although GUF1 was identified several years ago, its physiological role is still unclear. GUF1 had once been reported as prognostic markers for thyroid cancer (21). Furthermore, by using the least absolute shrinkage and selection operator (LASSO) regression model, Li and colleagues identified GUF1 for its contribution to a high probability of liver metastasis of CRC (22). We investigated the expression level of GUF1 in GC tissues through online bioinformatics tools and explored its relationship with the pathological features of GC. Analysis of data from Oncomine and The Cancer Genome Atlas datasets revealed that the expression of GUF1 in human GC samples was higher than in normal tissue. Similar result was obtained through our study in which GUF1 was expressed more in GC cells than in control cells. Moreover, GUF1 expression also correlated with clinical characteristics of the patients with GC. Our analysis using the Kaplan-Meier Plotter showed that a high GUF1 expression was significantly associated with OS, FP and PPS in all the patients with GC. In our Transwell assay, knockdown of the GUF1, EFTUD2 and GSPT1 could inhibit the migration ability of GC cells. Additionally, high GUF1 expression was significantly associated with the age of patients. Therefore, GUF1 may be a new target for GC treatment.
EFTUD2 is a highly conserved spliceosomal GTPase that plays a crucial role in diverse biological functions, including spliceosome activation (23) and immune responses (24,25). Meanwhile, it has been reported that deletion of EFTUD2 inhibits the association of endogenous proteins, leading to increased apoptosis in breast cancer cells (26). Spliceosome is responsible for removing noncoding introns from mRNA precursors and generating mature mRNA, therefore, it plays an important role in almost all life processes (27). A study (28) identified that EFTUD2 plays a key role in regulating microglial polarization and homeostasis possibly through the NF-κB signaling pathway. Previously, there was little known about the molecular function of EFTUD2 in GC development. In this study, we identified the exact role and related mechanism of EFTUD2 in GC. We observed that EFTUD2 was upregulated in GC tissues compared to adjacent nontumor. In addition, we found that EFTUD2 was an independent prognostic factor for GC patients. A high level of EFTUD2 expression predicted a shorter overall and recurrence-free survival time in GC patients.
Interestingly, we found that EFTUD2 likely plays a role in maintaining the survival of GC cell lines. Our results indicated that a transient knockdown of EFTUD2 with siRNA inhibited cell viability. Our data indicated that EFTUD2 knockdown decreased the cell proliferation and migration in vivo. Further study to investigate the molecular mechanism of possible link between cell proliferation and migration regulated by EFTUD2 in GC is warranted. A study of EFTUD2 revealed that it plays a pivotal role in hepatocellular carcinoma (HCC) cell proliferation and the cell cycle (29), meanwhile another reported (30) that EFTUD2 as a novel oncogene helps to maintain the survival of HCC cells and promotes HCC progression. The high level of expression of EFTUD2 in HCC tissues indicates shorter overall and recurrence-free survival in HCC patients. These findings have similarities to our observation that EFTUD2 had proliferation-promoting properties in GC cells, which suggests that EFTUD2 maybe a new target for GC therapy in the future.
GSPT1 and GSPT2 are small GTPases which were initially found essential for the G1 to S phase transition of the cell cycle and later reported to function as a polypeptide chain release factor 3 (eRF3). The functions of eRF3 include the regulation of cell cycle phase-shifting from G1 to S phase (31), involvement in mRNA degradation, and ribosomal recycling (32,33), and it occurs in two isoforms, eRF3a and eRF3b, encoded by GSPT1 and GSPT2 genes, respectively. GSPT1/2 associates with eRF1 to mediate stop codon recognition and nascent protein release from the ribosome (34). Malta-Vacas et al. previously reported (35) that the expression level of eRF3a/GSPT1 was significantly high in intestinal gastric tumors. In addition, Tian (36) explored the potential effect of eRF3a/GSPT1 on gastric carcinogenesis and found that the expression level of eRF3a/GSPT1 was significantly increased in GC tissues, which is similar to the result of our observation. In addition, GSPT1 has been found to be overexpressed and oncogenic in a number of cancers (37), including gastric (35) and breast cancers (38). A study (39) has shown that GSPT1 plays a tumor-promoting role in the occurrence and development of colon cancer through specific signaling pathways. In the present study, high GSPT1 expression was significantly correlated with OS and FP in all patients with GC, establishing the oncogenic role of GSPT1 in GC.
