
LncRNA AF117829.1 is correlated with prognosis and immune infiltration and facilitates tumor progression by targeting OR7C1 in colorectal cancer
Highlight box
Key findings
• Long non-coding RNA (lncRNA) gene AF117829.1 was significantly associated with the prognosis, immunological characteristics and immunotherapy response of colorectal cancer (CRC) patients and promoted malignant progression of CRC by targeting OR7C1.
What is known and what is new?
• Previous studies reported that immune-related lncRNA gene AF117829.1 could participate in the construction of clinical predictive signature in CRC patients. The biological functions and mechanism of AF117829.1 in CRC remain to be explored.
• In our study, the role of AF117829.1 in clinical implications, immune landscape and tumor progression of CRC was first explored in oncology research.
What is the implication, and what should change now?
• LncRNA AF117829.1 can be potentially used as a therapeutic target for CRC patients.
Introduction
According to global cancer statistics in 2022, colorectal cancer (CRC) is one of the most prevalent cancers, ranking third in incidence and second in mortality of all cancers worldwide (1,2). Due to the lack of noticeable clinical symptoms in the early stages, most of CRC patients already have advanced or metastatic lesions at initial diagnosis (3). Although the treatment techniques of CRC, such as the promising immunotherapy, have been continuously improved in recent years, the prognosis of advanced patients is still poor, mainly due to recurrence, metastasis and drug resistance (4). Therefore, more investigations are urgently needed to provide potential targets for treatment of advanced CRC patients.
Long non-coding RNAs (lncRNAs), defined as RNA transcripts longer than 200 nucleotides, have no or limited protein-coding capacity (5). LncRNAs are reported to exert a vital part in plentiful biological processes, including the tumorigenesis, tumor invasion and tumor metastasis (6). Studies have shown that lncRNAs can also impact cancer’s malignant progression by regulating the tumor immune microenvironment (TIME), such as the immune cell infiltration (ICI) and the expression of immune checkpoints (ICP) genes (7,8). The TIME of CRC is a complex system that includes various types of immune cells such as macrophages, neutrophils, dendritic cells, natural killer cells, and lymphocytes, as well as signaling molecules and immune regulatory factors (9). There is mounting evidence that the TIME takes important roles in determining tumor progression and immunotherapeutic strategies in CRC (10). For example, molecular subtyping of CRC was conducted based on expression characteristics and TIME, and the consensus molecular subtype (CMS) was released publicly, which divided CRC into four subtypes with different prognoses and treatment responses (11). Based on single-cell profiling of CRC, Khaliq et al. (12) proposed that patients having higher cancer-associated fibroblasts (CAFs) and complement 1 (C1) Q+ tumor-associated macrophages (TAMs) enrichments across the different CMS groups exhibited relatively poorer outcomes. Chen et al. (13) revealed that higher abundance of tumor-reactive-like CD8+ T (Ttr-like) cells suggested better efficacy of immune checkpoint blockade (ICB) in CRC. Henrich et al. (9) found an interplay between CAFs and CRC cells, influencing growth, invasiveness, angiogenesis, and immunogenicity of CRC. Using single-cell analyses, Becker et al. (14) found that advanced polyps contained increasing numbers of stem-like cells, regulatory T cells, while the cancerous state was associated with T cell exhaustion. Liu et al. (15) found that compared to left-side colon cancers, the relative abundance of myeloid cells and T lymphocytes increased in right-side colon cancers, while the relative abundance of B lymphocytes and plasma cells decreased, and they also proposed that ICB might be more active against right-side colon cancers. Recently, two independent bioinformatics studies reported that immune-related lncRNA gene AF117829.1 could participate in the construction of clinical predictive signature for CRC patients (16,17). In addition, Li et al. (18) found that lncRNA AF117829.1 could regulate CD8+ T lymphocyte function in severe aplastic anaemia patients by promoting receptor-interacting serine/threonine-protein kinase 2 (RIPK2) expression. We speculated that AF117829.1 might be involved in regulating the immune landscape and progression of CRC. However, the biological functions and mechanism of AF117829.1 in CRC progression still remain unexplored.
