
MiR-155-5p regulates autophagy and apoptosis of glioma cells through RICTOR
Highlight box
Key findings
• We investigated the relationship which miR-155-5p regulates autophagy and apoptosis of glioma cells by regulating RICTOR, and explored the possible mechanism of the action, which is beneficial to the provision of new therapeutic insights for glioblastoma (GBM).
What is known and what is new?
• By far, relevant research has shown that miR-155-5P plays a biological role in the proliferation, invasion, temozolomide resistance, and mesenchymal transition of gliomas.
• This study aimed to identify the relationship which miR-155-5p regulates autophagy and apoptosis of glioma cells by regulating RICTOR, exploring the possible mechanism in GBM.
What is the implication, and what should change now?
• Experiments in vitro and bioinformatics analysis were conducted. U87 cells were used for in vitro, including quantitative real-time polymerase chain reaction, western blotting, immune fluorescence, and flow cytometry. Bioinformatics analysis included obtaining relevant data by accessing Gene Expression Omnibus (GEO), Chinese Glioma Genome Atlas (CGGA), and The Cancer genome Atlas program (TCGA).
• Database for Annotation, Visualization and Integrated Discovery (DAVID), Gene Expression Profiling Interactive Analysis (GEPIA2), Encyclopedia of RNA Interactomes (ENCORI), Protein-protein Interaction Networks Functional Enrichment Analysis (STRING) databases, and analyzing and processing the data using software such as Rstudio, Draw Veen Diagram, and Cytoscape.
Introduction
Glioma is one of the most common primary malignant tumors of the brain, accounting for about 10.9% of the tumors (1). At present, the etiology of glioma is still unknown, mainly involving exposure to high-dose ionizing radiation, high penetrance genetic mutations, nitrite food, virus or bacterial infection and other factors. It is mainly classified into four types: ependymoma, astrocytoma, oligodendroglioma and mixed glioma. It has the characteristics of strong invasiveness, high malignancy and easy recurrence (2,3). At the same time, the conventional treatment of glioma mainly includes surgical treatment, postoperative radiotherapy and chemotherapy. However, due to the high degree of drug resistance of glioma, the prognosis of patients is poor. The median survival time is 15 months, and the 5-year average survival rate is only about 5% (4). Therefore, it is urgent to seek new therapeutic targets to enhance the therapeutic effect on postoperative recurrence, drug resistance and metastasis of glioma patients and prolong the survival prognosis of patients. MicroRNA (miRNA) plays a key role in the occurrence and development of glioma. Many studies have shown that miRNA is involved in a variety of biological behaviors of tumors, enhancing or inhibiting the occurrence and development of tumors (5-7).
MiRNAs are short non-coding RNAs with a length of 18–22 nucleotides (8). MiRNAs cause the degradation of target mRNAs by complete or partial complementary binding to target mRNAs or down-regulating the expression level of target proteins by inhibiting their transcription, and promoting the expression of related oncogenes or tumor suppressor genes (9), thus participating in the proliferation, invasion, metastasis, angiogenesis, mesenchymal epithelial transformation, drug resistance and other processes of tumor cells (10). A large number of studies have shown that miRNAs play an important role in various biological processes of tumors (11). MicroRNA-155 is located on chromosome 21q21.3, 1,421 bp in length, and its pre-miR-155 is cleaved into mature miR-155-5p (12,13). Studies have shown that miR-155-5p is involved in neuroinflammation (14), autoimmunity (15), atherosclerosis (16), Alzheimer’s disease (17), multiple sclerosis (18), breast cancer, oral cancer, leukemia, melanoma, cervical cancer, non-small cell lung cancer, tumor resistance and other pathophysiological processes (19,20). Furthermore, it plays an important role in the process of glioma metastasis, invasion, proliferation, methylation, mesenchymal transition and drug resistance (21-25). However, it is still unclear whether miR-155-5p regulates autophagy and apoptosis in glioma.
