
TCF4 as a potential prognostic biomarker and an anticancer target in gastric cancer
Highlight box
Key findings
• Transcription factor 4 (TCF4) has been implicated in the potential carcinogenesis of gastric cancer (GC) and may have utility as both a prognostic indicator and therapeutic target in the disease. Additionally, TCF4 may modulate the progression of GC through its interaction with myocyte enhancer factor 2C (MEF2C).
What is known and what is new?
• The Lymphoid enhancer-binding factor 1/T cell factor (LEF1/TCF) family of transcription factors play a crucial role in the development of malignant tumors. Specifically, in the progression of GC, TCF4 interacts with β-catenin to promote tumor cell proliferation both in laboratory settings and in animal models by upregulating the transcription of c-MYC and cyclin D1.
• The findings from protein-protein interaction analysis suggest that MEF2C may function as a regulatory gene for TCF4 and play a role in the progression of GC.
What is the implication, and what should change now?
• This study identified MEF2C as a potential therapeutic target for GC. Subsequent investigations are required to validate the interaction between MEF2C and TCF4 and to clarify the impact of TCF4 on MEF2C transcription.
IntroductionOther Section
Although the incidence and mortality rate of gastric cancer (GC) have declined among the Chinese population in the past two decades, GC ranks third for both incidence and mortality in China and has a 5-year survival rate of less than 50% (1,2). Therefore, there remain many challenges in managing GC therapy and prognosis. The progress in key areas, including potential therapeutic targets and prognostic markers, is needed to ensure that clinicians have the ability to attenuate the burden of GC.
Lymphoid enhancer-binding factor 1/T cell factor [LEF1/TCF family members comprise four nuclear factors, including LEF1, TCF1 or TCF7, TCF3 or TCF7L1, and TCF4 or TCF7L2], and function as the effectors of the Wnt signaling pathway (3). It has been reported that the individual TCFs exert distinct functions (4). LEF1 is an activator of transcription, while TCFs have been linked to messenger RNA (mRNA) alternative splicing and promoter usage (4). TCF4 was originally discovered to be a transcription activator, and TCF4 gene expression is partially overlapped with that of the LEF1 gene (5,6). Usually, the high expression of TCF4 is present in lymphocytes, esophageal squamous cell carcinoma, ovarian cancer, and lung adenocarcinoma (7-10). In GC progression, TCF4 interacts with CHAF1A to accelerate tumor cell growth in vitro and in vivo via activating the transcription of c-MYC and cyclin D1 (11). The disturbance of β-catenin and TCF4 dimerization can suppress GC proliferation, epithelial-mesenchymal transition, cancer stem cell proliferation, and metastasis (12-15). Nuclear translocation of TCF4 correlates with lymph node metastasis in GC (16). However, the prognostic significance and biological function of TCF4 in the progression of GC have not been extensively investigated.
In this study, we employed the online bioinformatics tool Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) (17) to assess the expression levels of the LEF1/TCF family members in GC tissues. Our analysis revealed that the transcripts and mRNA expression levels of LEF1, TCF3, TCF4, and TCF7 were significantly elevated in GC tissues compared to normal tissues. Specifically, a high expression of TCF4 was found to be associated with advanced tumor stage, overall survival (OS) in GC. Subsequent in vitro experiments were conducted to investigate the impact of TCF4 on GC cell proliferation, migration, and invasion. TCF4 knockdown will decrease GC cell proliferation, migration, and invasion in vitro. TCF4 may be a potential therapeutic target and prognostic marker for GC. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1290/rc).