Here, using the transcriptome data from The Cancer Genome Atlas (TCGA) database, we systematically investigated the expression patterns of AF117829.1 in pan-cancer and its associations with clinical characteristics and prognosis of CRC patients. Furthermore, the correlation of AF117829.1 expression with TIME in CRC was analyzed by bioinformatics tools and algorithms. Additionally, by utilizing Gene Set Enrichment Analysis (GSEA), CRC tissues and in vitro experiments, we confirmed that AF117829.1 promoted the malignant progression of CRC by targeting olfactory receptor family 7 subfamily C member 1 (OR7C1). We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-378/rc).
Methods
TCGA dataset
The messenger RNA (mRNA) expression data and clinical data of TCGA pan-cancer dataset including 11,057 samples and 60,484 genes were obtained from University of California Santa Cruz (UCSC) Xena Data Browser (https://xenabrowser.net/datapages/). RNA-sequencing (RNA-seq) data were then taken as log2 values. The latest TCGA colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) dataset with RNA-seq data and clinical data were obtained from Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov/). Then, TCGA COAD and READ dataset were combined into a TCGA-CRC dataset for the next analysis. After deleting cases with missing values, we obtained the transcriptome data of 618 CRC samples and 51 normal tissues. The RNA-seq data of AF117829.1 were converted to transcript per million (TPM) values and then transformed to log2 values.
Survival analysis
Patients were divided into high- and low-expression groups based on the median values of AF117829.1 expression. The Kaplan-Meier (KM) curve constructed between the two groups and the survival R package were applied to analyze the impact of AF117829.1 expression on the overall survival (OS), disease-specific survival (DSS) and disease-free interval (DFI) of patients. Furthermore, the optimal cut-off value for AF117829.1 expression stratification (low and high) was generated using the survminer R package. Differences between curves were examined using the log-rank test and a value of P<0.05 was considered significant.
Immunogenicity analysis
We analyzed some factors related to immunogenicity, such as tumor mutation burden (TMB) and microsatellite instability (MSI). TMB was calculated by VarScan2 from TCGA somatic mutation data and MSI was obtained from a previous study (19). The correlations between AF117829.1 expression and TMB and MSI were evaluated by Spearman correlation analysis.
Tumor microenvironment analysis
Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) is an algorithm for estimating stromal fractions and immune fractions in tumor tissues (20). Using the ESTIMATE R package, we calculated the values of overall stroma level (StromalScore), immunocyte infiltration (ImmuneScore), and combination (ESTIMATEScore) in each sample from the TCGA-CRC dataset. Based on the 1,000 permutations of 22-leukocyte signature matrix (LM22), the Cell-type Identification By Estimating Relative Subsets Of known RNA Transcripts (CIBERSORT) algorithm was used to calculate the infiltration levels of 22 different types of immune cells in the CRC TIME (21). The correlation between AF117829.1 expression and ICI was evaluated by Spearman correlation analysis. In addition, we obtained 47 ICP genes from previous studies and compared their correlation with AF117829.1 expression by Pearson correlation analysis (22,23). The distribution and expression of AF117829.1 in CRC tissues was analyzed by the single-cell RNA sequencing (scRNA-seq), which was obtained from Tumor Immune Single-cell Hub (TISCH) database (24).
Immunotherapy response prediction
The immunophenoscore (IPS) of CRC patients in TCGA was acquired from The Cancer Immunome Atlas (TCIA) database (https://tcia.at/). TCIA is a database based on TCGA and provides comprehensive immunogenomic analysis. IPS primarily includes four components (effector cells, immunosuppressive cells, major histocompatibility complex molecules, and immune modulators) that determine tumor immunogenicity (25). The IPS is calculated on a 0–10 scale, and higher IPS is associated with increased immunogenicity. Consequently, a higher IPS for a patient indicates that the patient can benefit from immunotherapy (25).
GSEA
After separating CRC patients into high- and low-AF117829.1 expression groups, we explored the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of AF117829.1. GSEA was performed using the clusterProfiler R package. The predefined gene sets “c5.go.v7.4.symbols.gmt” and “c2.cp.kegg.v7.4.symbols.gmt” were downloaded from the Molecular Signatures Database (MSigDB) (26).
Acquisition of clinical samples
All samples (including ten pairs of CRC and the corresponding adjacent normal tissues) were obtained from patients who were pathologically diagnosed with CRC at the First Affiliated Hospital of Nanjing Medical University. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (No. 2023-SR-206), and informed consent was obtained from all individual participants.
