
Construction of a prognostic model with exosome biogenesis- and release-related genes and identification of RAB27B in immune infiltration of pancreatic cancer
Highlight box
Key findings
• We constructed and validated a novel exosome biogenesis- and release-related risk signature and identified the pivotal role of RAB27B in promoting poor prognosis and regulating immune infiltration of pancreatic ductal adenocarcinoma (PDAC).
What is known and what is new?
• PDAC lacks exosome biogenesis- and release-related markers that can be used for prognosis prediction.
• The prognostic model was established to be associated with the prognosis of PDAC patients and RAB27B is correlated with immune infiltration in PDAC.
What is the implication, and what should change now?
• RAB27B participates in PDAC malignancy and could be a potential target for PDAC treatment.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) remains the most lethal malignant neoplasm and a leading cause of cancer death globally, with only a 10% 5-year survival rate in the United States (1,2). The latest epidemiological data demonstrated that there were 495,773 new pancreatic cancer cases and nearly 466,003 deaths worldwide in 2020 (3). Hence, it is urgent to construct an effective prognostic signature and develop new biomarkers for promoting early detection and guiding clinical decision-making of PDAC.
Exosomes are small membrane vesicles (40–100 nm) carrying a variety of complex contents, such as proteins, lipids, DNA, and RNA, from the parent cells. It was first discovered in rat reticulocytes in 1983 (4) and named “exosome” by Rose M. Johnstone in 1987 (5). The beginning process of exosome biogenesis and release is the formation of early endosomes (EEs) composed of plasma membrane invagination and primary endocytic vesicle fusion. EEs recycle the majority of their internalized cargo back to the plasma membrane through recycling endosomes (6) and are discharged from cells. However, EEs can also convert to multivesicular bodies (MVBs), also called late endosomes (LEs). During this process, the invagination of the endosomal membrane generates intraluminal vesicles (ILVs) in the organelle lumen, which are secreted as exosomes through plasma fusion and exocytosis of MVBs. Various molecules and complex regulatory pathways are involved in exosome biogenesis and release processes. EE-to-LE conversion is regulated and sorted by endosomal sorting complex required for transport (ESCRT) (7,8), Alix (8,9), tetraspanins (9,10), syndecan-syntenin-Alix (11,12) and the RAB7/hVPS34/p150 complex through phosphatidylinositol 3’-kinase cycling (13,14). The RAB27A and RAB27B genes were found to play a crucial role in the exosome release process, which occurred by inducing MVBs to transfer to the cell periphery and fuse with plasma membranes (15). A previous study showed that extracellular vesicles produced by pancreatic cancer cells can lead to gemcitabine resistance, while GW4869, an exosome biogenesis and release inhibitor, can reverse this effect (16), implying the potential for the regulation of biogenesis and release as new treatment options for overcoming chemoresistance of pancreatic cancer (17) alongside improving long-term survival of PDAC patients (18). Therefore, we constructed a prognostic signature based on exosome biogenesis- and release-related genes to predict the prognosis and optimize clinical treatment for pancreatic cancer patients.
In this study, we created a risk signature based on genes related to the process of exosome biogenesis and release to predict the prognosis of PDAC patients, and verified by tissue microarray and in vitro experiments. Furthermore, the potential correlation of exosome biogenesis- and release-related prognostic model with immune cell infiltration, somatic mutations, and DNA methylation was also explored, further validating the predictive value of the novel model. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-54/rc).
Methods
Construction and validation of exosome biogenesis- and release-related gene signature
First, the univariate Cox analysis was performed to identify the prognostic genes with a P value <0.05 in the total cohort. Second, we identified genes that promote exosome release while contributing to poor prognosis as well as those inhibit exosome release while contributing to good prognosis. Then, the least absolute shrinkage and selection operator (LASSO)-Cox dimension reduction analysis, and bi-directional stepwise multivariate Cox analysis were applied to further narrow down the optimal gene combination for constructing a risk signature. Then, all PDAC patients were randomly divided into the training and testing cohorts at a ratio of 1:1. According to the selected gene expression and their coefficients, the prognostic risk score was calculated as follows: risk score = (exprgene1 × Coefgene1) + (exprgene2 × Coefgene2) + … + (exprgenen × Coefgenen). Based on the median risk score in the training cohort, PDAC patients were divided into low-risk (risk score < median risk score) and high-risk (risk score > median risk score) groups in the training, testing, and total cohorts. The effectiveness of the risk signature was evaluated by the survival point diagram, survival heatmap, Kaplan-Meier (K-M) survival curve, and time-dependent receiver operating characteristic (ROC) curve in these three cohorts. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Peking Union Medical College Hospital (approval number: I-23PJ1544) and informed consent was obtained from all individual participants.
Tumor immune cell infiltration analysis
First, we used the CIBERSORT algorithm (https://github.com/Moonerss/CIBERSORT) to quantify the abundance of 22 different immune infiltrating cells between the two-risk groups, as well as exploring the correlation analysis between these 22 immune cells and the core prognostic genes. Furthermore, immune cell infiltration levels of different risk groups in the PDAC microenvironment were calculated by single sample gene set enrichment analysis (ssGSEA) algorithm. The immune, stromal, and estimate scores were calculated by the R package ‘estimate’.
Tumor mutation status analysis
The mutation data were acquired from The Cancer Genome Atlas (TCGA) database. Then, the R package ‘maftools’ was applied to analyze the mutation status between the low- and high-risk subgroups. We also compared the tumor mutation burden (TMB) scores for PDAC patient in the two risk groups.
DNA methylation status analysis in the CpG islands of the RAB27B gene
We used TCGA-pancreatic adenocarcinoma (PAAD) datasets to analyze the DNA methylation status of the RAB27B gene at CpG sites based on the MethSurv database (https://biit.cs.ut.ee/methsurv/). Then, we analyzed the association between RAB27B expression and the methylation level in highly methylated CpG sites. Furthermore, the CpG methylation status of RAB27B and overall survival (OS) of PAAD patients in the TCGA database were also evaluated.