MethodsOther Section
Clinical specimens
GC (n=55) and adjacent nontumor tissues (n=55) were collected from the Inner Mongolia Hospital Beijing Hospital of Traditional Chinese Medicine and maintained at −80 ℃ for the detection of mRNA expression. Prior to the collection of samples, informed consent forms were obtained from patients diagnosed with GC. The study received approval from the Ethics Committee of the Inner Mongolia Hospital Beijing Hospital of Traditional Chinese Medicine (No. 2017001045). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture and transfection
Normal gastric epithelial cell line GES-1 and GC cell lines (AGS, MGC-803, and SGC-7901) were procured from the Cell Bank of China Academy of Sciences and maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific) in a humidified incubator (Thermo Fisher Scientific). Small interfering RNA (siRNA) targeting TCF4 was synthesized by GenePharma (Shanghai, China) as si-TCF4, while that for the control was synthesized as si-Con. Lipofectamine 2000 (Invitrogen) was used to transfect si-Con and si-TCF4 into GC cells for 48 h at 37 ℃ following the manufacturer’s instructions. The cell experiment involved three biological replicates in each group. Real-time quantitative polymerase chain reaction (RT-qPCR) was utilized to confirm the transfection efficacy of si-TCF4, with si-TCF4 variants exhibiting inhibition rates exceeding 70% being selected for further cellular investigations. The mitigation of off-target effects was achieved through the refinement of transfection parameters, purification of si-RNA, and the inclusion of si-Con and RT-qPCR techniques. The sequences of si-TCF4 and si-Con were listed as follows: si-TCF4 sense (5'-3'): GAAAGGAAUCUGAAUCCGAAATT and antisense (5'-3'): UUUCGGAUUCAGAUUCCUUUCTT; si-Con sense (5'-3'): GCCAUGGCAAGGUCGGUAAGCTT and antisense (5'-3'): GCUUACCGACCUUGCCAUGGCTT. The optimal transfection conditions were determined to be a cell density of 5×103 cells for a duration of 48 h. Under these conditions, the gene expression of TCF was reduced by over 80% when the siRNA concentration was set at 30 nM. The cell experiment involved three biological replicates in each group.
RT-qPCR
The 7500 Real-Time PCR system (Applied Biosystems, Thermo Fisher Scientific) with TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) was used for the detection of mRNA expression. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was employed as the internal control, and the relative gene expression levels were determined using the 2−ΔΔCq method [20]. The primers were used as follows: TCF4 forward 5'-CAAGCACTGCCGACTACAATA-3', TCF4 reverse 5'-CCAGGCTGATTCATCCCACTG-3', myocyte enhancer factor 2C (MEF2C) forward 5'-CCAACTTCGAGATGCCAGTCT-3', and MEF2C reverse 5'-GTCGATGTGTTACACCAGGAG-3'.
Western blot
The experimental procedures involving Western blot analysis were conducted in accordance with previously published methods (18). Proteins were extracted using radioimmunoprecipitation assay (RIPA) buffer (Beyotime Institute of Biotechnology, Haimen, China). Following a blocking step with 5% skim milk, the membrane was incubated with primary antibodies at 37 ℃ for 2 h. The primary antibodies used for TCF4 (catalog number: ab217668; dilution: 1:1,000) and MEF2C (catalog number: ab227085; dilution: 1:1,000) were procured from Abcam (Cambridge, UK). After incubation with primary antibodies, the membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (cat. no. ab205718; dilution: 1:10,000; Abcam) at 37 ℃ for 1 h. Protein bands were visualized using an enhanced chemiluminescence kit (Thermo Fisher Scientific). Signals were analyzed with Quantity One® software version 4.5 (Bio Rad Laboratories, Inc., Hercules, CA, USA).
Cell proliferation, migration, and invasion, and apoptosis
Cell viability was assessed using a methylthiazolyldiphenyl-tetrazolium bromide (MTT) Cell Proliferation/Viability Assay kit from R&D Systems, Inc. (Minneapolis, MN, USA). Transwell migration assays were conducted using Transwell plates with an 8-µm pore size and without Matrigel, while Transwell invasion assays were carried out using Transwell plates with an 8-µm pore size and Matrigel in MGC-803 and SGC-7901 cells. Cell apoptosis was detected using a TdT-mediated dUTP nick end labeling (TUNEL) assay from the Beyotime Institute of Biotechnology following the manufacturer’s instructions.
