
S100A1 overexpression stimulates cell proliferation and is predictive of poor outcome in ovarian cancer
Highlight box
Key findings
• Elevated S100A1 protein levels in ovarian cancer (OC) tissues are associated with poor clinical outcomes. S100A1 knockout reduces OC cell proliferation and migration, and inhibits ferroptosis, lowering lipid reactive oxygen species levels.
What is known and what is new?
• Members of the S100 gene family are frequently dysregulated in various cancers, including OC. The prognostic significance of individual S100 genes, however, remains poorly understood.
• This study specifically highlights the prognostic value of S100A1 in OC. It demonstrates that high S100A1 levels are linked to adverse clinical outcomes and identifies S100A1 as a potential therapeutic target.
What is the implication, and what should change now?
• S100A1 has potential as a prognostic marker for OC and may be a promising target for therapeutic intervention. Further research should focus on validating S100A1 as a target for treatment and exploring its role in cancer progression.
Introduction
Ovarian cancer (OC) is a common gynecological malignancy with high incidence and mortality around the world (1). Epithelial OC more commonly occurs in postmenopausal females, while malignant germ cell tumors are more frequent in young females (2). The pathogenic factors of OC are not clear enough while according to data it may be related to genetic and endocrine factors (3). Most patients with OC are diagnosed at advanced stage, while some patients with malignant germ cell tumors can be diagnosed in the early stage (4). To date, surgery combined with chemotherapy is the major treatment for ovarian tumors (5). Targeted therapy, endocrine therapy and radiotherapy also have some therapeutic effects on OC (6,7). OC often relapses and a considerable number of patients die after multiple relapses and chemotherapy (8,9). One of the causes of OC relapse is due to the occurrence of chemotherapy resistance (10,11). Therefore, it is essential to discover novel therapeutic targets for OC treatment.
There are over 20 members in the S100 gene family. These genes encode calcium-binding proteins that operate both extracellularly and intracellularly as signaling factors and Ca2+ sensors (12), and have been implicated in different processes, such as proliferation, transformation, invasion, and migration (13,14). A number of S100 proteins, including S100A4 (15,16), S100A8 (17), S100A9 (18), S100A12 (19), and S100A13 (20), have been tied to the advancement and progression of thyroid cancer, suggesting a possible relationship between these proteins and OC.
The association of S100 expression with different types of cancer suggests that they may be useful as cancer biomarkers. High levels of S100A4 have been related to poor outcome in thyroid cancer (16) and silencing of S100A4 retards both invasion and survival in anaplastic thyroid cancer cells as well as potentiating the effect of vemurafenib in treating thyroid tumors (21). In contrast, overexpression of S100A4 has been closely related to the progression and metastasis of papillary thyroid carcinoma. Thus, it is possible that S100A4 may have a similar role in OC and may be a potential target for OC therapy (22). In addition, S100A12 levels are also raised in OC in which they are associated with cancer progression, including tumor size and stage, and lymph node metastasis, while silencing of the gene reverses these effects (19).
S100A1, also known as S100-α, was the first member of the S100 family being identified. It is present in many tissues, for instance, in heart and skeletal muscle. Silencing the gene diminishes cardiomyocyte contractility, while overexpression increases both contractility and energy production (23,24) and is also able to enhance recovery from ischemia-related damage (25,26). S100A1 is also strongly associated with cancer, for example, melanoma, in which it appears to participate in various pathophysiological activities (27). Abnormal S100A1 expression is also observed in OC, in which it has been reported to be an independent factor for the prediction of relapse-free survival in endometroid OC (28,29). However, the relationship between S100A1 and OC is not well understood. Here, the expression of S100A1 in OC tissue was examined to assess its use as a biomarker for OC diagnosis and prognostic prediction. Experiments, both in vitro and in vivo, were also performed to determine its role as a potential oncogene.
The current study can provide some new insights into understanding the biological functions of S100A1 in OC. We present this article in accordance with the MDAR and ARRIVE reporting checklists (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-430/rc).