Cell culture and transfection
We purchased the human CRC cell lines HCT116, SW480, SW620, and DLD-1 and the human normal intestinal epithelial cell line FHC from the Chinese Academy of Sciences (Shanghai, China). All cells were maintained in medium supplemented with 10% fetal bovine serum (FBS, Gibco, New York, USA) and were incubated at 37 ℃ in a humidified atmosphere of 5% CO2. The small interfering RNAs (siRNAs) specifically targeting AF117829.1 (si-AF117829.1) and OR7C1 (si-OR7C1) and the overexpression plasmid pcDNA3.1-AF117829.1 (oe-AF117829.1) were purchased from GeneChem (Shanghai, China). GeneChem Company also synthesized the negative controls (NCs) including non-specific siRNA and empty vector. Lipofectamine 3000 (Invitrogen, Carlsbad, USA) was used for the transfection of cells according to the manufacturer’s instructions. The siRNA sequences are listed in Table 1.
Table 1
| siRNA | Sense (5'–3') | Antisense (5'–3') |
|---|---|---|
| si-AF117829.1 | 5'-CAGCAGACUUGUAGACCAAUG-3' | 5'-UUGGUCUACAAGUCUGCUGUG-3' |
| si-OR7C1 | 5'-CAGUGGUCACCUUGUUCUAUG-3' | 5'-UAGAACAAGGUGACCACUGAG-3' |
| si-NC | 5'-UUCUCCGAACGUGUCACGUTT-3' | 5'-ACGUGACACGUUCGGAGAATT-3' |
siRNA or si, small interfering RNA; OR7C1, olfactory receptor family 7 subfamily C member 1; NC, negative control.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Total RNA was extracted and reverse-transcribed into complementary DNA (cDNA). Then qRT-PCR was performed using primers for amplification of AF117829.1 and OR7C1. The primers sequences used are listed in Table 2. Relative expression was normalized to β-actin and quantified using the 2−ΔΔCt method.
Table 2
| Gene name of primer | Forward primer | Reverse primer |
|---|---|---|
| AF117829.1 | 5'-TGTCGGATTGGTTAGCGACC-3' | 5'-CTTAAGTGTGGAGCCCTCGG-3' |
| OR7C1 | 5'-AGCTCTGTGGACTGCTGGTT-3' | 5'-GGACGCCAGTTGCAAAGTAT-3' |
| β-actin | 5'-GTGGACATCCGCAAAGAC-3' | 5'-AAAGGGTGTAACGCAACTA-3' |
qRT-PCR, quantitative real-time polymerase chain reaction; OR7C1, olfactory receptor family 7 subfamily C member 1.
Cell proliferation assays
Cell proliferation was evaluated after 24, 48, 72 and 96 h using a cell counting kit-8 (CCK-8) assay (Beyotime, Shanghai, China). Absorbance was measured at 450 nm by a microplate reader. In addition, a cell proliferation assay was also performed with the colony formation assay according to the manufacturer’s protocols. The detailed methods were described in our previous study (27).
Wound healing assays
Cells (3×105 cells/well) were seeded into six-well plates and cultured for 48 h till 90% confluent. Each well was scratched longitudinally with a 10-µL pipette tip. Then, the cells were washed and cultured in an incubator at 37 ℃, and the wound region was photographed at 0 and 24 h. The extent of wound healing was measured by ImageJ.
Transwell assays
Transwell assays were performed to detect cell migration and invasion. 24-well Millicell hanging cell culture inserts (8.0 µm, Millipore, Bedford, MA, USA) were used according to manufacturer’s protocols. To perform the migration assays, 4×104 cells (per well) were seeded onto the upper chambers in 200 µL serum-free medium. For the invasion assays, the upper chambers were coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA), and then 8×104 cells (per well) in serum-free medium were added to the upper chamber. Next, 500 µL complete medium containing 10% FBS in the bottom chamber served as a chemoattractant. After 24 h of incubation, cells attached to the bottom of the membrane were fixed and stained, followed by visualization using a light microscopy.
Immunohistochemistry (IHC)
IHC was used to detect the expression of OR7C1. The embedded CRC tissues were cut into 4-µm-thick slides. Next, dewaxing, hydration, antigen retrieval and endogenous peroxidase blocking were conducted. After being blocked with normal serum for 60 min, the tissues were incubated with rabbit polyclonal antibody for OR7C1 (HPA047127; Sigma-Aldrich, St. Louis, MO, USA) overnight, followed by the horseradish peroxidase (HRP)-conjugated secondary antibody (Maixin, Fuzhou, China) for 60 min. Then, the sections were stained with diaminobenzidine (DAB) kit (BL732A, Biosharp, Hefei, China) and hematoxylin. Finally, the sections were observed under a light microscope. The staining intensity was assessed as follows: absent =0, weak =1, moderate =2 and strong =3. The percentage of positive cells was graded as follows: 0 (negative), 1 (<25%), 2 (25–50%), 3 (>50–75%) and 4 (>75%). The staining score for each tissue was multiplied by the two scores and therefore calculated on a 0–12 scale.