Bioinformatics analysis
A GEPIA dataset was used to detect LEF/TCF mRNA based on The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases as described previously (19). TCF4-related genes (top 50) in GC were predicted based on the TCGA dataset using spearman correlation coefficient with gene expression levels (19). The functions of TCF4 and the related genes were predicted using Gene Ontology (GO) in the Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/summary.jsp). A protein-protein interaction (PPI) network was integrated into the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (version 11.0, updated on January 19, 2019) using Cytoscape software (version 3.7.8). The size of the circles representing gene symbols, as well as the thickness and depth of the connecting lines, were analyzed using Cytoscape software visualization to elaborate the strength of correlation. A GEPIA dataset online tool was used to analyze the OS and recurrence-free survival (RFS) of patients with GC based on the expression of LEF1/TCF and MEF2C mRNA expression in GC tissues (20).
Statistical analysis
Statistical analysis was conducted using SPSS version 19.0 (IBM Corp., Armonk, NY, USA). The Student t-test or the Wilcoxon rank-sum test was used to assess variances between two groups, while intergroup differences were evaluated via one-way analysis of variance. The Kaplan-Meier method was used to analyze OS and RFS. Spearman correlation analysis is used to analyze the correlation of genes. The chi-squared test and Fisher’s exact test were employed for categorical factors. A P value below 0.05 denoted statistical significance.
ResultsOther Section
LEF1/TCF expression in GC tissues
First, we investigated LEF1, TCF4, TCF7, and TCF3 mRNA expression in GC tissues using the GEPIA dataset. The results demonstrated that LEF1, TCF4, TCF7, and TCF3 transcripts and mRNA expression were significantly increased in GC tissues compared with normal tissues (Figure 1A,1B).

The association between LEF1/TCFs and tumor stages, OS, and RFS
As shown in Figure 2A, only LEF1 and TCF4 were significantly correlated with tumor stages for GC, whereas TCF7 and TCF3 did not show a significant difference. To further explore the correlation between the mRNA levels of LEF1/TCFs and the survival of patients with GC, the Kaplan-Meier Plotter tool was used to analyze the prognostic significance of LEF1/TCFs. As shown in Figure 2B, high TCF4 expression was associated with poorer OS in patients with GC. As shown in Figure 2C, those patients with high or low LEF1, TCF4, TCF7, and TCF3 expression had on obvious effect on RFS. These findings suggest that only high TCF4 expression may be a risk factor predictive of poor prognosis OS for patients with GC. Baseline information of patients with GC (n=375) in TCGA database indicated that high TCF4 expression was significantly associated with poor T and pathologic stage, histological type, and histologic grade (Table 1). In our cohort, no correlation was observed between TCF4 expression and variables such as age, gender, pathologic stage, or Helicobacter pylori (H. pylori) infection (Table 2).