The mechanistic target of rapamycin complex 2 (mTORC2) primarily composed of mTOR kinase, RICTOR, MAPKAP1 (or mSIN1), and MLST88 has been identified as a key signaling molecule in autophagy (26). Relevant studies have shown that silencing RICTOR as the key protein in mTORC2 complex can affect the aging and differentiation of fibroblasts by regulating autophagy (27-29). Second, in traumatic optic nerve injury, mTORC2 activates autophagy to protect the retinal ganglion (30). In addition, recent studies have shown that miR-155 aggravated pancreatic acinar cell damage by targeting RICTOR (31). The pro-osteoarthritic effect of 2,2',4,4',5,5'-hexachlorobiphenyl (PCB153) could inhibit autophagy by up-regulating miR-155 and inhibiting RICTOR/AKT/mTOR in rat primary chondrocytes (32). The latest study shows that RICTOR amplification in targeted therapy, drug resistance, and glioma autophagy plays an important role (33,34), but the specific mechanism is still unclear.
Autophagy is a self-digestive intracellular catabolic process that plays a crucial role in cell homeostasis under starvation, oxidative stress, and genotoxic stress conditions (35). Its main mechanism involves PI3K/AKT, MEK/ERK1/2, energy metabolism and other signaling pathways and related biological processes (36). Autophagy has two sides in the occurrence and development of glioma, and the related targets and mechanisms are still unclear. Related studies have shown that under hypoxic conditions, miR-155 expression is increased to target multiple autophagy-related genes to enhance autophagy flux (37), while miR-155-5p inhibits PDK1 and promotes autophagy through mTOR in cervical cancer (38), but whether it regulates autophagy in gliomas remains unclear.
Therefore, we present this article that expression difference of miR-155-5p between U87 glioma cells and normal cells, and verified that miR-155-5p is involved in autophagy and apoptosis of glioma cells, and further explored that miR-155-5p regulates autophagy and apoptosis of glioma cells by regulating the expression of RICTOR protein. Our results will help to understand the regulation of miR-155-5p/RICTOR axis in glioma and provide new insights for the treatment of glioma. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-543/rc).
Methods
Cell culture
Human glioma U87MG, U251 and HA1800 (Human Astrocytes; NHA) were purchased from Yimo Biotechnology Co., Ltd. (Xiamen, China). The more malignant U87MG cell line was selected for further experiment. These cells were cultured in summate basal medium (DMEM) medium containing 10% fetal bovine serum in a humidified incubator at 37 ℃ and 5% CO2, and the medium was changed every 2–3 days or during cell passage until cell confluence reached 70% to 90%.
Cell transfection and drug treatment
MiR-155-5p mimic negative control (NC)/mimic/inhibitor NC/inhibitor was purchased from Nantong Sowing Biotechnology Co., Ltd., riboFECTTM CP Transfection Kit Transfection reagent (Catalog No C10511-05, RiboBio, Suzhou, China), 3-MA (Catalog No. HY-19312, MedChemExpress, Shanghai, China), U87MG cells in good condition were digested, centrifuged, resuspended, evenly spread into 6-well plates to reach 30%, and left overnight. According to the plasmid transfection instructions of Ribobio Corporation, miR-155-5p mimic and inhibitor were mixed with transfection reagent and placed at room temperature for 15 minutes, then added to the complete medium. qPCR was used to detect the knockdown overexpression effect at 48 hours, and the protein level was detected at 72 hours. The pretransfected cells were treated with 3-MA (1 mM/L) for 24 hours and then harvested for protein extraction.
Western blot analysis
The glioma cells of each group were collected and washed with phosphate buffered saline (PBS) for 2–3 times, and then the cells were lysed on ice for 5 minutes. The cells were scraped and rotated in a refrigerator at 4 ℃ for 45 minutes, and then placed in a centrifuge at 15,000 rmp for about 20 minutes. After sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE), membrane was transferred with 20% methanol, blocked with 5% milk at room temperature for 2 hours, washed 3 times with Tris-buffered saline with Tween-20 (TBST), and added P62 (catalog number: 18420-1-AP, Proteintech Group, Wuhan, China), LC-3 (catalog number: 14600-1-AP, Proteintech Group, Wuhan, China), Bax (50599-2-Ig, Proteintech Group, Wuhan, China), Bcl-2 (catalog number: 68103-1-Ig, Proteintech Group, Wuhan, China), Caspase-3 (catalog number: 19677-1-AP, Proteintech Group, Wuhan, China) primary antibody dilution (diluted concentration: 1: 800) overnight at 4 ℃, and the cells were washed 3 times with TBST for 10 minutes each time. The secondary antibody dilution (dilution concentration 1:500) was added and incubated at room temperature for 2 hours. The membrane was washed by TBST, and the protein expression level was analyzed by visualization.