Cell culture
We purchased all pancreatic cancer cell lines from the American Type Culture Collection (ATCC). All cell lines were identified by short tandem repeat (STR) sequencing, and mycoplasma testing was performed every two months. The HPNE, MIACaPa-2, T3M4, and PANC-1 cell lines were cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM; Corning, USA), and the AsPC-1 and BxPC-3 cell lines were cultured in RPMI-1640 medium (Corning). Ten percent fetal bovine serum (FBS) (HyClone, Logan, UT, USA) was added to all culture media. All cell lines were placed in a humidified incubator setting temperature at 37 degrees with 5% carbon dioxide.
Transfection assay
Transfections were conducted by using OPTI-MEM (Invitrogen, USA) and Lipofectamine 3000 (Invitrogen). siRAB27B-1 (sense: 5'-GACUUAAUCAUGAAGCGAA-3'; antisense: 5'-UUCGCUUCAUGAUUAAGUC-3'), siRAB27B-2 (sense: 5'-CAAAUUCAUCACUACAGUA-3'; antisense: 5'-UACUGUAGUGAUGAAUUUG-3') and siNC were purchased from Tsingke (Beijing, China). The RAB27B-overexpression plasmid (RAB27B-OE) and negative control plasmid (RAB27B-VC) were purchased from GeneChem (Shanghai, China).
RNA isolation and quantitative real-time polymerase chain reaction (RT-PCR)
Total RNA was extracted from treated pancreatic cancer cell lines using TRIzol reagent (Ambion, Life Technologies, California, USA, 15596026). cDNA was synthesized using a cDNA Reverse Transcription kit (Thermo Scientific, Massachusetts, USA, K1622). Quantitative RT-PCR was performed using PowerUpTM SYBRTM Green Master Mix (Applied Biosystems, Massachusetts, USA, A25742) in StepOnePlusTM (Applied Biosystems) according to the manufacturer’s protocols. Primers were purchased from Sangon Biotech (Shanghai, China). The 2−ΔΔCT method was applied to quantify the fold change by normalizing expression values to GAPDH. The primer sequences were as follows: RAB27B: forward: 5'-GGATGAGCCAACTGCAAGCAAATG-3', reverse: 5'-CTGATCTGGTAGGTCTGCCTTGTTG-3'; GAPDH: forward: 5'-GTCTCCTCTGACTTCAACAGCG-3', reverse: 5'-ACCACCCTGTTGCTGTAGCCAA-3'.
Cell proliferation and migration experiment
At 24 h after transfection, T3M4 or MIAPaCa-2 cells were seeded in the 96-well plates (3,000 cells per well). Then, the cell proliferation was measured at 0, 24, 48, 72 and 96 h using the sulforhodamine B (SRB) assay (Sigma-Aldrich, Missouri, USA). The optical density at a wavelength of 564 nm (OD564) was measured by a microplate reader. For the migration experiment, 3×104 cells after transfection were seeded into the Transwell chamber (Corning) with FBS-free medium. The Transwell chambers were placed in 24 well plates cultured with 10% FBS medium. After 24 h incubation, the cells were fixed by methanol and stained by hematoxylin-eosin staining.
Apoptosis assay
T3M4 or MIAPaCa-2 cells were seeded into 6-well plates. At 48 h after transfection, these cells were resuspended in the binding buffer. These cells were next stained with propidium iodide (PI) and Annexin V-fluorescein isothiocyanate (FITC) according to the manufacturer’s instructions (Yishan Biotechnology, Shanghai, China). Subsequent analysis was performed through the FlowJo V10.8.1.
Western blotting
Western blotting assays were performed according to previously described protocols (19). The primary antibodies used were anti-RAB27B polyclonal antibody (Proteintech, Illinois, USA, 13412-1-AP), anti-Vimentin (Proteintech, 10366-1-AP), anti-GAPDH (Proteintech, 10494-1-AP), anti-CD9 (Abcam, Cambridge, UK, ab223052), anti-Alix (Abcam, ab76608), anti-calnexin (Abcam, ab22595), and anti-TSG101 (Abcam, ab30871).
Immunohistochemistry (IHC) and tissue microarray
IHC was manually performed as the protocol described in our previous research (20). For primary antibody incubation, three sequential sections of each patient were incubated with rabbit monoclonal anti-CD8 antibody (1:200) (Abcam, ab4055), rabbit monoclonal anti-CD3 antibody (1:1,000) (Proteintech, 17617-1-AP), rabbit monoclonal anti-NCR1 antibody (1:500) (Abcam, ab224703), rabbit monoclonal anti-CD226 antibody (1:1,000) (Abcam, ab212076), and anti-RAB27B polyclonal antibody (1:100) (Proteintech, 13412-1-AP) for 120 min. Secondary antibody polymer (ORIGENE, Maryland, USA, PV-9000) was incubated for 30 min at room temperature. For tissue microarray, immunohistochemical staining area of less than 50% was defined low quality and excluded for further study. Finally, a total of 81 PDAC patients with survival information who underwent surgical resection at Peking Union Medical College Hospital were included. Staining results were photographed by microscopy (Nikon ECLIPSETs2R, Tokyo, Japan). The results of staining were determined independently by two experienced pathologists. Staining score = tumor cell proportion × staining intensity. Proportion: 0 (no stained tumor cells), 1 (<25% stained tumor cells), 2 (25%≤ stained tumor cells <50%), 3 (50%≤ stained tumor cells <75%), and 4 (≥75% stained tumor cells). Intensity: 0 (negative particles), 1 (lightly yellow particles), 2 (brownish-yellow particles) and 3 (brown particles). Patients were divided into high and low RAB27B expression groups according to the median staining score.
Extracellular vesicles isolation and bicinchoninic acid (BCA) protein quantification
Total extracellular vesicles were extracted from PDAC cell lines by differential ultracentrifugation. The samples were centrifuged at 600 ×g (4 ℃) for 10 min and 2,000 ×g (4 ℃) for 10 min. Then, they were centrifuged at 100,000 ×g (4 ℃) for 70 min twice successively and resuspended in PBS. Samples were then stored at −80 ℃ for further analysis. Nanoparticle tracking analysis were performed by Particle Metrix’ ZetaView to analyze the size distribution of extracellular vesicles. BCA method was performed to measure protein concentration by restricting both the cell supernatant and cell number to the same.