Statistical analysis
All experiments were repeated at least three times. Data were presented as mean ± standard deviation (SD) and analyzed using R software (version 4.3.1) and GraphPad Prism 8.0 (GraphPad Software, CA, USA). To compare the differences between two groups or multiple groups, Student’s t-test or one-way analysis of variance (ANOVA) was conducted for normally distributed data. The Wilcoxon test was conducted for the data that were not normally distributed, and the Chi-squared test was used for categorical variables. P<0.05 was considered statistically significant.
Results
AF117829.1 overexpression was identified in pan-cancer and positively associated with high tumor-node-metastasis (TNM) stage and poor prognosis in CRC
To explore the possible role of AF117829.1 in carcinogenesis, we first analyzed the expression of the AF117829.1 gene in 33 human cancers using the TCGA pan-cancer dataset (Figure 1A). The results showed that AF117829.1 was remarkably upregulated in the tissues of COAD (P<0.001) and READ (P=0.004) compared with the respective normal samples. In addition, AF117829.1 was also found to be significantly upregulated in the majority of tumors compared with the respective control tissues, including bladder urothelial carcinoma (BLCA) (P<0.001), cholangiocarcinoma (CHOL) (P<0.001), glioblastoma multiforme (GBM) (P=0.002), head and neck squamous cell carcinoma (HNSC) (P<0.001), kidney renal clear cell carcinoma (KIRC) (P<0.001), kidney renal papillary cell carcinoma (KIRP) (P=0.001), liver hepatocellular carcinoma (LIHC) (P<0.001), lung adenocarcinoma (LUAD) (P<0.001), prostate adenocarcinoma (PRAD) (P<0.001), stomach adenocarcinoma (STAD) (P<0.001) and uterine corpus endometrial carcinoma (UCEC) (P=0.02). In contrast, AF117829.1 was significantly downregulated in the tissues of thyroid carcinoma (THCA) when compared to the normal samples (P=0.002).
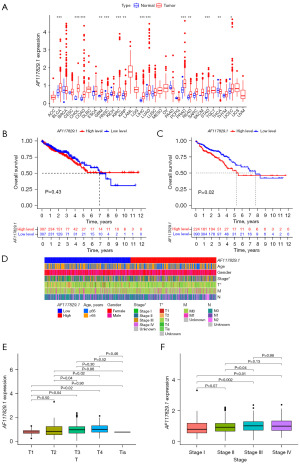
When three cases with insufficient clinical data were deleted from the TCGA-CRC dataset, we displayed the clinical characteristics of 615 CRC patients in Table 3. Next, the prognostic value of AF117829.1 on OS of CRC patients was estimated with KM plotter. When the cut-off was the median of AF117829.1 expression, there was no significant difference in OS between the high- and low-AF117829.1 expression groups (P=0.43, Figure 1B). Meanwhile, we divided the CRC samples into high- and low-AF117829.1 expression groups according to the optimal cut-off value of AF117829.1 expression [log2 (TPM+1) =1.10], which was calculated using the survminer package. Interestingly, the KM survival analysis showed that CRC patients with higher AF117829.1 expression had remarkably shorter OS time than those with lower AF117829.1 expression (P=0.02, Figure 1C). As shown in Figures S1-S3, the prognostic value of AF117829.1 on OS, DSS and DFI of pan-cancer was also evaluated using KM plotter tool, respectively. Subsequently, we compared the relationship between AF117829.1 expression and the clinical characteristics of CRC patients. The results indicated that the expression of AF117829.1 tended to increase along with the progression of tumor (T) stage (T3 versus T1, P=0.04; T4 versus T1, P=0.02; T3 versus T2, P=0.04; T4 versus T2, P=0.02) and TNM stage (stage III versus stage I, P=0.002; stage IV versus stage I, P=0.01; stage III versus stage II, P=0.04) in CRC (Figure 1D-1F). Then, the relationship between AF117829.1 expression and TNM stage of pan-cancer was also investigated (Figure S4). The above findings clarified that dysregulated AF117829.1 expression might play a crucial role in the progression of CRC.