Quantitative real-time polymerase chain reaction (qRT-PCR)
In this experiment, the mRNA levels of miR-155-5p and RICTOR in each group were analyzed by qRT-PCR. The miR-155-5p primers were purchased from Suzhou Gemma Company, and other primer sequences are shown in Table 1. Total RNA was extracted from samples in each group (n=3 each) using total RNA extract (catalog number: R1100, Solarbio, Beijing, China). The amount of total RNA was determined using a Thermo NanoDrop one (Thermo Fisher, Waltham, MA, USA). Reverse transcription of complementary DNA (cDNA) was achieved using PrimeScriptTM RT Master Mix (catalog number: RR036A-1, TakaraBio, Kusatsu, Shiga, Japan). The qRT-PCR was then performed using BeyoFastTM SYBR Green qPCR Mix (2×, Low ROX) master mixture (catalog number: D7262, Beyotime, Shanghai, China) in a qRT-RCR instrument 7500 (Thermo Fisher, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control to normalize the data.
Table 1
| Name | Sequence |
|---|---|
| RICTOR | F: CTGGATCTGACCCGAGAACC |
| R: TTGATACTCCCTGCAATCTGGC | |
| EEF2 | F: ACCTTGTGGAGATCCAGTGTC |
| R: CCCCGCTTCCTGTTCAAAAC | |
| RAP1B | F: GTGACAGCGTGAGAGGTACT |
| R: GACTTTCCAACGCCTCCTGA | |
| PKN2 | F: GTCCATACCGGAGCGCAA |
| R: CGGGAATCCCCCTGCAGT | |
| TCF4 | F: ACAGAAAGGGGCTCATACTCA |
| R: AGGAGACTCTGCTGGTGG | |
| KRAS | F: ACACAAAACAGGCTCAGGACT |
| R: AGCATCCTCCACTCTCTGTCT | |
| CARD11 | F: TTGAGAGGAACACGAGGAAC |
| R: ATCTGGACTTGCAGTGCTTG | |
| CBL | F: GGAACTGCTTCTAAGGCTGC |
| R: GAAGTGTGGGAGGTACTGGC | |
| BDNF | F: GGCGGTACCCCCAGTTCC |
| R: CTTTCATGGGGGCAGCCTTC |
F, forward primer; R, reverse primer.
Cytofluorometry
After the stimulation, the 24-well plates were placed on ice, washed 3 times, fixed with 4% polylysine at 20 ℃ for 20 minutes, washed 3 times with PBS, and blocked at room temperature for 2 hours. Diluted antibody LC-3 (1:200) was added overnight at 4 ℃ and washed 3 times with PBS. Fluorescent secondary antibody (diluted concentration 1:1,000) was added and incubated at room temperature for 2 hours in the dark, and washed 3 times with PBS. Then, DAPI was added (diluted concentration 1:2,000) and incubated at room temperature in the dark for 20 minutes, and washed 3 times with PBS for 5 minutes each time. Then, the small discs were picked out and placed on the blocking solution, and the slides were made.
Biological information analysis
RNA-sequencing data were extracted by visiting the Gene Expression Omnibus (GEO) biological database, and the data were imported into the Database for Annotation, Visualization and Integrated Discovery (DAVIA; https://davidbioinformatics.nih.gov) database to obtain the functional enrichment analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) of related genes. The visualize functional enrichment analysis was plotted by https://www.bioinformatics.com.cn (last accessed on 20 May 2024), an online platform for data analysis and visualization. The survival prognosis curve of related genes was obtained by visiting The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov), Gene Expression Profiling Interactive Analysis (GEPIA2; http://gepia2.cancer-pku.cn) and Chinese Glioma Genome Atlas (CGGA; http://www.cgga.org.cn) databases. The target genes were predicted and intersected by TargetScan 8.0, MIRBD and Mirtarbase target gene websites. Then, the Draw Veen Diagram website was used to intersect the significantly different genes, and the Protein-protein Interaction Networks Functional Enrichment Analysis (STRING; https://cn.string-db.org) protein interaction website was used to obtain the protein interaction network diagram. The protein interaction data were imported into Cytoscape to screen the key genes and carry out experimental verification. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
All data were presented as mean ± standard deviation. The t-test was used for pairwise comparison, and one-way analysis of variance (ANOVA) was used for comparison among multiple groups. Images were processed using ImageJ and analyzed using GraphPad Prism version 9 software; P<0.05 was considered statistically significant.