Statistical analysis
All statistical analyses and visualization were performed using R software (version 4.1.2; R Foundation for Statistical Computing, Vienna, Austria). A P value of <0.05 (two-sided) was considered statistically significant.
Results
Construction of protein-protein interaction (PPI) network
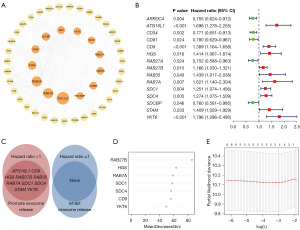
Forty-eight genes involved in the process of exosome biogenesis and release were extracted and summarized from the Gene Ontology database (http://geneontology.org), Gene Set Enrichment Analysis (GSEA) website (http://www.gsea-msigdb.org/gsea/index.jsp), and a review (21). The functional PPI network of the 48 genes was analyzed by the STRING database (https://string-db.org/) and visualized by Cytoscape software 3.9.1, as shown in Figure 1A. The size of the circle indicates the value of betweenness centrality.
Data collection and processing
Gene expression profile, mutation profile, and the relevant prognostic and clinical information were downloaded from TCGA database (https://portal.gdc.cancer.gov/; ≤ February 1, 2023). The original STAR-counts expression profile was transferred to Transcripts Per Kilobase Million (TPM). Meanwhile, GSE183795 (22), GSE78229 (23), GSE57495 (24) datasets was extracted from Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/; ≤ February 1, 2023). We merged these three datasets and eliminate the batch effects by applying the “Combat” algorithm, as shown in Figure S1. This combined GEO expression profile was further merged with TCGA dataset and used throughout our analysis unless otherwise mentioned.
We removed cases with non-PDAC type in TCGA dataset and OS less than 30 days. Finally, 148 cases from TCGA-PAAD, 63 cases from GSE57495, 49 cases from GSE78229, and 134 cases from GSE183795 with complete follow-up information were included (detailed in Tables S1-S4).
Establishment and validation of exosome biogenesis- and release-related gene signature
First, we screened 15 prognosis-related genes (P<0.05 in univariate Cox analysis) out of 48 exosome biogenesis- and release-related genes (Figure 1B). Among them, ARRDC4 (25), ATG16L1 (26), CD34 (27), CD81 (9), CD9 (28), HGS (29), RAB27A, RAB27B (15), RAB35 (30), RAB7A (31), SDC1 (32), SDC4 (33), SDCBP (34), STAM (35), YKT6 (36) can promote the release of exosomes, while none of them inhibit exosome release. Then, 10 genes (ATG16L1, CD9, HGS, RAB27B, RAB35, RAB7A, SDC1, SDC4, STAM, and YKT6) that promote exosome release while causing poor prognosis [hazard ratio (HR) >1] were selected. Because our hypothesis and most of the previous literature showed that exosome-mediated intercellular communication promotes tumor progression, so 10 genes that promote exosomes and were associated with poor prognosis were selected (Figure 1C). Additionally, a single-cell cohort CRA001160 was analyzed in the Tumor Immune Single-cell Hub database (http://tisch.comp-genomics.org/). As shown in Figure S2, RAB35, ATG16L1, and STAM were lowly expressed in cancer cells, so they were excluded form subsequent analysis. Random forest models demonstrate the ordering of gene importance with RAB27B ranking the highest (Figure 1D). LASSO regression (Figure 1E) and bi-directional stepwise multivariate Cox analysis were further adopted to select the optimal gene signature, detailed in Table S5. Two hub genes (RAB27B, YKT6) were finally screened out to construct the risk signature. Since RAB27B ranked the highest in the random forest models and showed lowest P value in the multivariate Cox regression, we selected RAB27B for the subsequent study. Meanwhile, the total cohort were randomly divided into the training and testing cohort at a ratio of 1:1. The risk score for each PDAC patient in the training, testing and entire cohorts were calculated as: risk score = expression level of RAB27B × 0.240 + expression level of YKT6 × 0.352. Furthermore, these three exploration sets were divided into low-risk and high-risk groups based on the training set’s median risk score, detailed in Tables S6-S8.
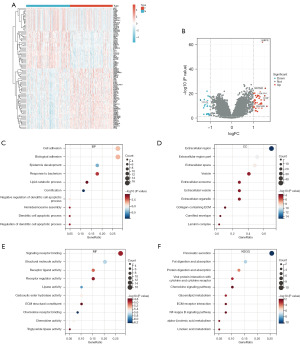
Differentially expressed genes (DEGs) between the low- and high-risk groups in total cohort were identified using the Limma package and visualized as a heatmap and volcano map (Figure 2A,2B). To further explore the functional enrichment of the risk signature, we implemented GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses between the low- and high-risk groups of the above DEGs. GO analysis was divided into three categories: biological process (BP), cellular component (CC), and molecular function (MF). As shown in Figure 2C-2E, the top component of BP was cell and biological adhesion, CC was extracellular region, and MF was signaling receptor binding. The KEGG analysis showed that DEGs were significantly enriched in pancreatic secretion, which further verified the validity of the prognostic model (Figure 2F).
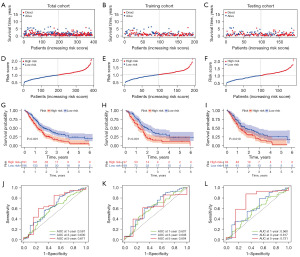
The risk score distribution and OS days of PDAC samples in the total, training, and testing cohorts are shown in Figure 3A-3F, respectively. Kaplan-Meier analysis demonstrated that patients in the high-risk group had significantly poorer survival than those in the low-risk group in these cohorts (Figure 3G-3I). Time-dependent ROC curves showed that our risk model had good predictive ability (Figure 3J-3L). The AUCs for 1-, 3- and 5-year survival of PDAC in the total cohort were 0.581, 0.630, 0.671, those in the training cohort were 0.607, 0.648, 0.604, and those in the testing cohort were 0.560, 0.617, 0.731, respectively.