Table 3
| Characteristic | Number of patients | Percentage (%) |
|---|---|---|
| Age (years) | ||
| ≤65 | 264 | 42.9 |
| >65 | 351 | 57.1 |
| Gender | ||
| Female | 288 | 46.8 |
| Male | 327 | 53.2 |
| Overall survival information | ||
| Known | 614 | 99.8 |
| Unknown | 1 | 0.2 |
| Stage | ||
| I | 103 | 16.7 |
| II | 228 | 37.1 |
| III | 177 | 28.8 |
| IV | 87 | 14.1 |
| Unknown | 20 | 3.3 |
| T classification | ||
| T1 | 19 | 3.1 |
| T2 | 104 | 16.9 |
| T3 | 421 | 68.4 |
| T4 | 69 | 11.2 |
| Tis | 1 | 0.2 |
| Unknown | 1 | 0.2 |
| N classification | ||
| N0 | 349 | 56.7 |
| N1 | 147 | 23.9 |
| N2 | 116 | 18.9 |
| Unknown | 3 | 0.5 |
| M classification | ||
| M0 | 456 | 74.1 |
| M1 | 86 | 14.0 |
| Unknown | 73 | 11.9 |
TCGA, The Cancer Genome Atlas; CRC, colorectal cancer; T, tumor; N, node; M, metastasis.
Correlation analysis between AF117829.1 expression and TMB, MSI and ICP genes expression in pan-cancer
Researchers Sun et al. (17) and Ma et al. (16) obtained the immune-related lncRNA gene AF117829.1 by the Immport database (https://www.immport.org) and the co-expression analysis. Additionally, Li et al. (18) reported that AF117829.1 could regulate lymphocyte function. TMB and MSI in cancers have been proved to be closely associated with antitumor immunity and can predict the therapeutic efficacy of immunotherapy (28,29). To investigate the role of AF117829.1 in the immunogenicity of tumor microenvironment, we conducted the Spearman correlation analysis between AF117829.1 expression and TMB (Figure 2A) and MSI (Figure 2B) across all cancer types from TCGA. Here, we observed that AF117829.1 expression was remarkably correlated with TMB and MSI in many types of cancers (all P<0.05). For example, AF117829.1 expression was significantly and negatively correlated with MSI in COAD (P=0.02).
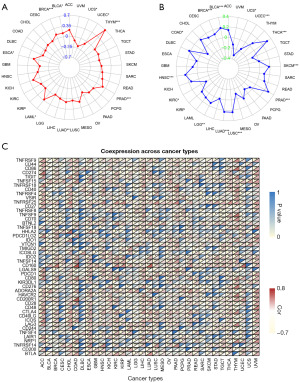
Studies have proven that ICP genes can modulate the signaling pathways in the regulation of immune response and play a vital part in the area of anti-cancer immunotherapy (30,31). Consequently, the correlation between the expression of AF117829.1 and 47 ICP genes which were obtained from the research of Topalian et al. (22) was explored in pan-cancer in our study. According to the results shown in Figure 2C, Pearson correlation analysis revealed that AF117829.1 expression was remarkably correlated with more than 30 ICP genes expression in eleven types of cancers, including COAD (all P<0.05). From the above results, we speculated that AF117829.1 might affect tumor immunity by regulating the immune composition and process in the TIME.