Results
The expression of miR-155-5p was increased in glioma cells
In order to explore the biological role of miR-155-5p in glioma, we visited two data sets of GEO database GSE165937 and GSE138764, and utilized the limma package to identify genes with differential expression linked to miR-155-5p expression, applying a threshold of |log2 fold change| ≥2 and an adjusted P value of <0.05 (Figure 1A). The intersection of GSE165937 and GSE138764 was taken through the online Wayne map website, and it was found that miR-155-5p was significantly highly expressed in glioma. Subsequently, we obtained the box diagram of the expression of miR-155-5p in gliomas with different degrees of malignancy in the CCGA database (Figure 1B). Then by visiting the TCGA database and the CGGA database, it was found that the expression of miR-155-5p was negatively correlated with survival prognosis (Figure 1C). Then we used qRT-PCR to detect the expression of miR-155-5p in U87, U251 glioma cell lines and NHA. It was found that the expression of miR-155-5p in U87 and U251 was significantly higher than that in NHA, while there was no significant difference between U87 and U251 (Figure 1D). Then, in order to further explore its related mechanism, we used Target Scan 8.0, MIRBD, miRTarBase target gene prediction software to predict the related target genes of miR-155-5p and take their intersection (Figure 1E). The key target genes were screened by STRING protein interaction website and different algorithms in Cytoscape software, including RAP1B, CBL, BDNF, EEF2, SMAD2, KRAS, PKN2, RICTOR, FGF7, and YWHAZ (Figure 1F).
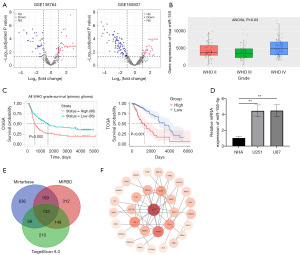
GO and KEGG analysis of miR-155-5p target genes
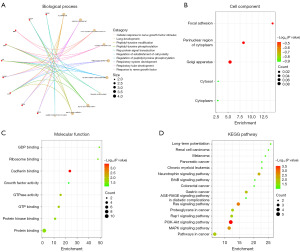
The key target genes were screened by different algorithms in Cytoscape software and the key target genes were imported into the DAVID database to obtain the GO and KEGG enrichment analysis results of the key genes, and then the relevant results were imported into the online visualization website: https://www.bioinformatics.com.cn, to obtain the functional enrichment map of the key target genes (Figure 2). Figure 2A shows that biological processes are mainly enriched in signal transduction, protein phosphorylation, positive regulation of gene expression, negative regulation of apoptotic process, lung development and other biological processes; Figure 2B shows that the cell components are mainly enriched in cytosol, cytoplasm, focal adhesion, perinuclear region of cytoplasm, and Golgi apparatus; Figure 2C shows that molecular functions are mainly enriched in protein binding, cadherin binding, GTPase activity, GTP binding, protein kinase binding and other molecular functions; Figure 2D shows that the key target genes are mainly enriched in PI3K-Akt, Ras, MAPK, cancer, neurotrophin and other related signaling pathways. The PI3K-Akt, Ras, and MAPK-related signaling pathways play a key role in the autophagy process of glioma cells (39-42). Therefore, we further verified that miR-155-5p regulates the autophagy biological process of glioma by regulating related target genes.