Risk signature is associated with immune activity and mutation landscape
CIBERSORT algorithm was applied to evaluate the correlation between risk score and the abundance of 22 immune cells. As shown in Figure 4A, the risk score was positively associated with M0 macrophages, M2 macrophages, dendritic cells, and neutrophils, while negatively correlated with CD8+ T cells, naive B cells, and monocytes. Additionally, we examined how the two core genes in our risk model relate to the immune cell abundance (Figure 4B). Furthermore, the immune cell infiltration levels in the PDAC tumour microenvironment (TME) were also determined by ssGSEA algorithm (Figure 4C), which also revealed that CD8+ T cells and natural killer (NK) cells were negatively correlated with the risk score.
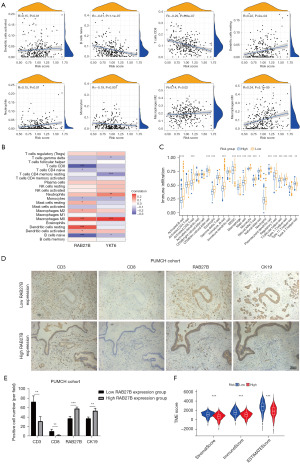
Then, IHC was manually performed to verify CD8+ T cells, NK cells, and RAB27B expression in 10 pancreatic cancer paraffin samples from Peking Union Medical College Hospital (PUMCH). NCR1 expression is by far the most reliable marker providing information about the presence of NK cells (37). However, NCR1 expression is relatively weaker than that of other specific genes. CD226 is widely expressed in cytotoxic lymphocytes, including NK cells (38). Therefore, we applied anti-NCR1 and anti-CD226 to explore the relationship between NK cells and RAB27B expression. Also, we used anti-CD3 and anti-CD8 to mark CD8+ T cells, and anti-CK19 to mark PDAC cells. We divided these ten patients into high and low RAB27B expression groups. Representative IHC images of CD3, CD8, CK19, NCR1, CD226, and RAB27B are shown in Figure 4D and Figure S3. Patients with higher expression of RAB27B presented lower CD3, CD8, NCR1 and CD226 expression (Figure 4E and Figure S3), which was consistent with previous bioinformatics analysis. Lastly, based on the R package “ESTIMATE”, a higher risk score was related with a significantly lower stromal score, immune score, and ESTIMATE score (Figure 4F).
The mutation landscapes of each PDAC patient in the TCGA cohort were analyzed based on the two risk groups (Figure 5A,5B). A higher mutation rate in the high-risk group was observed compared to the low-risk group (94% vs. 78%). For the gene mutation frequency, KRAS, TP53, and SMAD4 were the most frequently mutated genes in the high-risk group compared with the low-risk group (79% vs. 45%, 71% vs. 52%, and 29% vs. 15%, respectively). Also, tumor mutation analysis also revealed a higher TMB score in the high-risk group (Figure 5C).
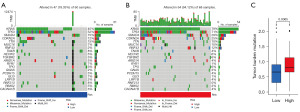
CpG island methylation levels of RAB27B predict the prognosis of PDAC patients
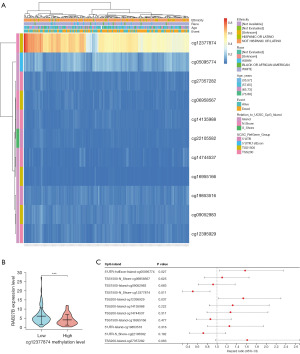
DNA methylation levels and the prognostic value of the CpG islands in the RAB27B gene were analyzed using the MetSurv tool. Methylated CpG islands cg12377874 showed elevated levels of DNA methylation in RAB27B (Figure 6A). Then, we verified that RAB27B was highly expressed in the low cg12377874 methylation group (Figure 6B), implying that targeted detection of cg12377874 methylation levels could effectively predict RAB27B expression. The methylation levels of three CpG islands, namely, cg12377874, cg12395929, and cg05095774, were related to prognosis (P<0.05) (Figure 6C). Elevated levels of RAB27B methylation in cg12377874 were associated with better OS of PDAC patients, while cg12395929 and cg05095774 were associated with poor prognosis.
Quality control of exosome extraction
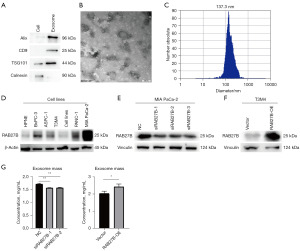
Western blotting showed that the expression of membrane protein CD9, and intra-membrane protein TSG101 and Alix in exosome were higher than cell samples. Also, negative marker Calnexin was not expressed in exosome sample (Figure 7A). Transmission electron microscopy captured the membrane ultrastructure of exosome’s double-layer capsule (Figure 7B). Nanoparticle tracking analysis also revealed that the peak concentration presented in the 137.3 nm which is consistent with the known exosome size (Figure 7C).
Knockdown or overexpression of RAB27B regulates exosome secretion of PDAC in vitro
To explore the role of RAB27B in regulating exosome release and biogenesis of PC, the expression of RAB27B in five pancreatic cancer cell lines (BxPC-3, AsPC-1, T3M4, PANC-1, and MIA PaCa-2) and one normal pancreatic ductal cell line, HPNE, was first analyzed. The expression of RAB27B was lower in T3M4 cells and higher in MIA PaCa-2 cells than in other cell lines (Figure 7D). Hence, we detected the efficiency of RAB27B-siRNA in the MIA PaCa-2 cell line and RAB27B-OE plasmid in the T3M4 cell line using western blotting (Figure 7E,7F). Then, we perform BCA to measure exosome protein concentration. With the same amount of cell supernatant and cell number, overexpression of RAB27B extracted more exosomes while knockdown of RAB27B would decrease exosomes release (Figure 7G).
RAB27B inhibits apoptosis while promotes PDAC cell proliferation and migration
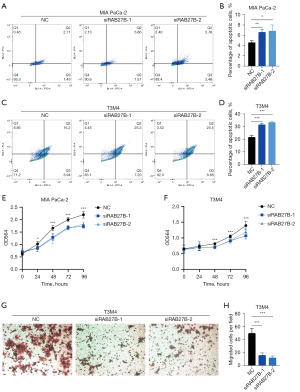
We examined the apoptosis levels of MIA PaCa-2 and T3M4 cells after RAB27B knockdown by flow cytometry. Consistently, the number of apoptotic cells (including early and late apoptotic cells) was significantly increased after downregulating RAB27B (Figure 8A-8D). The pancreatic cancer cells’ proliferation was detected by SRB assay, which showed that the cell proliferation was significantly decreased after RAB27B downregulation in MIA PaCa-2 and T3M4 cells (Figure 8E,8F). Moreover, using the Transwell assay, we discovered that the PDAC cells’ migration rate after RAB27B knockdown was significantly lower than the control cells in T3M4 cells (Figure 8G,8H).