Methods
Databases
A protocol was prepared before the study without registration. We obtained data from the GEPIA2 (Gene Expression Profiling Interactive Analysis) database (http://gepia.cancer-pku.cn), which allows the systematic analysis of cancer-associated gene expression (30). The database includes information from The Cancer Genome Atlas (TCGA) program and the Genotype Tissue Expression (GTEx) program. We used the database to develop plots of S100A1 expression in a variety of cancer types and their associated normal tissues.
Data download
The GSE14407 (31) gene set was downloaded from the public GEO (Gene Expression Omnibus) database (https://www.ncbi.nlm.nih.gov/geo/).
UALCAN database
We also used the UALCAN (the University of ALabama at Birmingham Cancer Data Analysis Portal) database (http://ualcan.path.uab.edu/), which comprises RNA-Seq data and clinical information from a variety of different cancers from the TCGA (32). The database allows the in-depth analysis of the expression of disease-related genes. We used the function module in the TCGA analysis for extracting data on S100A1 expression in different cancers.
Tissue samples
Samples of OC tumors and their adjoining normal tissues were achieved from 60 patients admitted to Shaanxi Provincial People’s Hospital between January 2012 and January 2014. The tumor diagnosis was confirmed by two independent pathologists. The patients were followed up for a median of 26.5 months following surgery; the maximum follow-up period was 72.7 months. The investigation was executed in compliance with the principles of the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of Shaanxi Provincial People’s Hospital (approval number: 2024R028). The patients gave written informed consent for participating by providing tissue samples.
Immunohistochemistry (IHC)
Tissues were embedded in paraffin, heated to 60 ℃ for 1 h, deparaffinized in xylene, and rehydrated in a 100–95–90–80–70–50% ethanol gradient. Antigen retrieval was executed by employing 0.1 M citrate buffer, pH 6.0. Endogenous peroxidase was inhibited using methanol with 0. 3% H2O2 for 30 min, and the sections were incubated with a mouse anti-S100A1 monoclonal antibody (Cell Signaling Technology, MA, Cat. 64095, dilution 1:100) at 4 ℃ overnight. The LSAB+ kit (Dako, USA) was used, following the instructions of the manufacturer. The sections were then stained with hematoxylin and assessed by two independent blinded investigators, using the semiquantitative histochemistry scoring method (H-SCORE). Final agreements on scoring were obtained for each specimen, despite occasional earlier discrepancies in immunostaining results (22). The H-SCORE reflects the positivity and intensity of staining by employing the formula: “H-SCORE=Σ (PI × I) = (percentage of cells of weak intensity ×1) + (percentage of cells of moderate intensity ×2) + (percentage of cells of strong intensity ×3)” with PI representing the percentage of positively-stained cells as a function of the total number of cells, and I representing the staining intensity. The scoring was as follows: I=0: blue, I=1: light yellow, I=2: brown, and I=3: dark brown staining. The H-SCORE ranges between 0 and 300, with greater scores indicative of more potent positivity.
Cell culture, vector construction, siRNAs and reagents
The human OC cell lines CAOV3, SK-OV3, HEY, and A2780 were acquired from central laboratory of the third affiliated hospital of Xi’an Jiaotong University, China. All cells were grown in RPMI 1640 containing 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA) and antibiotics at 37 ℃ and 5% CO2. The S100A1 overexpression vector was constructed and cloned into the BamHI and HindIII sites of the GV141 vector, yielding GV141-S100A1. The primers used in this experiment were as follows: forward: 5'-CGGGATCCATGGGCTCTGAGCTGGAGACGGCGA-3' (underline represents the BamHI enzyme cleavage site), reverse: 5'-CCCAAGCTTACTGTTCTCCCAGAAGAAATTGTTA-3' (underline represents the HindIII enzyme cleavage site). CAOV3 cells were transfected with either an empty vector or the GV141-S100A1 vector (GeneChem, China) implementing Lipofectamine 3000 (Thermo Fisher Scientific, MA), in accordance with the instructions of the manufacturer. The transfected cells were subsequently cultured for 48 h and subsequently analyzed for protein expression.