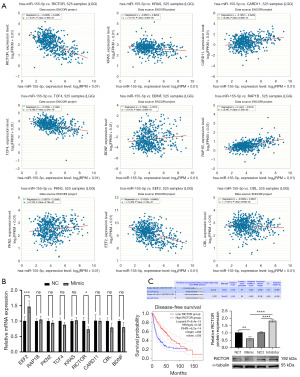
MiR-155-5p regulates RICTOR and inhibits its expression in glioma
By accessing the Encyclopedia of RNA Interactomes (ENCORI) database, we obtained the correlation analysis between miR-155-5p and EEF2, RAP1B, PKN2, TCF4, KRAS, RICTOR, CARD11, CBL, BDNF genes in glioma, and found that miR-155-5p was significantly negatively correlated with the expression of RICTOR (Figure 3A). Subsequently, qRT-PCR verification showed that RICTOR could be significantly inhibited after overexpression of miR-155-5p (Figure 3B). Subsequently, we knocked down miR-155-5p and found that RICTOR expression increased. Finally, we further verified its regulatory relationship by Western blotting. It was found that knockdown of miR-155-5p could up-regulate the protein expression of RICTOR, and overexpression of miR-155-5p could inhibit the expression of RICTOR protein, which was consistent with the results of qPCR. The target location information of miR-155-5p and RICTOR gene and the survival prognosis curve of RICTOR were obtained by visiting TargetScan 8.0 and GEPIA2 database (Figure 3C).
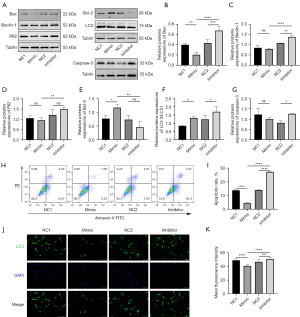
Effects of miR-155-5p on autophagy and apoptosis in glioma cells U87
Subsequently, to explore the effects of miR-155-5p on autophagy and apoptosis in glioma cells, we detected the effects of miR-155-5p knockdown and overexpression on the expression of autophagy and apoptosis-related proteins in U87 cells (Figure 4A). The results of Figure 4 showed that overexpression of miR-155-5p could reduce the expression of Bax protein in U87 glioma cells (Figure 4B), and increase the expression of Bcl-2 protein (Figure 4E). However, knockdown could increase the expression of Bax, LC3II/LC3I ratio, Beclin-1, P62, and Caspase-3 (Figure 4B-4D,4F,4G), while the expression of Bcl-2 protein decreased (Figure 4E). The results of apoptotic cells detected by flow cytometry suggested that inhibition of miR-155-5p expression increased the apoptosis of U87 cells (Figure 4H,4I). Therefore, we preliminarily concluded that knockdown of miR-155-5p could promote autophagy and apoptosis in U87 glioma, and cell fluorescence also confirmed that knockdown of miR-155-5p could enhance autophagy-related flux (Figure 4J,4K).
Correlation between autophagy and apoptosis in glioma cells
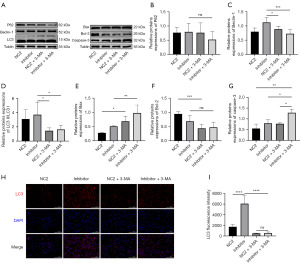
Then, to clarify the correlation between autophagy and apoptosis in glioma, miR-155-5p knockdown was followed by the addition of autophagy inhibitor 3-MA (1 mM). Western blot experiments showed that knockdown of miR-155-5p combined with autophagy inhibitor could further increase the expression of apoptosis-related proteins in glioma cells (Figure 5A-5G) and inhibit the progression of glioma. And, the efficacy of 3-MA was briefly verified by immunofluorescence single staining of LC3 (Figure 5H,5I). The cell fluorescence results verified that the autophagy inhibitor significantly inhibited the expression of autophagy-related protein LC3 (Figure 5H,5I).
Discussion
Autophagy and apoptosis of tumor cells are closely related to the occurrence and development of tumors. MiRNAs can regulate many key cellular processes, such as proliferation, invasion, angiogenesis, apoptosis, stress response, differentiation and development. This provides a certain idea and direction for the treatment of tumors.