RAB27B is related to shorter survival of PDAC patients in a PUMCH clinical cohort
We further employed 81 clinical PDAC samples and examined the RAB27B expression levels in PDAC tissues by a tissue microarray (Figure 9A). Representative images were shown in Figure 9B. The results showed that the PDAC patients in the high RAB27B expression group had significantly shorter OS than the low RAB27B expression group (Figure 9C).
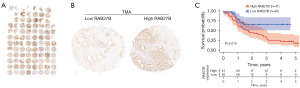
Discussion
PDAC is a highly lethal disease that is expected to surpass lung cancer as the second most common cause of cancer-related death worldwide by 2030 (39). One of the major reasons for this dismal situation is the lack of reliable risk prediction models and early diagnostic biomarkers, which impedes PDAC patients’ individualized treatment. Recently, many studies have revealed the crucial role of exosomes in pancreatic cancer development, metastasis and drug resistance (40-46). Therefore, we summarized all genes related to the exosome biogenesis-release process and explored a predictive model to evaluate PDAC patient prognosis and provide potential therapeutic options. Since RAB27B accounted for the highest value in the random forest model and displayed lowest P value in multivariate Cox analysis, we paid special attention to RAB27B and conducted more in-depth research in the following exploration.
Exosome biogenesis and release process consists of cargo sorting, MVB formation, MVB transportation, and MVB-plasma membrane fusion, which are regulated by some key molecules. RNA binding proteins, such as hnRNPA2B1, hnRNPK, major vault protein (MVP), and FMR1 are considered to participate in the exosome sorting process in different cancer models (47-49). MVB formation is the center to exosome biogenesis, and ESCRT-dependent/independent pathways mediated membrane budding and ILVs generation are key components in this process (50,51). In the regulation of exosome transportation, many RABs are proposed to participate in this process, including RAB2B, RAB5A, RAB9A, RAB27A and RAB27B (15). Also, Hsu et al. proposed that docking of MVBs to the plasma membrane is mediated by the RAB35, which promotes exosome secretion (52). Besides, RAB7 is found to be a critical gene in determining MVB degradation (53,54).
RAB27B, a small guanosine-5’-triphosphatase from the RAB family, is a key regulator of extracellular vesicles (EVs) fusion and trafficking. MVBs release EVs into the extracellular environment by regulating RAB27B (15). Among the 4-gene signature we identified, RAB27B was the most widely researched. Downregulation of RAB27B can decrease exosome secretion in diabetic kidney disease (55), hepatocellular carcinoma (56), and renal cell carcinoma (57). However, the relationship between RAB27B and exosome biogenesis and release in pancreatic cancer cells still lacks literature reports. In this article, we verified that knockdown and overexpression of RAB27B in the pancreatic cancer cell lines MIA PaCa-2 and T3M4, respectively, altered the amount of exosome secretion, demonstrating its role in the regulation of exosome biogenesis and release of pancreatic cancer cells. Furthermore, downregulating RAB27B could enhance drug sensitivity to cisplatin, induce apoptosis in the human PDAC cell lines ASPC-1 and PANC-1, and prevent PDAC invasion and proliferation (58). Overexpression of RAB27B could promote cell proliferation, migration and tumor invasion in pancreatic cancer (59). In mice, the zinc transporter ZIP4 activated CREB-regulated RAB27B expression, promoting the growth of orthotopic pancreatic tumors and the loss of muscle mass (60). Notably, the rescue experiment performed in many articles (61,62), normally through suppressing cellular exosome release, was achieved by stable knockdown or knockout of RAB27A. Our results confirmed that RAB27B not only had the potential to serve as a ‘Tool Gene’ to regulate exosome secretion similar to RAB27A but also as a potential therapeutic target to improve the prognosis of PDAC patients. In addition to RAB27B, Jaé et al. (31) found that exosome secretion of miR-143 was strongly inhibited by silencing RAB7A and RAB27B. NOTCH4hiCD8+ Treg cells suppress the expression of RAB7A and vesicular secretion of NADPH oxidase 2, resulting in the accumulation of early and recycling endosomes (63). RAB9A has been reported to play an oncogenic role in human liver cancer cells by promoting their invasion and migration through AKT/mTOR signaling pathways (64). ATP9A, a phospholipid flippase, is involved in the regulation of endosomal recycling (65), exosome release and the lipid composition of exosomes (66). Unfortunately, no studies have confirmed the associations of these genes with pancreatic cancer carcinogenesis and prognosis.
The PDAC TME contains tumour cells, stromal cells, immune cells, extracellular matrix and a variety of soluble molecules. Among them, the extracellular components play an essential role in the process of tumour development, invasion and metastasis. In PDAC TME, tumour cells only account for a small part, and most of the rest consists of stromal components, which contain a variety of immunosuppressive cell infiltration, such as myeloid-derived suppressor cells (MDSCs), tumor-associated macrophages (TAMs), and cancer-associated fribroblasts (CAFs), with immunosuppression as a prominent feature (67,68). In contrast, anti-tumor immune cells such as CD8+ T cells and NK cells are infiltrated to a lesser extent. Tumor-derived extracellular vesicles are involved in regulating the immune microenvironment (69). Various tumor-derived molecular signals are able to reprogram tumor-infiltrating immune cells via EVs (70,71). Tumor-associated proteins could be acquired within released EVs due to microenvironmental acidosis, which increases EVs release from tumor cells (72-74). Therefore, secreted EVs can connect with tumor malignancy and be seen as diagnostic biomarkers for cancer treatment. Tumor-released EVs are also recognized as major carriers of immunosuppressive factors that help tumor cells escape immune detection. For example, exosomal programmed cell death ligand 1 (PD-L1) has been shown to suppress CD8+ T cells, facilitate tumor immune escape, and promote drug resistance (62,75). Additionally, exosomes expressing NKG2DLs can selectively downregulate NK cell surface NKG2D expression, resulting in the impairment of NKG2D-dependent cytotoxic functions and expansion of tumor immune evasion (76). Marcon et al. discovered that peripheral blood NK cells of pancreatic cancer patients displayed high expression of CD16, CD57, and IL-10 with significant downregulation of NKG2D and IFN-γ (77). In contrast, NK cells were almost absent in tumor tissue and showed distinct downregulation of both CD16 and CD57, leading to a reduction in cytotoxic activity against tumor tissue. CD8+ T cells and NK cells are cytotoxic lymphocytes with the highest tumor killing capacity. In the present study, we found that PDAC patients in the high-risk group showed higher proportion of immune infiltrating cells, such as CD8+ T cells and NK cells, which might lead to poor prognosis. Besides, immune, stromal, and ESTIMATE score were lower in the high-risk group, further demonstrating their immunosuppressive status of TME.