Table 1
| Characteristics | Low expression of TCF4 | High expression of TCF4 | P value |
|---|---|---|---|
| N | 187 | 188 | |
| Age (years), mean ± SD | 66.63±10.4 | 65.02±10.87 | 0.15 |
| Gender, n (%) | 0.78 | ||
| Female | 65 (17.3) | 69 (18.4) | |
| Male | 122 (32.5) | 119 (31.7) | |
| Race, n (%) | 0.19 | ||
| Asian | 41 (12.7) | 33 (10.2) | |
| Black or African American | 7 (2.2) | 4 (1.2) | |
| White | 108 (33.4) | 130 (40.2) | |
| Age, n (%) | 0.28 | ||
| ≤65 years | 77 (20.8) | 87 (23.5) | |
| >65 years | 110 (29.6) | 97 (26.1) | |
| T stage, n (%) | 0.007 | ||
| T1 | 15 (4.1) | 4 (1.1) | |
| T2 | 43 (11.7) | 37 (10.1) | |
| T3 | 89 (24.3) | 79 (21.5) | |
| T4 | 39 (10.6) | 61 (16.6) | |
| N stage, n (%) | 0.07 | ||
| N0 | 64 (17.9) | 47 (13.2) | |
| N1 | 51 (14.3) | 46 (12.9) | |
| N2 | 37 (10.4) | 38 (10.6) | |
| N3 | 28 (7.8) | 46 (12.9) | |
| M stage, n (%) | 0.43 | ||
| M0 | 164 (46.2) | 166 (46.8) | |
| M1 | 15 (4.2) | 10 (2.8) | |
| Pathologic stage, n (%) | 0.004 | ||
| Stage I | 36 (10.2) | 17 (4.8) | |
| Stage II | 59 (16.8) | 52 (14.8) | |
| Stage III | 61 (17.3) | 89 (25.3) | |
| Stage IV | 22 (6.2) | 16 (4.5) | |
| Histological type, n (%) | 0.001 | ||
| Diffuse type | 21 (5.6) | 42 (11.2) | |
| Mucinous type | 5 (1.3) | 14 (3.7) | |
| Not otherwise specified | 109 (29.1) | 98 (26.2) | |
| Papillary type | 4 (1.1) | 1 (0.3) | |
| Signet ring type | 4 (1.1) | 7 (1.9) | |
| Tubular type | 44 (11.8) | 25 (6.7) | |
| Histologic grade, n (%) | <0.001 | ||
| G1 | 5 (1.4) | 5 (1.4) | |
| G2 | 88 (24) | 49 (13.4) | |
| G3 | 90 (24.6) | 129 (35.2) | |
| H. pylori infection, n (%) | 0.17 | ||
| No | 78 (47.9) | 67 (41.1) | |
| Yes | 6 (3.7) | 12 (7.4) |
Owing to the incomplete clinical data for certain patients, the aggregate counts for subgroups including race, age, T/N/M stage, pathologic stage, histological type, histologic grade, and H. pylori infection did not total 375. TCGA, The Cancer Genome Atlas; TCF4, transcription factor 4; SD, standard deviation; H. pylori, Helicobacter pylori.
Table 2
| Characteristics | Low expression of TCF4 | High expression of TCF4 | P value |
|---|---|---|---|
| N | 27 | 28 | |
| Age (years), mean ± SD | 62.44±12.12 | 64.51±14.93 | 0.32 |
| Gender, n (%) | 0.49 | ||
| Female | 11 (20.0) | 14 (25.5) | |
| Male | 16 (29.0) | 14 (25.5) | |
| Age, n (%) | 0.86 | ||
| ≤65 years | 10 (18.2) | 11 (20.0) | |
| >65 years | 17 (30.9) | 17 (30.9) | |
| Pathologic stage, n (%) | 0.21 | ||
| Stage I | 10 (18.2) | 5 (9.1) | |
| Stage II | 9 (16.4) | 7 (12.7) | |
| Stage III | 6 (10.9) | 12 (21.8) | |
| Stage IV | 2 (3.6) | 4 (7.3) | |
| H. pylori infection, n (%) | 0.67 | ||
| No | 22 (40.0) | 24 (43.6) | |
| Yes | 5 (9.1) | 4 (7.3) |
TCF4, transcription factor 4; SD, standard deviation; H. pylori, Helicobacter pylori.
Analysis of TCF4-associated genes
Figure 3A shows the top 50 genes most associated with TCF4 in a coexpressive heatmap based on an RNA-sequencing dataset from TCGA, suggesting that MEF2C is the most relevant TCF4-assocatied gene in GC. In addition, GO analysis suggested that TCF4-associated genes were closely related to cell migration (Figure 3B). As shown in Figure 3C, the PPI network analysis also revealed that MEF2C was the most relevant TCF4-associated gene.