AF117829.1 expression was significantly correlated with the TIME of CRC and predicted immunotherapy response in CRC
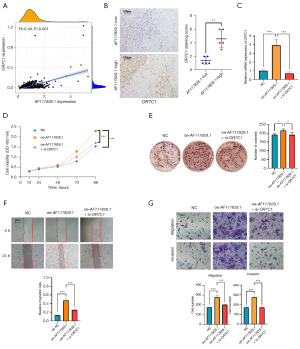
Then, to clarify the relationship between AF117829.1 expression and the TIME of CRC, we obtained the StromalScore, ImmuneScore and ESTIMATEScore of the two groups with high- and low-AF117829.1 expression by the ESTIMATE algorithm. We found that the high-AF117829.1 expression group had a significantly higher StromalScore compared with the low-AF117829.1 expression group in CRC (P=0.008, Figure 3A). To investigate the relationship of AF117829.1 expression with the ICI levels in the TIME of CRC, the infiltration of various tumor infiltrating immune cells was identified by the CIBERSORT algorithm. Our results showed that there was a significant difference in the infiltration of 6 out of 22 immune cells between the high- and low-AF117829.1 expression groups (all P<0.05, Figure 3B). Meanwhile, the correlation analysis was performed and revealed that the infiltration levels of naive B cells (R=0.13, P=0.002), M0 macrophages (R=0.12, P=0.007), resting mast cells (R=0.11, P=0.01) and T follicular helper cells (R=0.098, P=0.02) were significantly and positively correlated with AF117829.1 expression, while the infiltration levels of CD8 T cells (R=−0.13, P=0.003), plasma cells (R=−0.16, P<0.001) and T regulatory cells (R=−0.19, P<0.001) were significantly and negatively correlated with AF117829.1 expression (Figure 3C,3D). Additionally, the correlation between the expression of AF117829.1 and ICP genes was also explored in CRC. As shown in Figure 3E, the expression of ICP genes including NRP1, CD160, TNFSF4, CD80, ADORA2A, CD200R1, CD28, TNFSF15 and BTLA was significantly and positively correlated with AF117829.1 expression, while the expression of TNFRSF14 and LGALS9 was significantly and negatively correlated with AF117829.1 expression (all |R| ≥0.3, all P<0.001). We also studied the expression of AF117829.1 at the single-cell level. In database CRC_GSE166555, single-cell cluster map described the expression profiles of AF117829.1 in the single cells obtained from CRC tissues (Figure 3F). Violin plot showed the AF117829.1 expression was the most abundant in the CRC malignant cells, and then in the stromal cells and immune cells (Figure 3G). The above results indicated that AF117829.1 might be involved in the immune response in the TIME of CRC by affecting ICI levels and ICP genes expression.
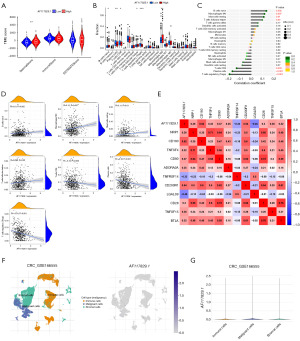
Considering the noticeable association between AF117829.1 expression and TIME in CRC, we speculated that AF117829.1 expression could predict immunotherapy response in CRC. Here we obtained the IPS scores of TCGA-CRC dataset using TCIA database and divided all the CRC patients into four subgroups depending on the usage of anti-programmed cell death protein-1 (PD-1) and anti-cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4) immunotherapies: CTLA-4-negative, PD-1 negative (Figure 4A); CTLA-4-negative, PD-1-positive (Figure 4B); CTLA-4-positive, PD-1-negative (Figure 4C); and CTLA-4-positive, PD-1-positive (Figure 4D). Interestingly, our results showed that in all of the four subgroups, the IPS scores of low-AF117829.1 group were higher than those of high-AF117829.1 group (all P<0.001), suggesting that CRC patients with low-AF117829.1 expression could benefit more from the immunotherapy based on anti-CTLA-4 and anti-PD-1.

AF117829.1 overexpression promoted the proliferation, migration and invasion of CRC cells in vitro
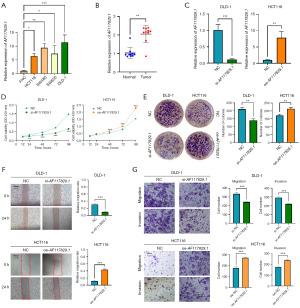
AF117829.1 expression was verified to be significantly higher in CRC cell lines (HCT116, SW480, SW620 and DLD-1) than in the normal intestinal epithelial cell line FHC using qRT-PCR (all P<0.05, Figure 5A). Consistently, the results of qRT-PCR showed that AF117829.1 expression was significantly increased in ten cases of CRC tissues when compared with the paired adjacent normal tissues (P=0.002, Figure 5B). To determine the potential function of AF117829.1 in CRC, the expression of AF117829.1 in CRC cell lines was downregulated by transfecting siRNA into DLD-1 cells or upregulated by transfecting overexpression plasmid into HCT116 cells, and the transfection efficiencies were examined by qRT-PCR (all P<0.01, Figure 5C). CCK-8 assays (Figure 5D) and colony formation assays (Figure 5E) revealed that AF117829.1 knockdown suppressed the proliferation of DLD-1 cells, whereas AF117829.1 overexpression promoted the proliferation of HCT116 cells (all P<0.05). Additionally, wound healing assays (Figure 5F) and transwell assays (Figure 5G) showed that AF117829.1 knockdown reduced the migration and invasion of DLD-1 cells, and AF117829.1 overexpression increased the migration and invasion of HCT116 cells (all P<0.001). Consequently, the results of our in vitro experiments revealed that AF117829.1 functioned as an oncogenic lncRNA in CRC.