The present study has shown that glioma stem cells (GSCs-derived) exosomes exhibit higher miR-155-5p expression levels (22). Moreover, many studies have shown that miR155-5p promotes the progression of cancer in a variety of tumors, including liver cancer, squamous cell carcinoma and colon cancer (43,44). As recently reported, exosome miR-155-5p can promote CXCL12/CXCR7-induced colorectal cancer metastasis through cross-talk with cancer-associated fibroblasts (45). Reactive oxygen species reprogram macrophages through the exosome miR-155-5p/PD-L1 pathway to inhibit anti-tumor immune responses (46). In our study, we searched the CGGA database and found that miR-155 expression was negatively correlated with survival prognosis (Figure 1B). We then examined the expression of miR-155-5p in U87 and U251 glioma cell lines and NHA. It was found that the expression of miR-155-5p in U87 and U251 was significantly higher than that in normal brain tissues, while there was no significant difference between U87 and U251 (Figure 1D).
Glioma is a common primary malignant tumor of the central nervous system. With the development of science and technology, major breakthroughs have been made in the treatment of many tumors, but the prognosis of patients with aggressive glioma is still very poor. Therefore, further studies on glioma are urgently needed to improve the prognosis of glioma patients. A recent study reported that miR-155-5p enhanced glioma migration and invasion by directly targeting ACOT12 (22). In this work, the key target gene RICTOR was screened by String and Cytoscape software, and the targeted regulatory relationship was further verified by Western blot and qPCR. We found that knocking down miR-155-5p up-regulated the protein expression of RICTOR. Overexpression of miR-155-5p inhibited the expression of RICTOR protein. Further study showed that the low expression of miR-155-5p promoted the apoptosis of U87 cells, and the specific inhibition of miR-155-5p also increased the expression of autophagy-related proteins in U87 cells, thereby enhancing autophagy and homeostatic regulation (Figure 4). Subsequent study has shown that knockdown expression of miR-155-5p combined with autophagy inhibitors may further promote the apoptosis of glioma cell line U87, thereby enhancing the therapeutic effect of glioma (Figure 5).
This study has several limitations. An in vivo animal model was not established for further study. Due to the time and the source of patients, not enough human tumor specimens were obtained for further research and verification. The cell lines studied were relatively homogeneous.
Our results identify a novel miR-155-5p/RICTOR regulatory axis in glioma cells and provide new insights into the regulation of apoptosis and autophagy in glioma. We suggest that miR-155-5p may be a novel therapeutic target for glioma.
Conclusions
MiR-155-5p regulates autophagy and apoptosis-related proteins in glioma cells through RICTOR, affecting the occurrence and development of glioma.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-543/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-543/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-543/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-543/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Austin CP. Opportunities and challenges in translational science. Clin Transl Sci 2021;14:1629-47. [Crossref] [PubMed]
- Sadowski K, Jażdżewska A, Kozłowski J, et al. Revolutionizing Glioblastoma Treatment: A Comprehensive Overview of Modern Therapeutic Approaches. Int J Mol Sci 2024;25:5774. [Crossref] [PubMed]
- Porter KR, McCarthy BJ, Berbaum ML, et al. Conditional survival of all primary brain tumor patients by age, behavior, and histology. Neuroepidemiology 2011;36:230-9. [Crossref] [PubMed]