TCF4 was positively correlated with MEF2C in GC tissues
In our cohort population, we aimed to clarify the association of TCF4 and MEF2C expression in the progression of GC. Fifty-five pairs of GC tissues and paracancerous tissues were collected to assess TCF4 and MEF2C mRNA expression, and our findings revealed a significant up-regulation of TCF4 (Figure 4A) and MEF2C (Figure 4B) mRNA expression in GC tissues as compared to paracancerous tissues. Spearman rank correlation corroborated the significant positive correlation between TCF4 and MEF2C mRNA expression in GC tissues in our cohort population (r=0.470; P<0.001; Figure 4C) and TCGA dataset (r=0.880; P<0.001; Figure 4D). In addition, the 55 patients with GC were divided into two groups, a low TCF4 expression group (n=27) and a high TCF4 expression group (n=28) according to a fold change of TCF4 mRNA expression greater than 2. Patients with high TCF4 expression demonstrated a decreased OS compared to those with low TCF4 expression (Figure 4E). Analysis of the GEPIA dataset revealed a significant upregulation of MEF2C mRNA in GC tissues relative to normal tissues (Figure 4F). Furthermore, elevated MEF2C expression was strongly associated with advanced tumor stages (Figure 4G) in patients with GC. However, no significant correlation was observed between MEF2C expression and OS in patients with GC (Figure 4H).

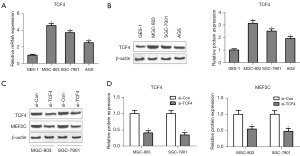
TCF4 expression was upregulated in GC cell lines
To investigate the function of TCF4 in vitro, the mRNA and protein expression of TCF4 was detected in GC cell lines. Compared with that in human normal gastric epithelial cell line GES-1, the mRNA and protein expression of TCF4 was markedly increased in GC cell lines, AGS, MGC-803, and SGC-7901 (Figure 5A,5B). Following the transfection of si-TCF4 into MGC-803 and SGC-7901 cells, TCF4 mRNA expression was significantly reduced by over 80% compared to the si-Con group (Figure S1). After transfection of si-TCF4 into MGC-803 and SGC-7901 cells, TCF4 protein expression was significantly reduced (Figure 5C,5D), and the MEF2C protein level was inhibited after si-TCF4 transfection (Figure 5C,5D).
TCF4 knockdown suppressed proliferation, migration, and invasion and induced the apoptosis of GC cells
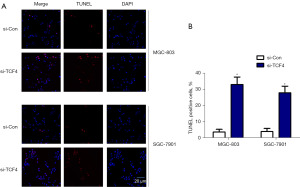
MTT assay suggested that knockdown of TCF4 led to significant inhibition of cell proliferation of MGC-803 and SGC-7901 cells after 24, 48, and 72 h on a growth medium (Figure 6A). After 24-h incubation with growth medium, transfection with si-TCF4 markedly suppressed the migration and invasion of MGC-803 and SGC-7901 cells (Figure 6B,6C). TUNEL staining indicated that transfection with si-TCF4 resulted in a significant increase in the number of TUNEL-positive cells, suggesting that TCF4 loss of function could induce the apoptosis of GC cells (Figure 7A,7B).

DiscussionOther Section
Using the GEPIA dataset analysis platform and experiments on our own specimens, we confirmed a significant upregulation of TCF4 mRNA expression in GC tissues compared to normal nontumor tissues. Additionally, elevated levels of TCF4 mRNA expression were observed in GC cell lines. In vitro experiments demonstrated that the suppression of TCF4 resulted in a marked inhibition of proliferation, migration, and invasion in MGC-803 and SGC-7901 cells. These findings suggest that TCF4 plays a vital role in maintaining the malignant properties of GC. In addition, patients with GC with high TCF4 expression were associated with poor T and pathologic stage, histologic grade, OS, and RFS, indicating that TCF4 might be a potential prognostic marker of GC.