Notably, it has been reported that GSPT1 promotes the proliferation, invasion, and migration of non-small cell lung cancer (NSCLC) cells, enhances tumorigenicity, and promotes the progression of lung cancer (40,41). In GC tissues, GSPT1 is highly expressed, miRNA-144 expression is down-regulated, and GSPT1 expression is significantly increased. By inhibiting miRNA-144, GSPT1 over-expression can promote the proliferation, invasion, and migration of GC cells, thereby promoting GC progression, which is consistent with the role of GSPT1 in tumors (42). It has also been reported that the overexpression of GSPT1 is related to the specific expression of GGCn alleles in various cancer cells, and that it is a potential oncogene (43). It could be argued that because all cells require protein synthesis, inhibitors of protein elongation would have substantial cytotoxic effects on normal tissues. However, rapidly growing tumor tissue may be more sensitive to decreases in protein synthesis than normal tissue because of the added burdens of an increased proliferation rate. Perhaps, when eEF1A2-inactivating agents are found, they might have efficacy in GC.
Tumor microenvironment (TME) refers to a complex and rich environment composed of multiple cells, which plays an important role in the growth and development of tumors. TME includes primary tumor cells, a variety of tissue cells (such as endothelial cells and fibroblasts), various immune cells, adipocytes, and extracellular fluid, which make TME a highly complex local environment. TME is an important component of tumor growth and development, and has an important impact on the growth, metastasis, and drug resistance of tumor cells (44). In this study, GSPT1 GUF1, EFTUD2, and GSPT1 were highly expressed in GC tissues, and the knockdown of GUF1, EFTUD2, and GSPT1 could inhibit the proliferation, migration and invasion of GC cells. We hypothesize that the GSPT1 gene plays a promoting role in the growth of GC tumor cells. The goal of future drug development can be to inhibit the expression of GUF1, EFTUD2, GSPT1, which may overcome many of the current limitations in the diagnosis and prognosis of cancer patients, and can improve the specificity and effectiveness of current tumor treatments, and provide new strategies and ideas for personalized treatment and comprehensive treatment of cancer patients.
Conclusions
In summary, we determined that GUF1, EFTUD2 and GSPT1 have diverse functions and complex regulatory mechanisms in cancer. The mechanism in tumors has not been studied to a great degree yet. However, there is no doubt that GUF1, EFTUD2 and GSPT1 play an important role in GC tumorigenesis and development and may be potential targets for clinical diagnosis and treatment of GC.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-125/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-125/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-125/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-125/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Meng X-Y, Jiang Q-Q, Yu X-D, et al. Eukaryotic translation elongation factor 1 alpha (eEF1A) inhibits Siniperca chuatsi rhabdovirus (SCRV) infection through two distinct mechanisms. J Virol 2023;97:e0122623. [Crossref] [PubMed]
- Condeelis J. Elongation factor 1 alpha, translation and the cytoskeleton. Trends Biochem Sci 1995;20:169-70. [Crossref] [PubMed]
- Shiina N, Gotoh Y, Kubomura N, et al. Microtubule severing by elongation factor 1 alpha. Science 1994;266:282-5. [Crossref] [PubMed]