- Gatto L, Di Nunno V, Franceschi E, et al. Pharmacotherapeutic Treatment of Glioblastoma: Where Are We to Date? Drugs 2022;82:491-510. [Crossref] [PubMed]
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol 2014;9:287-314. [Crossref] [PubMed]
- Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 2015;15:321-33. [Crossref] [PubMed]
- Fu P, Guo Y, Luo Y, et al. Visualization of microRNA therapy in cancers delivered by small extracellular vesicles. J Nanobiotechnology 2023;21:457. [Crossref] [PubMed]
- Floyd D, Purow B. Micro-masters of glioblastoma biology and therapy: increasingly recognized roles for microRNAs. Neuro Oncol 2014;16:622-7. [Crossref] [PubMed]
- Zhou Q, Liu J, Quan J, et al. MicroRNAs as potential biomarkers for the diagnosis of glioma: A systematic review and meta-analysis. Cancer Sci 2018;109:2651-9. [Crossref] [PubMed]
- Prabhu KS, Raza A, Karedath T, et al. Non-Coding RNAs as Regulators and Markers for Targeting of Breast Cancer and Cancer Stem Cells. Cancers (Basel) 2020;12:351. [Crossref] [PubMed]
- Jiménez-Morales JM, Hernández-Cuenca YE, Reyes-Abrahantes A, et al. MicroRNA delivery systems in glioma therapy and perspectives: A systematic review. J Control Release 2022;349:712-30. [Crossref] [PubMed]
- Gines G, Menezes R, Xiao W, et al. Emerging isothermal amplification technologies for microRNA biosensing: Applications to liquid biopsies. Mol Aspects Med 2020;72:100832. [Crossref] [PubMed]
- Mashima R. Physiological roles of miR-155. Immunology 2015;145:323-33. [Crossref] [PubMed]
- Zingale VD, Gugliandolo A, Mazzon E. MiR-155: An Important Regulator of Neuroinflammation. Int J Mol Sci 2021;23:90. [Crossref] [PubMed]
- Xu WD, Feng SY, Huang AF. Role of miR-155 in inflammatory autoimmune diseases: a comprehensive review. Inflamm Res 2022;71:1501-17. [Crossref] [PubMed]
- Bruen R, Fitzsimons S, Belton O. miR-155 in the Resolution of Atherosclerosis. Front Pharmacol 2019;10:463. [Crossref] [PubMed]
- Liu JJ, Long YF, Xu P, et al. Pathogenesis of miR-155 on nonmodifiable and modifiable risk factors in Alzheimer's disease. Alzheimers Res Ther 2023;15:122. [Crossref] [PubMed]
- Maciak K, Dziedzic A, Miller E, et al. miR-155 as an Important Regulator of Multiple Sclerosis Pathogenesis. A Review. Int J Mol Sci 2021;22:4332. [Crossref] [PubMed]
- Chen Z, Ma T, Huang C, et al. The pivotal role of microRNA-155 in the control of cancer. J Cell Physiol 2014;229:545-50. [Crossref] [PubMed]
- Kalkusova K, Taborska P, Stakheev D, et al. The Role of miR-155 in Antitumor Immunity. Cancers (Basel) 2022;14:5414. [Crossref] [PubMed]
- Wu X, Wang Y, Yu T, et al. Blocking MIR155HG/miR-155 axis inhibits mesenchymal transition in glioma. Neuro Oncol 2017;19:1195-205. [Crossref] [PubMed]
- Bao Z, Zhang N, Niu W, et al. Exosomal miR-155-5p derived from glioma stem-like cells promotes mesenchymal transition via targeting ACOT12. Cell Death Dis 2022;13:725. [Crossref] [PubMed]
- Gu J, Lu Z, Ji C, et al. Melatonin inhibits proliferation and invasion via repression of miRNA-155 in glioma cells. Biomed Pharmacother 2017;93:969-75. [Crossref] [PubMed]
- Yang L, Li C, Liang F, et al. MiRNA-155 promotes proliferation by targeting caudal-type homeobox 1 (CDX1) in glioma cells. Biomed Pharmacother 2017;95:1759-64. [Crossref] [PubMed]
- Wang R, Chen Y, Kuang W, et al. Valproic acid regulates the miR-155/Jarid2 axis by affecting miR-155 promoter methylation in glioma. Acta Biochim Biophys Sin (Shanghai) 2024;56:174-83. [Crossref] [PubMed]
- Ballesteros-Álvarez J, Andersen JK. mTORC2: The other mTOR in autophagy regulation. Aging Cell 2021;20:e13431. [Crossref] [PubMed]