CRISPR-Cas 9 knockout of S100A1
An sgRNA sequence was designed for use with the CRISPR/Cas 9 system. This sequence was 5'-CAACGTGTTCCACGCCCACT-3'. Oligos comprising the sequence were cloned into the PX459 vector (SpCas9(BB)-2A-Puro V2.0 Addgene #62988), and transfected into A2780 cells by employing 1,000 ng DNA and Lipofectamine 3000. The cells were then incubated with 1 µg/mL puromycin for 72 h. T7 endonuclease evaluation was conducted to determine the plasmid efficiency as previously described, and the cells were resuspended to low densities and were seeded in 96-well plates with each well containing a single cell. The resulting clones were collected, lysed, and analyzed by western blotting using an anti-S100A1 antibody, while the A100A1 genomic region was amplified through PCR and sequenced. Controls included the empty PX459 vector.
Western blotting (WB)
Cells and tissues were lysed with RIPA buffer including protease inhibitors (Beyotime Institute of Biotechnology, China). The total protein was evaluated using the BCA (bicinchoninic acid) Protein Assay Kit (Pierce, Dallas, TX). Thirty micrograms of total protein per well were electrophoresed on 10% SDS-PAGE gels and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore Corp., Bedford, MA, USA). Following blocking with 5% fat-free milk in Tris-buffered saline (TBST), the incubation of the membranes was accomplished with the primary antibodies overnight at 4 ℃. The antibodies used were in Table S1. After three washes (15 min each in TBST), the blots were probed with secondary antibodies and visualized by enhanced. Chemiluminescence (Pierce, Rockford, IL, USA), in accordance with the manufacturer’s protocol.
Cell Counting Kit-8 (CCK8) assay
The cells were seeded in plates containing 96 wells at 3×103 cells/well. Ten microliters of CCK8 solution (Dojindo, Kumamoto, Japan) were added at 24-h intervals to each of the wells, allowed to react for 2 h, and the absorbances at 450 nm were read. Five replicate wells were used and the assessment was repeated a minimum of three times.
Colony-formation assay
Serial dilutions of cells (1,000–2,000 cells/mL) were plated in plates containing 6 wells and cultured for 2 weeks until colonies were discernible. The colonies were fixed in methanol (1 mL/well) for 10 min at ambient temperature and subsequently stained with 0.1% crystal violet for 5 min at ambient temperature. The cells were thoroughly washed under running water to minimize background staining, and the numbers of colonies were counted.
Transwell migration assay
Cells (2×105 cells/mL) in serum-free RPMI 1640 were seeded in the upper chamber of a Transwell apparatus (8 µm; Corning, Santa Barbara, CA, USA). Five hundred microliters of the milieu with 10% FBS were included in the lower chamber, and the apparatus was incubated for 24 h. Cells on the top surface of the membrane were moderately removed, fixed in 95% methanol, stained with crystal violet as above, and counted under light microscopy (Nikon, Japan).
LinkedOmics analysis
LinkedOmics (http://www.linkedomics.org) was used for the analysis of omics and clinical data from TCGA (33). The analysis provides information on the associations between genes and clinical parameters, including outcomes. The query involved the selection of tumor type (32 cancer types are available), selection of the data type (RNA-Seq in this case), selection of data attributes, selection of the target data type, and, finally, the selection of the statistical methods required. The output shows the correlation between the gene of interest and clinicopathological data, such as survival outcome, TNM (tumor, node, metastasis) staging, and ethnicity, as well as producing heatmaps showing genes positively and negatively associated with the expression of the gene of interest. We analyzed S100A1 RNA-Seq data from 303 OC patients using Spearman correlations. Plots for individual genes were obtained using LinkFinder and volcano plots and heatmaps were used for visualizing co-expression data. Genes that showed strong positive or negative correlations with S100A1 were submitted to the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases to determine annotations and pathway enrichment, respectively.