TCF4, also known as TCF7L2, is a transcriptional activator that can enhance the transcriptional activity of Wnt/beta-catenin signaling axis, which is a notorious signaling cascade contributing to cancer initiation, growth, and metastasis via mediation of its target genes cyclin D1 and c-Myc (21-24). TCF4 serves as an oncogene to facilitate cancer migration, invasion, and drug resistance and is upregulated in several cancers, including colorectal cancer, acute myelocytic, leukemia, and breast cancer (25-27). In addition, TCF4 is a risk factor associated with poor prognosis in various cancers (7-9,28). Previous findings (11) and our present results indicate that the upregulation of TCF4 in GC tissues is positively correlated with poor prognosis. Our findings also suggest that TCF4 loss of function has the ability to inhibit the proliferation, migration, and invasion of GC cells in vitro.
According to TCGA dataset analysis, MEF2C was found to be the most relevant TCF4-associated gene in GC. MEF2C is a transcription factor uniquely present in hematopoietic, muscle, and neuronal lineages (29). It is commonly upregulated in leukemia and is associated with chemotherapy resistance and poor outcome (29,30). Upregulation of brain MEF2C is associated with breast cancer brain metastasis (31). MEF2C is an independent prognostic factor for predicting the OS of H. pylori-induced GC (32). According to our research and analysis of TCGA database, MEF2C exhibited a notable upregulation in GC tissues in comparison to normal nontumor tissues. Moreover, its expression was positively correlated with tumor stage and OS in patients with GC. Furthermore, we conducted an initial exploration to ascertain the potential role of TCF4 in regulating MEF2C expression in GC cells. The transfection of si-TCF4 resulted in a significant reduction in MEF2C protein levels in vitro. Nevertheless, the precise interaction between TCF4 and MEF2C in the progression of GC remains to be fully elucidated.
Based on the Wnt signaling pathway, upstream Wnt signaling facilitates the accumulation of β-catenin in the cytoplasm. Subsequently, β-catenin translocates into the nucleus where it associates with TCF-4. The β-catenin/TCF-4 complexes activate the transcription of Wnt target genes, thereby promoting the proliferation of tumor cells (33). These findings indicate that TCF-4 is critically involved in tumor development. An immunohistochemical analysis of 107 lung cancer samples revealed that 80.3% exhibited high TCF-4 expression (33). Mesoderm posterior basic helix-loop-helix (bHLH) transcription factor 2 competitively binds to TCF4, thereby suppressing GC progression through the inhibition of the TCF4/β-catenin transcriptional complex. This interaction results in a reduced occupancy of the complex on the S-phase kinase-associated protein 2 promoter and promotes the accumulation of p27 (15). The promotion of TCF4 transcription and the subsequent expression of its downstream target, MMP7, have the potential to enhance the invasive capabilities of GC cells (34). These findings indicate that TCF4 may play a role in both the metastasis of GC and the β-catenin-mediated progression of the disease. While targeted therapeutics for TCF4 are not yet available in clinical practice, the present research offers a robust theoretical basis for the development of such drugs in the future.
Despite our detailed exploration of the potential interaction between TCF4 and MEF2C in mediating GC progression, our study has several limitations. Firstly, we lack evidence to determine whether TCF4 and MEF2C interact directly or indirectly. Secondly, our conclusions are not supported by in vivo evidence. Thirdly, we need to collect a sufficient number of clinical samples to analyze the expression levels of TCF4 and MEF2C comprehensively.
ConclusionsOther Section
Our results suggest that TCF4 is upregulated in GC tissues and potentially exerts a carcinogenic role in the progression of GC. TCF4 might serve as a prognostic indicator and therapeutic target for GC. Furthermore, our findings indicate a strong correlation between the expression of TCF4 and MEF2C, suggesting that simultaneous inhibition of these two factors could serve as a promising therapeutic approach for GC. Future research endeavors will focus on investigating the direct or indirect interplay between TCF4 and MEF2C, as well as elucidating the specific mechanisms through which these proteins contribute to the progression of GC.