- Bernard M, Yang B, Migneault F, et al. Autophagy drives fibroblast senescence through MTORC2 regulation. Autophagy 2020;16:2004-16. [Crossref] [PubMed]
- Bernard M, Dieudé M, Yang B, et al. Autophagy fosters myofibroblast differentiation through MTORC2 activation and downstream upregulation of CTGF. Autophagy 2014;10:2193-207. [Crossref] [PubMed]
- Dai N, Christiansen J, Nielsen FC, et al. mTOR complex 2 phosphorylates IMP1 cotranslationally to promote IGF2 production and the proliferation of mouse embryonic fibroblasts. Genes Dev 2013;27:301-12. [Crossref] [PubMed]
- Wen YT, Zhang JR, Kapupara K, et al. mTORC2 activation protects retinal ganglion cells via Akt signaling after autophagy induction in traumatic optic nerve injury. Exp Mol Med 2019;51:1-11. [Crossref] [PubMed]
- Zhang X, Chu J, Sun H, et al. MiR-155 aggravates impaired autophagy of pancreatic acinar cells through targeting Rictor. Acta Biochim Biophys Sin (Shanghai) 2020;52:192-9. [Crossref] [PubMed]
- Liu Z, Huang Y, Jin X, et al. PCB153 suppressed autophagy via PI3K/Akt/mTOR and RICTOR/Akt/mTOR signaling by the upregulation of microRNA-155 in rat primary chondrocytes. Toxicol Appl Pharmacol 2022;449:116135. [Crossref] [PubMed]
- Vehlow A, Cordes N. DDR1 (discoidin domain receptor tyrosine kinase 1) drives glioblastoma therapy resistance by modulating autophagy. Autophagy 2019;15:1487-8. [Crossref] [PubMed]
- Zhao D, Jiang M, Zhang X, et al. The role of RICTOR amplification in targeted therapy and drug resistance. Mol Med 2020;26:20. [Crossref] [PubMed]
- Chun Y, Kim J. Autophagy: An Essential Degradation Program for Cellular Homeostasis and Life. Cells 2018;7:278. [Crossref] [PubMed]
- Kondo Y, Kanzawa T, Sawaya R, et al. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer 2005;5:726-34. [Crossref] [PubMed]
- Wan G, Xie W, Liu Z, et al. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy 2014;10:70-9. [Crossref] [PubMed]
- Wang F, Shan S, Huo Y, et al. MiR-155-5p inhibits PDK1 and promotes autophagy via the mTOR pathway in cervical cancer. Int J Biochem Cell Biol 2018;99:91-9. [Crossref] [PubMed]
- Liu X, Zhao P, Wang X, et al. Celastrol mediates autophagy and apoptosis via the ROS/JNK and Akt/mTOR signaling pathways in glioma cells. J Exp Clin Cancer Res 2019;38:184. [Crossref] [PubMed]
- Luo M, Liu Q, He M, et al. Gartanin induces cell cycle arrest and autophagy and suppresses migration involving PI3K/Akt/mTOR and MAPK signalling pathway in human glioma cells. J Cell Mol Med 2017;21:46-57. [Crossref] [PubMed]
- Xu Y, Sun Q, Yuan F, et al. RND2 attenuates apoptosis and autophagy in glioblastoma cells by targeting the p38 MAPK signalling pathway. J Exp Clin Cancer Res 2020;39:174. [Crossref] [PubMed]
- Zhong C, Shu M, Ye J, et al. Oncogenic Ras is downregulated by ARHI and induces autophagy by Ras/AKT/mTOR pathway in glioblastoma. BMC Cancer 2019;19:441. [Crossref] [PubMed]
- Baba O, Hasegawa S, Nagai H, et al. MicroRNA-155-5p is associated with oral squamous cell carcinoma metastasis and poor prognosis. J Oral Pathol Med 2016;45:248-55. [Crossref] [PubMed]
- Lu M, Zhu WW, Wang X, et al. ACOT12-Dependent Alteration of Acetyl-CoA Drives Hepatocellular Carcinoma Metastasis by Epigenetic Induction of Epithelial-Mesenchymal Transition. Cell Metab 2019;29:886-900.e5. [Crossref] [PubMed]
- Wang D, Wang X, Song Y, et al. Exosomal miR-146a-5p and miR-155-5p promote CXCL12/CXCR7-induced metastasis of colorectal cancer by crosstalk with cancer-associated fibroblasts. Cell Death Dis 2022;13:380. [Crossref] [PubMed]
- Li X, Wang S, Mu W, et al. Reactive oxygen species reprogram macrophages to suppress antitumor immune response through the exosomal miR-155-5p/PD-L1 pathway. J Exp Clin Cancer Res 2022;41:41. [Crossref] [PubMed]