Determination of lipid ROS levels
To quantify lipid-ROS levels, cells were seeded in 6-well plates and transitioned to serum-free media supplemented with 10 µmol/L 2',7'-dichlorodihydrofluorescein diacetate (DCFH-DA, Sigma-Aldrich, St. Louis, MO, USA), followed by a 30-minute incubation in darkness with gentle agitation every 5 minutes. Subsequently, cells were harvested by centrifugation at 1,000 rpm for 5 minutes, washed thrice with serum-free media, resuspended in serum-free media, and then incubated in darkness with 5 µL of 7-aminoactinomycin D (Beyotime Institute of Biotechnology, Jiangsu, China) for 5 minutes. Fluorescence emission was measured using an excitation wavelength of 488 nm and an emission wavelength of 525 nm. The mean fluorescence intensity per group reflected the intracellular ROS levels.
Xenograft mouse model
Animal assessments were executed under a project license granted by the ethics committee of Health Science Center of Xi’an Jiaotong University (approval number: XJTUAE2020-1056) and were conducted according to the Animal Care guidelines of Xi’an Jiaotong University for the care and use of animals. Six-week-old female BALB/c nude mice (18–20 g, Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were subcutaneously injected with A2780: S100A1 KO A2780 cells, or control A2780 cells (5×106/mouse; n=6/group). The development of tumors was monitored and the tumor volumes were evaluated by employing the equation: Tumor volume = length × width × width/2. The humane endpoints were a tumor diameter ≥20 mm, together with other symptoms including weight loss (up to 16 g), slow, shallow, or labored breathing, reduced activity, social behavior, and grooming, and muscle atrophy. Twenty-four days after injection, the mice were sacrificed by inhalation of carbon dioxide (where the flow rate did not displace more than 30% of the chamber volume per minute), and the tumors excised, fixed, and frozen at -80°C for further analysis.
Statistical analysis
Data were presented as means ± standard deviation (SD) and were compared between groups by implementing Student’s t-test. The curves of Kaplan-Meier and log-rank assessments were employed to compare survival outcomes. P<0.05 was considered significant, and SPSS 19.0 (IBM Corp., Armonk, NY, USA) was employed for all statistical testing.
Results
S100A1 expression and association with OC survival
S100A1 levels in various tumors were examined using the UALCAN and GEPIA2 databases. The highest mRNA levels were seen in OC, SKCM, THCA, THYM, UCEC, and UCS cancer tissues (Figure S1A,S1B) compared with healthy tissue (Figure 1A). OC patient survival outcomes were assessed using GSE14407 and S100A1 is overexpression in OC patient (Figure 1B). S100A1 expression was significantly elevated in OC, as shown in Figure 1A, largely in patients with advanced-stage disease and distant metastases (Figure 1C), supporting the TCGA information.

We verified these observations in 60 pairs of OC tumor tissues and their normal adjoining tissues using IHC and WB. IHC showed that 71. 67% (43/60) of patients had high S100A1 levels in their tumor tissues (Figure 1D,1E). Raised levels were also linked to unfavorable outcomes, specifically, reduced OS (Figure 1F). WB confirmed the elevated S100A1 levels (Figure 1G). These results indicate that S100A1 expression is considerably upregulated in OC tumor tissue.
Downregulation of S100A1 represses proliferation and migration in OC cells
S100A1 expression was then analyzed in OC cell lines using WB. This showed relatively high levels of S100A1 in A2780 cells (Figure 2A). We then used CRISPR-Cas9 editing to generate S100A1 mutations in A2780 cells, introducing a frameshift mutation, resulting in a frameshift mutation in exon 2 and the resulting lack of expression was confirmed by WB (Figure 2B). The knockout cells showed significantly reduced ability to proliferate and migrate compared to the control transfected cells (Figure 2C-2E).

Enrichment of S100A1-co-expressed genes in OC
We used LinkedOmics to examine genes that might be co-expressed with S100A1 in the TCGA data of 3030 OC patients. The volcano plot (Figure 3A) demonstrates the positive relationship between 1,752 genes (dark-red dots) and S100A1, while 2,884 genes (dark-green dots) were negatively correlated [false discovery rate (FDR) <0.01]. The heatmap (Figure 3B,3C) illustrates the top 50 negatively and positively correlated genes.