DNA methylation is a common method of gene epigenetic modification. Alterations in the methylation levels of certain genes have been linked with tumorigenesis and cancer progression (78,79). We explored the relationship between the methylation status of RAB27B and the prognosis of PDAC patients. Hypermethylation of the CpG site cg12377874 was associated with prolonged OS and good prognosis. We further demonstrated that a lower cg12377874 methylation level of the RAB27B gene was related to higher RAB27B expression in PDAC patients, implying its significance in predicting prognosis and providing new therapeutic targets for pancreatic cancer.
To the best of our knowledge, this study is the first attempt to developed and validated a risk prediction model associated with the process of exosome biogenesis and release in PDAC patients, which showed a great efficacy on survival prediction. We believe these could serve as a preliminary exploration and lay foundation for subsequent high-quality research. Furthermore, we identified RAB27B as the core gene in our risk signature and demonstrated its inhibitory effect on the release of exosomes from PDAC cells. Also, we found RAB27B could inhibit PDAC cellular apoptosis while facilitate proliferation and migration by in vitro experiments. However, there are still several limitations in our study. First and foremost, this study mainly collected data from retrospective public databases, and thus further prospective research is necessary to validate our results. Also, an independent external validation cohort with greater sample size and long-term follow-up is needed in the future. Besides, there may be undiscovered genes affecting exosome biogenesis and release processes that have not been included due to the hysteresis of scientific research. In addition, the role of YKT6 in the exosome biogenesis-release process and tumor progression in PDAC has not been studied. Moreover, due to the limited time and experimental techniques, we only studied NK cells and CD8+ T cells by IHC and did not further explore other immune cells. Lastly, we did not perform in vivo experiments to validate our results.
Conclusions
In summary, we created an exosome biogenesis and release related prognostic model with good predictive capability. Also, we revealed the pivotal role of RAB27B in tumor progression, immune microenvironment and exosome biogenesis and release process.
Acknowledgments
We thank the support from the National Multidisciplinary Cooperative Diagnosis and Treatment Capacity Building Project for Major Diseases.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-54/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-54/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-54/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-54/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Peking Union Medical College Hospital (approval number: I-23PJ1544) and informed consent was obtained from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. Erratum in: CA Cancer J Clin 2021;71:359. [Crossref] [PubMed]
- Wu W, Miao Y, Yang Y, et al. Real-world study of surgical treatment of pancreatic cancer in China: annual report of China Pancreas Data Center (2016-2020). Journal of Pancreatology 2022;5:1-9. [Crossref]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol 1983;97:329-39. [Crossref] [PubMed]
- Johnstone RM, Adam M, Hammond JR, et al. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 1987;262:9412-20. [Crossref] [PubMed]
- Huotari J, Helenius A. Endosome maturation. EMBO J 2011;30:3481-500. [Crossref] [PubMed]
- Christ L, Raiborg C, Wenzel EM, et al. Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem Sci 2017;42:42-56. [Crossref] [PubMed]
- Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol 2013;5:a016766. [Crossref] [PubMed]
- Larios J, Mercier V, Roux A, et al. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J Cell Biol 2020;219:e201904113. [Crossref] [PubMed]
- Jankovičová J, Sečová P, Michalková K, et al. Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction. Int J Mol Sci 2020;21:7568. [Crossref] [PubMed]
- Matsui T, Osaki F, Hiragi S, et al. ALIX and ceramide differentially control polarized small extracellular vesicle release from epithelial cells. EMBO Rep 2021;22:e51475. [Crossref] [PubMed]
- Baietti MF, Zhang Z, Mortier E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol 2012;14:677-85. [Crossref] [PubMed]
- Stein MP, Feng Y, Cooper KL, et al. Human VPS34 and p150 are Rab7 interacting partners. Traffic 2003;4:754-71. [Crossref] [PubMed]
- Stein MP, Cao C, Tessema M, et al. Interaction and functional analyses of human VPS34/p150 phosphatidylinositol 3-kinase complex with Rab7. Methods Enzymol 2005;403:628-49. [Crossref] [PubMed]
- Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 2010;12:19-30; sup pp 1-13.