- Hassan MK, Kumar D, Naik M, et al. The expression profile and prognostic significance of eukaryotic translation elongation factors in different cancers. PLoS One 2018;13:e0191377. [Crossref] [PubMed]
- Beyer S, Müller L, Mitter S, et al. High RIG-I and EFTUD2 expression predicts poor survival in endometrial cancer. J Cancer Res Clin Oncol 2023;149:4293-303. [Crossref] [PubMed]
- Cheng Y, Wang S, Mu X. Long non-coding RNA LINC00511 promotes proliferation, invasion, and migration of non-small cell lung cancer cells by targeting miR-625-5p/GSPT1. Transl Cancer Res 2021;10:5159-73. [Crossref] [PubMed]
- Huang Y, Wang Y, Liu S, et al. An integrative analysis of the tumor suppressors and oncogenes from sexual dimorphism and gene expression alteration features in thyroid cancer. Cancer Biomark 2023;38:1-16. [Crossref] [PubMed]
- Lam WW, Millichap JJ, Soares DC, et al. Novel de novo EEF1A2 missense mutations causing epilepsy and intellectual disability. Mol Genet Genomic Med 2016;4:465-74. [Crossref] [PubMed]
- Giudici F, Petracci E, Nanni O, et al. Elevated levels of eEF1A2 protein expression in triple negative breast cancer relate with poor prognosis. PLoS One 2019;14:e0218030. [Crossref] [PubMed]
- Newbery HJ, Loh DH, O'Donoghue JE, et al. Translation elongation factor eEF1A2 is essential for post-weaning survival in mice. J Biol Chem 2007;282:28951-9. [Crossref] [PubMed]
- Kebaabetswe LP, Haick AK, Gritsenko MA, et al. Proteomic analysis reveals down-regulation of surfactant protein B in murine type II pneumocytes infected with influenza A virus. Virology 2015;483:96-107. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Jou E, Rajdev L. Current and emerging therapies in unresectable and recurrent gastric cancer. World J Gastroenterol 2016;22:4812-23. [Crossref] [PubMed]
- Zhang XY, Zhang PY. Gastric cancer: somatic genetics as a guide to therapy. J Med Genet 2017;54:305-12. [Crossref] [PubMed]
- Wei S, Wang D, Li H, et al. Fatty acylCoA synthetase FadD13 regulates proinflammatory cytokine secretion dependent on the NF-κB signalling pathway by binding to eEF1A1. Cell Microbiol 2019;21:e13090. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Cao W, Chen HD, Yu YW, et al. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 2021;134:783-91. [Crossref] [PubMed]
- Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015;385:977-1010. [Crossref] [PubMed]
- Booth S, Lewis RJ. Structural basis for the coordination of cell division with the synthesis of the bacterial cell envelope. Protein Sci 2019;28:2042-54. [Crossref] [PubMed]
- Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Li W, Guo L, Tang W, et al. Identification of DNA methylation biomarkers for risk of liver metastasis in early-stage colorectal cancer. Clin Epigenetics 2021;13:126. [Crossref] [PubMed]
- Guo R, Zheng L, Park JW, et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol Cell 2014;56:298-310. [Crossref] [PubMed]
- Zhu C, Xiao F, Hong J, et al. EFTUD2 Is a Novel Innate Immune Regulator Restricting Hepatitis C Virus Infection through the RIG-I/MDA5 Pathway. J Virol 2015;89:6608-18. [Crossref] [PubMed]
- Hu P, Li Y, Zhang W, et al. The Spliceosome Factor EFTUD2 Promotes IFN Anti-HBV Effect through mRNA Splicing. Mediators Inflamm 2023;2023:2546278. [Crossref] [PubMed]
- Sato N, Maeda M, Sugiyama M, et al. Inhibition of SNW1 association with spliceosomal proteins promotes apoptosis in breast cancer cells. Cancer Med 2015;4:268-77. [Crossref] [PubMed]