The complete information on these genes is listed in Tables S2,S3. The top three positively correlated genes were S100A13 (r=0.776504, P=2.49E−62), SLPI (r=0.525189, P=7E−23) and S100A5 (r=0.511494, P=1.33E−21) (Table S2, Figure 3B), and the top three negatively correlated genes were TIMELESS (r=−0.45119, P=1.33E−16), MURC (r=−0.44827, P=2.20E−16), and KIF7 (r=−0.44223, P=6.13E−16) (Table S3, Figure 3C). These genes are involved in proliferation, cell adhesion, and ferroptosis. GO analysis indicated associations that the differentially expressed genes were associated with “DNA replication”, “NADH dehydrogenase complex assembly”, “double-strand break repair”, “mitochondrial protein complex”, “structural constituents of ribosome”, and “helicase activity” (Figure 4A, Figure S2). KEGG pathway enrichment showed links with “Fanconi anemia pathway”, “microRNAs in cancer”, “Hedgehog signaling pathway”, “Ribosome”, “Oxidative phosphorylation”, and “ferroptosis” (Figure 4B). In Figure 4C, we used Spearman-correlation analysis and found a positive correlation between S100A1 and GPX4 (r=0.3165, P=1.773e−08), with GPX4 being a key gene in ferroptosis. From the Figure 4D, we observe a correlation between S100A1 and ferroptosis. The levels of lipid ROS in CAOV3 cells were initially quantified, which indicated that the lipid ROS was significantly elevated after S100A1 and Erastin treatment compared to untreated CAOV3 cells (Figure 4E). Furthermore, S100A1 knockout reduced the levels of proliferating cell nuclear antigen (PCNA), Survivin, and GPX4 (Figure 2C, Figure S3), while overexpression had the opposite effect (Figure 4F). Collectively, these findings suggest that S100A1 is responsible for target protein expression levels in ferroptosis.

S100A1 knockout suppresses tumor growth in vivo
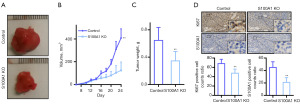
S100A1-knockout and control A2780 cells were injected into nude mice, resulting in reduced tumor growth in the knockout group (**P<0.01) (Figure 5A,5B). After 24 days, the tumor weights in the knockout group were significantly lower (**P<0.01) (Figure 5C) with reduced numbers of Ki67-positive cells (Figure 5D) indicating that reduced S100A1 inhibits tumor growth in vivo.
Discussion
The S100A gene family includes 16 members that have been shown to be involved in a wide spectrum of biological procedures, such as proliferation, calcium regulation, and inflammation (34). In the latter, S100A proteins work together with several proinflammatory cytokines, including interleukin 1α and interleukin 33, to regulate inflammation (35,36). S100A proteins are secreted from the cell into the extracellular environment where they interact with specific cell-surface receptors to modulate both innate and acquired immunity, as well as cell migration and tissue development (34,35,37).
S100A1 family members have been found to be strongly expressed in a variety of cancers, in comparison with normal tissue, and, in a comprehensive analysis of numerous patients, S100A2 was suggested as a potential biomarker for the diagnosis and outcome prediction of breast cancer (28), where S100A2 was observed to inhibit cancer progression through regulation of BRCA (BReast-CAncer susceptibility gene)/p63 and stabilizing p53 (38). S100A2, however, shows different expression in the squamous cell carcinoma lines, FADU and RPMI 2650, and has been shown to modulate various cellular functions (39). Activation of S100A2 and glucose transporter type 1 (GLUT1) promotes the progression of colon cancer through regulation of glycolysis (40).
The presence of various S100 family members has also been reported in OC, specifically, elevated S100A1 levels in serious OC in which it is associated with increased Silverberg grade, although not with cancer stage, while in endometrial OC, there are no relationship between S100A1 expression and clinical stage or grade, but the protein level is linked to reduced relapse-free survival (29). The concentration of S100A6 in the serum is related to both peritoneal tumor burden and advanced staging (41). S100A4, S100B, S100A14, and S100P levels have been linked to poor outcomes (28,42), as does strong cytoplasmic S100A10 staining in serious OC (43,44).