- Rimmer E, Rashid S, Kraev I, et al. Extracellular Vesicles Inhibit the Response of Pancreatic Ductal Adenocarcinoma Cells to Gemcitabine and TRAIL Treatment. Int J Mol Sci 2022;23:7810. [Crossref] [PubMed]
- Richards KE, Zeleniak AE, Fishel ML, et al. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene 2017;36:1770-8. [Crossref] [PubMed]
- Catalano M, O'Driscoll L. Inhibiting extracellular vesicles formation and release: a review of EV inhibitors. J Extracell Vesicles 2019;9:1703244. [Crossref] [PubMed]
- Qin C, Li T, Wang Y, et al. CHRNB2 represses pancreatic cancer migration and invasion via inhibiting β-catenin pathway. Cancer Cell Int 2022;22:340. [Crossref] [PubMed]
- Chen Y, Xu R, Ruze R, et al. Construction of a prognostic model with histone modification-related genes and identification of potential drugs in pancreatic cancer. Cancer Cell Int 2021;21:291. [Crossref] [PubMed]
- Gurung S, Perocheau D, Touramanidou L, et al. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun Signal 2021;19:47. [Crossref] [PubMed]
- Yang S, Tang W, Azizian A, et al. Dysregulation of HNF1B/Clusterin axis enhances disease progression in a highly aggressive subset of pancreatic cancer patients. Carcinogenesis 2022;43:1198-210. [Crossref] [PubMed]
- Wang J, Yang S, He P, et al. Endothelial Nitric Oxide Synthase Traffic Inducer (NOSTRIN) is a Negative Regulator of Disease Aggressiveness in Pancreatic Cancer. Clin Cancer Res 2016;22:5992-6001. [Crossref] [PubMed]
- Chen DT, Davis-Yadley AH, Huang PY, et al. Prognostic Fifteen-Gene Signature for Early Stage Pancreatic Ductal Adenocarcinoma. PLoS One 2015;10:e0133562. [Crossref] [PubMed]
- Foot NJ, Gonzalez MB, Gembus K, et al. Arrdc4-dependent extracellular vesicle biogenesis is required for sperm maturation. J Extracell Vesicles 2021;10:e12113. [Crossref] [PubMed]
- Guo H, Sadoul R, Gibbings D. Autophagy-independent effects of autophagy-related-5 (Atg5) on exosome production and metastasis. Mol Cell Oncol 2018;5:e1445941. [Crossref] [PubMed]
- Sun KT, Huang YN, Palanisamy K, et al. Reciprocal regulation of γ-globin expression by exo-miRNAs: Relevance to γ-globin silencing in β-thalassemia major. Sci Rep 2017;7:202. [Crossref] [PubMed]
- Mathieu M, Névo N, Jouve M, et al. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat Commun 2021;12:4389. [Crossref] [PubMed]
- Chen L, Chen R, Kemper S, et al. Pathways of production and delivery of hepatocyte exosomes. J Cell Commun Signal 2018;12:343-57. [Crossref] [PubMed]
- Zhang T, Zhao L, Han L, et al. TRX2/Rab35 Interaction Impairs Exosome Secretion by Inducing Rab35 Degradation. Int J Mol Sci 2022;23:6557. [Crossref] [PubMed]
- Jaé N, McEwan DG, Manavski Y, et al. Rab7a and Rab27b control secretion of endothelial microRNA through extracellular vesicles. FEBS Lett 2015;589:3182-8. [Crossref] [PubMed]
- Stepp MA, Pal-Ghosh S, Tadvalkar G, et al. Syndecan-1 and Its Expanding List of Contacts. Adv Wound Care (New Rochelle) 2015;4:235-49. [Crossref] [PubMed]
- Furini G, Schroeder N, Huang L, et al. Proteomic Profiling Reveals the Transglutaminase-2 Externalization Pathway in Kidneys after Unilateral Ureteric Obstruction. J Am Soc Nephrol 2018;29:880-905. [Crossref] [PubMed]
- Kugeratski FG, Hodge K, Lilla S, et al. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat Cell Biol 2021;23:631-41. [Crossref] [PubMed]
- Colombo M, Moita C, van Niel G, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci 2013;126:5553-65. [Crossref] [PubMed]
- Sun C, Wang P, Dong W, et al. LncRNA PVT1 promotes exosome secretion through YKT6, RAB7, and VAMP3 in pancreatic cancer. Aging (Albany NY) 2020;12:10427-40. [Crossref] [PubMed]
- Cózar B, Greppi M, Carpentier S, et al. Tumor-Infiltrating Natural Killer Cells. Cancer Discov 2021;11:34-44. [Crossref] [PubMed]
- Shibuya A, Shibuya K. DNAM-1 versus TIGIT: competitive roles in tumor immunity and inflammatory responses. Int Immunol 2021;33:687-92. [Crossref] [PubMed]
- Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913-21. [Crossref] [PubMed]
- Sun W, Ren Y, Lu Z, et al. The potential roles of exosomes in pancreatic cancer initiation and metastasis. Mol Cancer 2020;19:135. [Crossref] [PubMed]
- McAndrews KM, Xiao F, Chronopoulos A, et al. Exosome-mediated delivery of CRISPR/Cas9 for targeting of oncogenic KrasG12D in pancreatic cancer. Life Sci Alliance 2021;4:e202000875. [Crossref] [PubMed]
- Xie Z, Gao Y, Ho C, et al. Exosome-delivered CD44v6/C1QBP complex drives pancreatic cancer liver metastasis by promoting fibrotic liver microenvironment. Gut 2022;71:568-79. [Crossref] [PubMed]
- Marin AM, Mattar SB, Amatuzzi RF, et al. Plasma Exosome-Derived microRNAs as Potential Diagnostic and Prognostic Biomarkers in Brazilian Pancreatic Cancer Patients. Biomolecules 2022;12:769. [Crossref] [PubMed]
- Pan Y, Tang H, Li Q, et al. Exosomes and their roles in the chemoresistance of pancreatic cancer. Cancer Med 2022;11:4979-88. [Crossref] [PubMed]
- Guo Y, Li H, Sun C. Exosomal miR-125b-5p derived from cancer-associated fibroblasts promotes the growth, migration, and invasion of pancreatic cancer cells by decreasing adenomatous polyposis coli (APC) expression. J Gastrointest Oncol 2023;14:1064-76. [Crossref] [PubMed]
- Tamura T, Yoshioka Y, Sakamoto S, et al. Extracellular vesicles as a promising biomarker resource in liquid biopsy for cancer. Extracell Vesicles Circ Nucleic Acids 2021;2:148-74. [Crossref]
- Wozniak AL, Adams A, King KE, et al. The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J Cell Biol 2020;219:e201912074. [Crossref] [PubMed]