- Papasaikas P, Valcárcel J. The Spliceosome: The Ultimate RNA Chaperone and Sculptor. Trends Biochem Sci 2016;41:33-45. [Crossref] [PubMed]
- Yang GC, Shi Y, Fan CN, et al. Spliceosomal GTPase Eftud2 regulates microglial activation and polarization. Neural Regen Res 2023;18:856-62. [Crossref] [PubMed]
- Lv C, Li XJ, Hao LX, et al. Over-activation of EFTUD2 correlates with tumor propagation and poor survival outcomes in hepatocellular carcinoma. Clin Transl Oncol 2022;24:93-103. [Crossref] [PubMed]
- Tu M, He L, You Y, et al. EFTUD2 maintains the survival of tumor cells and promotes hepatocellular carcinoma progression via the activation of STAT3. Cell Death Dis 2020;11:830. [Crossref] [PubMed]
- Dubovaia VI, Kolosov PM, Alkalaeva EZ, et al. Influence of individual domains of the translation termination factor eRF1 on induction of the GTPase activity of the translation termination factor eRF3. Mol Biol (Mosk) 2006;40:310-6. [PubMed]
- Cosson B, Berkova N, Couturier A, et al. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol Cell 2002;94:205-16. [Crossref] [PubMed]
- Hosoda N, Kobayashi T, Uchida N, et al. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J Biol Chem 2003;278:38287-91. [Crossref] [PubMed]
- Hoshino S, Imai M, Mizutani M, et al. Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF). Its identification as eRF3 interacting with eRF1. J Biol Chem 1998;273:22254-9. [Crossref] [PubMed]
- Malta-Vacas J, Aires C, Costa P, et al. Differential expression of the eukaryotic release factor 3 (eRF3/GSPT1) according to gastric cancer histological types. J Clin Pathol 2005;58:621-5. [Crossref] [PubMed]
- Tian QG, Tian RC, Liu Y, et al. The role of miR-144/GSPT1 axis in gastric cancer. Eur Rev Med Pharmacol Sci 2018;22:4138-45. [PubMed]
- Malta-Vacas J, Ferreira P, Monteiro C, et al. Differential expression of GSPT1 GGCn alleles in cancer. Cancer Genet Cytogenet 2009;195:132-42. [Crossref] [PubMed]
- Malta-Vacas J, Chauvin C, Gonçalves L, et al. eRF3a/GSPT1 12-GGC allele increases the susceptibility for breast cancer development. Oncol Rep 2009;21:1551-8. [PubMed]
- Long X, Zhao L, Li G, et al. Identification of GSPT1 as prognostic biomarker and promoter of malignant colon cancer cell phenotypes via the GSK-3β/CyclinD1 pathway. Aging (Albany NY) 2021;13:10354-68. [Crossref] [PubMed]
- Sun W, Zhang L, Yan R, et al. LncRNA DLX6-AS1 promotes the proliferation, invasion, and migration of non-small cell lung cancer cells by targeting the miR-27b-3p/GSPT1 axis. Onco Targets Ther 2019;12:3945-54. [Crossref] [PubMed]
- Nair S, Bora-Singhal N, Perumal D, et al. Nicotine-mediated invasion and migration of non-small cell lung carcinoma cells by modulating STMN3 and GSPT1 genes in an ID1-dependent manner. Mol Cancer 2014;13:173. [Crossref] [PubMed]
- Xiao R, Li C, Chai B. miRNA-144 suppresses proliferation and migration of colorectal cancer cells through GSPT1. Biomed Pharmacother 2015;74:138-44. [Crossref] [PubMed]
- Zhang C, Zou Y, Dai DQ. Downregulation of microRNA-27b-3p via aberrant DNA methylation contributes to malignant behavior of gastric cancer cells by targeting GSPT1. Biomed Pharmacother 2019;119:109417. [Crossref] [PubMed]
- Shi Q, Han S, Liu X, et al. Integrated single-cell and transcriptome sequencing analyses determines a chromatin regulator-based signature for evaluating prognosis in lung adenocarcinoma. Front Oncol 2022;12:1031728. [Crossref] [PubMed]