Accumulating evidence shows that S100A proteins stimulate OC progression (28). Exosome-associated S100A9 has been observed to promote inflammation and inhibit steroidogenesis through the activation of NFκB signaling, resulting in increased inflammation and dysfunctional steroidogenesis in polycystic ovary syndrome (45). Lv et al. reported that S100A4 acts as an autocrine/paracrine factor in promoting OC progression and is associated particularly with aggressive tumors (46). These findings support our results which showed that S100A1 advanced both proliferation and migration in OC. Ferroptosis is a new form of cell death distinguished from apoptosis, necrosis, and autophagy, and it is caused by iron-dependent lipid ROS accumulation. Many studies support that ferroptosis regulates the migration and proliferation of tumor cells. Lu et al. found that KLF2 deficiency impaired GPX4 transcriptional repression, promoting renal cell carcinoma cell migration and invasion by inhibiting ferroptosis (47). In addition, we observed that S100A1 blocked ferroptosis. Similarly, a report by Zou et al. indicated the involvement of both GPX4 and ferroptosis in clear-cell OC, characterized by clear cytoplasm due to lipid and glycogen accumulation, where GPX4 was found to block ferroptosis (48). GPX4 is a phospholipid hydroperoxidase that provides protection for membranes against lipid peroxidation using glutathione as a cofactor, and reduction in GPX4 levels, either by genetic manipulation or direct inhibition, is known to result in ferroptosis (49).
Conclusions
We demonstrated overexpression of S100A1 in OC and observed that this was associated with unfavorable clinical outcome and the stimulation of proliferation and migration in OC cells through blocking ferroptosis. This suggests that S100A1 can be a useful biomarker and a potential target for treating OC.
Acknowledgments
Thanks to Central Laboratory of The Third Affiliated Hospital of Xi’an Jiaotong University for providing us with the cells.
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-430/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-430/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-430/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-430/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The investigation was executed in compliance with the principles of the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of Shaanxi Provincial People’s Hospital (approval number: 2024R028). The patients gave written informed consent to participate by providing tissue samples. Animal assessments were executed under a project license granted by the ethics committee of Health Science Center of Xi’an Jiaotong University (approval number: XJTUAE2020-1056), and were conducted according to the Animal Care guidelines of Xi’an Jiaotong University for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang Z, Meng F, Zhong Z. Emerging targeted drug delivery strategies toward ovarian cancer. Adv Drug Deliv Rev 2021;178:113969. [Crossref] [PubMed]
- Rickard BP, Conrad C, Sorrin AJ, et al. Malignant Ascites in Ovarian Cancer: Cellular, Acellular, and Biophysical Determinants of Molecular Characteristics and Therapy Response. Cancers (Basel) 2021;13:4318. [Crossref] [PubMed]
- Nowicki A, Kulus M, Wieczorkiewicz M, et al. Ovarian Cancer and Cancer Stem Cells-Cellular and Molecular Characteristics, Signaling Pathways, and Usefulness as a Diagnostic Tool in Medicine and Oncology. Cancers (Basel) 2021;13:4178. [Crossref] [PubMed]
- Yang J, Huang S, Cheng S, et al. Application of Ovarian Cancer Organoids in Precision Medicine: Key Challenges and Current Opportunities. Front Cell Dev Biol 2021;9:701429. [Crossref] [PubMed]