- Teng Y, Ren Y, Hu X, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun 2017;8:14448. [Crossref] [PubMed]
- Robinson H, Ruelcke JE, Lewis A, et al. Caveolin-1-driven membrane remodelling regulates hnRNPK-mediated exosomal microRNA sorting in cancer. Clin Transl Med 2021;11:e381. [Crossref] [PubMed]
- Hurley JH, Boura E, Carlson LA, et al. Membrane budding. Cell 2010;143:875-87. [Crossref] [PubMed]
- Han QF, Li WJ, Hu KS, et al. Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer. Mol Cancer 2022;21:207. [Crossref] [PubMed]
- Hsu C, Morohashi Y, Yoshimura S, et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol 2010;189:223-32. [Crossref] [PubMed]
- Jongsma ML, Bakker J, Cabukusta B, et al. SKIP-HOPS recruits TBC1D15 for a Rab7-to-Arl8b identity switch to control late endosome transport. EMBO J 2020;39:e102301. [Crossref] [PubMed]
- Pedersen NM, Wenzel EM, Wang L, et al. Protrudin-mediated ER-endosome contact sites promote MT1-MMP exocytosis and cell invasion. J Cell Biol 2020;219:e202003063. [Crossref] [PubMed]
- Zeng M, Wen J, Ma Z, et al. FOXO1-Mediated Downregulation of RAB27B Leads to Decreased Exosome Secretion in Diabetic Kidneys. Diabetes 2021;70:1536-48. [Crossref] [PubMed]
- Chen J, Lin Z, Liu L, et al. GOLM1 exacerbates CD8+ T cell suppression in hepatocellular carcinoma by promoting exosomal PD-L1 transport into tumor-associated macrophages. Signal Transduct Target Ther 2021;6:397. [Crossref] [PubMed]
- Tsuruda M, Yoshino H, Okamura S, et al. Oncogenic effects of RAB27B through exosome independent function in renal cell carcinoma including sunitinib-resistant. PLoS One 2020;15:e0232545. [Crossref] [PubMed]
- Li J, Jin Q, Huang F, et al. Effects of Rab27A and Rab27B on Invasion, Proliferation, Apoptosis, and Chemoresistance in Human Pancreatic Cancer Cells. Pancreas 2017;46:1173-9. [Crossref] [PubMed]
- Jiang P, Yin Y, Wu Y, et al. Silencing of long non-coding RNA SNHG15 suppresses proliferation, migration and invasion of pancreatic cancer cells by regulating the microRNA-345-5p/RAB27B axis. Exp Ther Med 2021;22:1273. [Crossref] [PubMed]
- Yang J, Zhang Z, Zhang Y, et al. ZIP4 Promotes Muscle Wasting and Cachexia in Mice With Orthotopic Pancreatic Tumors by Stimulating RAB27B-Regulated Release of Extracellular Vesicles From Cancer Cells. Gastroenterology 2019;156:722-734.e6. [Crossref] [PubMed]
- Cao M, Isaac R, Yan W, et al. Cancer-cell-secreted extracellular vesicles suppress insulin secretion through miR-122 to impair systemic glucose homeostasis and contribute to tumour growth. Nat Cell Biol 2022;24:954-67. [Crossref] [PubMed]
- Poggio M, Hu T, Pai CC, et al. Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory. Cell 2019;177:414-427.e13. [Crossref] [PubMed]
- Jin K, Wen Z, Wu B, et al. NOTCH-induced rerouting of endosomal trafficking disables regulatory T cells in vasculitis. J Clin Invest 2021;131:e136042. [Crossref] [PubMed]
- Sun P, Li L, Li Z. RAB9A Plays an Oncogenic Role in Human Liver Cancer Cells. Biomed Res Int 2020;2020:5691671. [Crossref] [PubMed]
- Tanaka Y, Ono N, Shima T, et al. The phospholipid flippase ATP9A is required for the recycling pathway from the endosomes to the plasma membrane. Mol Biol Cell 2016;27:3883-93. [Crossref] [PubMed]
- Xu X, Xu L, Zhang P, et al. Effects of ATP9A on Extracellular Vesicle Release and Exosomal Lipid Composition. Oxid Med Cell Longev 2020;2020:8865499. [Crossref] [PubMed]
- Ho WJ, Jaffee EM, Zheng L. The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat Rev Clin Oncol 2020;17:527-40. [Crossref] [PubMed]
- Li Y, Xiang S, Pan W, et al. Targeting tumor immunosuppressive microenvironment for pancreatic cancer immunotherapy: Current research and future perspective. Front Oncol 2023;13:1166860. [Crossref] [PubMed]
- Hou PP, Chen HZ. Extracellular vesicles in the tumor immune microenvironment. Cancer Lett 2021;516:48-56. [Crossref] [PubMed]
- Chen J, Fei X, Wang J, et al. Tumor-derived extracellular vesicles: Regulators of tumor microenvironment and the enlightenment in tumor therapy. Pharmacol Res 2020;159:105041. [Crossref] [PubMed]
- Whiteside TL. Exosomes and tumor-mediated immune suppression. J Clin Invest 2016;126:1216-23. [Crossref] [PubMed]
- Logozzi M, Capasso C, Di Raimo R, et al. Prostate cancer cells and exosomes in acidic condition show increased carbonic anhydrase IX expression and activity. J Enzyme Inhib Med Chem 2019;34:272-8. [Crossref] [PubMed]
- Logozzi M, Mizzoni D, Angelini DF, et al. Microenvironmental pH and Exosome Levels Interplay in Human Cancer Cell Lines of Different Histotypes. Cancers (Basel) 2018;10:370. [Crossref] [PubMed]
- Logozzi M, Spugnini E, Mizzoni D, et al. Extracellular acidity and increased exosome release as key phenotypes of malignant tumors. Cancer Metastasis Rev 2019;38:93-101. [Crossref] [PubMed]
- Chen G, Huang AC, Zhang W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018;560:382-6. [Crossref] [PubMed]
- Clayton A, Mitchell JP, Court J, et al. Human tumor-derived exosomes down-modulate NKG2D expression. J Immunol 2008;180:7249-58. [Crossref] [PubMed]
- Marcon F, Zuo J, Pearce H, et al. NK cells in pancreatic cancer demonstrate impaired cytotoxicity and a regulatory IL-10 phenotype. Oncoimmunology 2020;9:1845424. [Crossref] [PubMed]
- Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 2009;41:178-86. [Crossref] [PubMed]
- Stirzaker C, Song JZ, Ng W, et al. Methyl-CpG-binding protein MBD2 plays a key role in maintenance and spread of DNA methylation at CpG islands and shores in cancer. Oncogene 2017;36:1328-38. [Crossref] [PubMed]