AcknowledgmentsOther Section
Funding: None.
FootnoteOther Section
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1290/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1290/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1290/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1290/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Inner Mongolia Hospital Beijing Hospital of Traditional Chinese Medicine (No. 2017001045) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Wang FH, Zhang XT, Tang L, et al. The Chinese Society of Clinical Oncology (CSCO): Clinical guidelines for the diagnosis and treatment of gastric cancer, 2023. Cancer Commun (Lond) 2024;44:127-72. [Crossref] [PubMed]
- Jiang L, Wang A, Yang S, et al. The Burden of Gastric Cancer Attributable to High Sodium Intake: A Longitudinal Study from 1990 to 2019 in China. Nutrients 2023;15:5088. [Crossref] [PubMed]
- Vava A, Paccez JD, Wang Y, et al. DCUN1D1 Is an Essential Regulator of Prostate Cancer Proliferation and Tumour Growth That Acts through Neddylation of Cullin 1, 3, 4A and 5 and Deregulation of Wnt/Catenin Pathway. Cells 2023;12:1973. [Crossref] [PubMed]
- Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol 2012;4:a007906. [Crossref] [PubMed]
- van de Wetering M, Oosterwegel M, Dooijes D, et al. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J 1991;10:123-32. [Crossref] [PubMed]
- Hrckulak D, Kolar M, Strnad H, et al. TCF/LEF Transcription Factors: An Update from the Internet Resources. Cancers (Basel) 2016;8:70. [Crossref] [PubMed]
- Ishiguro H, Wakasugi T, Terashita Y, et al. Nuclear expression of TCF4/TCF7L2 is correlated with poor prognosis in patients with esophageal squamous cell carcinoma. Cell Mol Biol Lett 2016;21:5. [Crossref] [PubMed]
- Liu L, Zeng Z, Yi J, et al. Expression and clinical significance of transcription factor 4 (TCF4) in epithelial ovarian cancer. Cancer Biomark 2019;24:213-21. [Crossref] [PubMed]
- Rice SJ, Liu X, Hyland V, et al. Mutations in genes connected with the TCF7L2 transcription factor are associated with a poor prognosis in non-small cell lung cancer. Lung Cancer 2020;141:97-100. [Crossref] [PubMed]
- Sade-Feldman M, Yizhak K, Bjorgaard SL, et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 2018;175:998-1013.e20. [Crossref] [PubMed]
- Zheng L, Liang X, Li S, et al. CHAF1A interacts with TCF4 to promote gastric carcinogenesis via upregulation of c-MYC and CCND1 expression. EBioMedicine 2018;38:69-78. [Crossref] [PubMed]
- Ge Q, Hu Y, He J, et al. Zic1 suppresses gastric cancer metastasis by regulating Wnt/β-catenin signaling and epithelial-mesenchymal transition. FASEB J 2020;34:2161-72. [Crossref] [PubMed]
- Chen QY, Xu KX, Huang XB, et al. Circ-0075305 hinders gastric cancer stem cells by indirectly disrupting TCF4-β-catenin complex and downregulation of SOX9. Commun Biol 2024;7:545. [Crossref] [PubMed]
- Liu H, Zhang X, Fang C, et al. Resveratrol induces the growth inhibition of CDX-deficient gastric cancer cells using CDX2 and RUNX3 via the β-catenin/TCF4 signaling pathway. Transl Oncol 2023;35:101727. [Crossref] [PubMed]
- Ge L, Zhao G, Lan C, et al. MESP2 binds competitively to TCF4 to suppress gastric cancer progression by regulating the SKP2/p27 axis. Cell Death Discov 2023;9:79. [Crossref] [PubMed]