- Leary A, Tan D, Ledermann J. Immune checkpoint inhibitors in ovarian cancer: where do we stand? Ther Adv Med Oncol 2021;13:17588359211039899. [Crossref] [PubMed]
- Luo X, Xu J, Yu J, et al. Shaping Immune Responses in the Tumor Microenvironment of Ovarian Cancer. Front Immunol 2021;12:692360. [Crossref] [PubMed]
- Coughlan AY, Testa G. Exploiting epigenetic dependencies in ovarian cancer therapy. Int J Cancer 2021;149:1732-43. [Crossref] [PubMed]
- Eckert MA, Orozco C, Xiao J, et al. The Effects of Chemotherapeutics on the Ovarian Cancer Microenvironment. Cancers (Basel) 2021;13:3136. [Crossref] [PubMed]
- Palanisamy CP, Cui B, Zhang H, et al. Anti-ovarian cancer potential of phytocompound and extract from South African medicinal plants and their role in the development of chemotherapeutic agents. Am J Cancer Res 2021;11:1828-44. [PubMed]
- Fantone S, Piani F, Olivieri F, et al. Role of SLC7A11/xCT in Ovarian Cancer. Int J Mol Sci 2024;25:587. [Crossref] [PubMed]
- Tossetta G, Fantone S, Goteri G, et al. The Role of NQO1 in Ovarian Cancer. Int J Mol Sci 2023;24:7839. [Crossref] [PubMed]
- Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer 2015;15:96-109. [Crossref] [PubMed]
- Wright NT, Cannon BR, Zimmer DB, et al. S100A1: Structure, Function, and Therapeutic Potential. Curr Chem Biol 2009;3:138-45. [PubMed]
- Wang T, Huo X, Chong Z, et al. A review of S100 protein family in lung cancer. Clin Chim Acta 2018;476:54-9. [Crossref] [PubMed]
- Jiao X, Zhang H, Xu X, et al. S100A4 Knockout Sensitizes Anaplastic Thyroid Carcinoma Cells Harboring BRAFV600E/Mt to Vemurafenib. Cell Physiol Biochem 2018;49:1143-62. [Crossref] [PubMed]
- Zou M, Famulski KS, Parhar RS, et al. Microarray analysis of metastasis-associated gene expression profiling in a murine model of thyroid carcinoma pulmonary metastasis: identification of S100A4 (Mts1) gene overexpression as a poor prognostic marker for thyroid carcinoma. J Clin Endocrinol Metab 2004;89:6146-54. [Crossref] [PubMed]
- Reeb AN, Li W, Sewell W, et al. S100A8 is a novel therapeutic target for anaplastic thyroid carcinoma. J Clin Endocrinol Metab 2015;100:E232-42. [Crossref] [PubMed]
- Ito Y, Arai K. S100A9 expression is significantly linked to dedifferentiation of thyroid carcinoma. Pathol Res Pract 2005;201:551-6. [Crossref] [PubMed]
- Wang X, Sun Z, Tian W, et al. S100A12 is a promising biomarker in papillary thyroid cancer. Sci Rep 2020;10:1724. [Crossref] [PubMed]
- Zhong J, Liu C, Chen YJ, et al. The association between S100A13 and HMGA1 in the modulation of thyroid cancer proliferation and invasion. J Transl Med 2016;14:80. [Crossref] [PubMed]
- Shi Y, Zou M, Collison K, et al. Ribonucleic acid interference targeting S100A4 (Mts1) suppresses tumor growth and metastasis of anaplastic thyroid carcinoma in a mouse model. J Clin Endocrinol Metab 2006;91:2373-9. [Crossref] [PubMed]
- Ito Y, Yoshida H, Tomoda C, et al. S100A4 expression is an early event of papillary carcinoma of the thyroid. Oncology 2004;67:397-402. [Crossref] [PubMed]
- Yu J, Lu Y, Li Y, et al. Role of S100A1 in hypoxia-induced inflammatory response in cardiomyocytes via TLR4/ROS/NF-κB pathway. J Pharm Pharmacol 2015;67:1240-50. [Crossref] [PubMed]
- Kraus C, Rohde D, Weidenhammer C, et al. S100A1 in cardiovascular health and disease: closing the gap between basic science and clinical therapy. J Mol Cell Cardiol 2009;47:445-55. [Crossref] [PubMed]
- Jungi S, Fu X, Segiser A, et al. Enhanced Cardiac S100A1 Expression Improves Recovery from Global Ischemia-Reperfusion Injury. J Cardiovasc Transl Res 2018;11:236-45. [Crossref] [PubMed]
- Fan L, Liu B, Guo R, et al. Elevated plasma S100A1 level is a risk factor for ST-segment elevation myocardial infarction and associated with post-infarction cardiac function. Int J Med Sci 2019;16:1171-9. [Crossref] [PubMed]
- Xiong TF, Pan FQ, Li D. Expression and clinical significance of S100 family genes in patients with melanoma. Melanoma Res 2019;29:23-9. [Crossref] [PubMed]
- Bai Y, Li LD, Li J, et al. Prognostic values of S100 family members in ovarian cancer patients. BMC Cancer 2018;18:1256. [Crossref] [PubMed]
- DeRycke MS, Andersen JD, Harrington KM, et al. S100A1 expression in ovarian and endometrial endometrioid carcinomas is a prognostic indicator of relapse-free survival. Am J Clin Pathol 2009;132:846-56. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Bowen NJ, Walker LD, Matyunina LV, et al. Gene expression profiling supports the hypothesis that human ovarian surface epithelia are multipotent and capable of serving as ovarian cancer initiating cells. BMC Med Genomics 2009;2:71. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Gross SR, Sin CG, Barraclough R, et al. Joining S100 proteins and migration: for better or for worse, in sickness and in health. Cell Mol Life Sci 2014;71:1551-79. [Crossref] [PubMed]
- Holzinger D, Tenbrock K, Roth J. Alarmins of the S100-Family in Juvenile Autoimmune and Auto-Inflammatory Diseases. Front Immunol 2019;10:182. [Crossref] [PubMed]
- Donato R, Cannon BR, Sorci G, et al. Functions of S100 proteins. Curr Mol Med 2013;13:24-57. [Crossref] [PubMed]
- Wu Y, Li Y, Zhang C, et al. S100a8/a9 released by CD11b+Gr1+ neutrophils activates cardiac fibroblasts to initiate angiotensin II-Induced cardiac inflammation and injury. Hypertension 2014;63:1241-50. [Crossref] [PubMed]
- Buckley NE, D’Costa Z, Kaminska M, et al. S100A2 is a BRCA1/p63 coregulated tumour suppressor gene with roles in the regulation of mutant p53 stability. Cell Death Dis 2014;5:e1070. [Crossref] [PubMed]
- Nagy N, Brenner C, Markadieu N, et al. S100A2, a putative tumor suppressor gene, regulates in vitro squamous cell carcinoma migration. Lab Invest 2001;81:599-612. [Crossref] [PubMed]
- Li C, Chen Q, Zhou Y, et al. S100A2 promotes glycolysis and proliferation via GLUT1 regulation in colorectal cancer. FASEB J 2020;34:13333-44. [Crossref] [PubMed]
- Wei BR, Hoover SB, Ross MM, et al. Serum S100A6 concentration predicts peritoneal tumor burden in mice with epithelial ovarian cancer and is associated with advanced stage in patients. PLoS One 2009;4:e7670. [Crossref] [PubMed]
- Wang X, Tian T, Li X, et al. High expression of S100P is associated with unfavorable prognosis and tumor progression in patients with epithelial ovarian cancer. Am J Cancer Res 2015;5:2409-21. [PubMed]
- Lokman NA, Pyragius CE, Ruszkiewicz A, et al. Annexin A2 and S100A10 are independent predictors of serous ovarian cancer outcome. Transl Res 2016;171:83-95.e1-2.
- Nymoen DA, Hetland Falkenthal TE, Holth A, et al. Expression and clinical role of chemoresponse-associated genes in ovarian serous carcinoma. Gynecol Oncol 2015;139:30-9. [Crossref] [PubMed]
- Li H, Huang X, Chang X, et al. S100-A9 protein in exosomes derived from follicular fluid promotes inflammation via activation of NF-κB pathway in polycystic ovary syndrome. J Cell Mol Med 2020;24:114-25. [Crossref] [PubMed]
- Lv Y, Niu Z, Guo X, et al. Serum S100 calcium binding protein A4 (S100A4, metatasin) as a diagnostic and prognostic biomarker in epithelial ovarian cancer. Br J Biomed Sci 2018;75:88-91. [Crossref] [PubMed]
- Lu Y, Qin H, Jiang B, et al. KLF2 inhibits cancer cell migration and invasion by regulating ferroptosis through GPX4 in clear cell renal cell carcinoma. Cancer Lett 2021;522:1-13. [Crossref] [PubMed]
- Zou Y, Palte MJ, Deik AA, et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun 2019;10:1617. [Crossref] [PubMed]
- Yang WS. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014;156:317-31. [Crossref] [PubMed]