- Chen XY, Wang ZC, Li H, et al. Nuclear translocations of beta-catenin and TCF4 in gastric cancers correlate with lymph node metastasis but probably not with CD44 expression. Hum Pathol 2005;36:1294-301. [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Liu Z, Dou C, Yao B, et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget 2016;7:25350-65. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep 2021;11:6047. [Crossref] [PubMed]
- Gao J, Zhao C, Liu Q, et al. Cyclin G2 suppresses Wnt/β-catenin signaling and inhibits gastric cancer cell growth and migration through Dapper1. J Exp Clin Cancer Res 2018;37:317. [Crossref] [PubMed]
- Hou G, Yuan X, Li Y, et al. Cardamonin, a natural chalcone, reduces 5-fluorouracil resistance of gastric cancer cells through targeting Wnt/β-catenin signal pathway. Invest New Drugs 2020;38:329-39. [Crossref] [PubMed]
- García de Herreros A, Duñach M. Intracellular Signals Activated by Canonical Wnt Ligands Independent of GSK3 Inhibition and β-Catenin Stabilization. Cells 2019;8:1148. [Crossref] [PubMed]
- Wu J, Xie N, Xie K, et al. GPR48, a poor prognostic factor, promotes tumor metastasis and activates β-catenin/TCF signaling in colorectal cancer. Carcinogenesis 2013;34:2861-9. [Crossref] [PubMed]
- Kendziorra E, Ahlborn K, Spitzner M, et al. Silencing of the Wnt transcription factor TCF4 sensitizes colorectal cancer cells to (chemo-) radiotherapy. Carcinogenesis 2011;32:1824-31. [Crossref] [PubMed]
- Saenz DT, Fiskus W, Mill CP, et al. Mechanistic basis and efficacy of targeting the β-catenin-TCF7L2-JMJD6-c-Myc axis to overcome resistance to BET inhibitors. Blood 2020;135:1255-69. [Crossref] [PubMed]
- Sergio S, Coluccia AML, Lemma ED, et al. 3D-microenvironments initiate TCF4 expression rescuing nuclear β-catenin activity in MCF-7 breast cancer cells. Acta Biomater 2020;103:153-64. [Crossref] [PubMed]
- Xiang J, Hu Q, Qin Y, et al. TCF7L2 positively regulates aerobic glycolysis via the EGLN2/HIF-1α axis and indicates prognosis in pancreatic cancer. Cell Death Dis 2018;9:321. [Crossref] [PubMed]
- Tarumoto Y, Lu B, Somerville TDD, et al. LKB1, Salt-Inducible Kinases, and MEF2C Are Linked Dependencies in Acute Myeloid Leukemia. Mol Cell 2018;69:1017-1027.e6. [Crossref] [PubMed]
- Brown FC, Still E, Koche RP, et al. MEF2C Phosphorylation Is Required for Chemotherapy Resistance in Acute Myeloid Leukemia. Cancer Discov 2018;8:478-97. [Crossref] [PubMed]
- Sereno M, Haskó J, Molnár K, et al. Downregulation of circulating miR 802-5p and miR 194-5p and upregulation of brain MEF2C along breast cancer brain metastasization. Mol Oncol 2020;14:520-38. [Crossref] [PubMed]
- Liu D, Ma X, Yang F, et al. Discovery and validation of methylated-differentially expressed genes in Helicobacter pylori-induced gastric cancer. Cancer Gene Ther 2020;27:473-85. [Crossref] [PubMed]
- Xu HT, Wei Q, Liu Y, et al. Overexpression of axin downregulates TCF-4 and inhibits the development of lung cancer. Ann Surg Oncol 2007;14:3251-9. [Crossref] [PubMed]
- Chen Y, Yang L, Qin Y, et al. Effects of differential distributed-JUP on the malignancy of gastric cancer. J Adv Res 2020;28:195-208. [Crossref] [PubMed]
(English Language Editor: J. Gray)