AF117829.1 functioned as an oncogenic lncRNA by targeting OR7C1 in CRC
To further study the mechanism by which AF117829.1 mediated tumor biological behavior, we conducted the GSEA in CRC. From GO gene sets analysis, we found that among the top five signaling pathways, the detection of chemical stimulus, sensory perception of smell, odorant binding and olfactory receptor activity were significantly enriched in the AF117829.1 high expression group, whereas the cristae formation was significantly enriched in the AF117829.1 low expression group (Figure 6A). From KEGG gene sets analysis, the data suggested that among the top five signaling pathways, the olfactory transduction was significantly enriched in the AF117829.1 high expression group, whereas the aminoacyl-tRNA biosynthesis, citrate cycle/tricarboxylic acid cycle (TCA cycle), DNA replication and selenoamino acid metabolism were significantly enriched in the AF117829.1 low expression group (Figure 6B). The results of both GO-related and KEGG-related GSEA showed that olfactory transduction-related signaling pathways were enriched in the AF117829.1 high expression group and might be targeted by AF117829.1.

Morita et al. (32) reported that olfactory receptor family 7 subfamily C member 1, also called OR7C1, was a novel marker for CRC cancer-initiating cells (CICs) and could be a target of potent CIC-targeting immunotherapy. Considering our results of GSEA, we used the data from the TCGA-CRC dataset to analyze the correlation between AF117829.1 gene and OR7C1 gene. We observed that AF117829.1 expression was significantly and positively correlated with OR7C1 expression (R=0.49, P<0.001, Figure 7A). IHC of our ten CRC samples revealed that OR7C1 expression was significantly higher in the AF117829.1 high expression group than that in the AF117829.1 low expression group (P=0.001, Figure 7B). The results of qRT-PCR showed that OR7C1 mRNA levels in HCT116 cells were upregulated by transfecting AF117829.1 overexpression plasmid, whereas co-transfection with AF117829.1 overexpression plasmid and siRNA targeting OR7C1 reduced the upregulated expression of OR7C1 triggered by the overexpression of AF117829.1 (all P<0.001, Figure 7C). Furthermore, to validate whether AF117829.1 could regulate CRC cell proliferation, migration and invasion by upregulating OR7C1 expression, rescue assays were performed. CCK-8 assays and colony formation assays indicated that co-transfection counteracted the increased proliferation abilities resulted from AF117829.1 overexpression (all P<0.05, Figure 7D,7E). In addition, wound healing assays and transwell assays showed that the decreased OR7C1 expression counteracted the increased migration and invasion abilities induced by AF117829.1 overexpression (all P<0.001, Figure 7F,7G). These results indicated that the effect of AF117829.1 on CRC partially involved targeting OR7C1.
Discussion
CRC is an aggressive malignant tumor of the digestive system and is the second leading cause of cancer-related death worldwide (2). Despite progress in the current CRC therapies including surgery, chemotherapy, radiotherapy and immunotherapy, the prognosis for CRC patients remains poor. Therefore, identifying the key functional genes and exploring the underlying molecular mechanisms in regulating CRC occurrence and development are crucial to effectively treat CRC. Currently, accumulating evidence has proved that lncRNAs can participate in the tumorigenesis of cancers through multiple signaling pathways, especially the cross talk among cancer cells and tumor microenvironment (33,34). Liang et al. (35) reported that lncRNA RPPH1 promoted CRC cells metastasis, enhanced exosomes-mediated macrophages M2 polarization and changed the tumor microenvironment. Chen et al. (36) proved that extracellular vesicle-packaged lncRNA HISLA from TAMs regulated aerobic glycolysis of breast cancer cells. However, the biological function of lncRNA is not fully understood, and the clinical application of lncRNA is hindered by issues such as specificity, delivery methods, and immunogenicity (37). Therefore, it is necessary to conduct more extensive and in-depth research and clinical validation on the role and mechanism of lncRNA in cancer progression. In our study, we explored the expression of AF117829.1 in pan-cancer including CRC and its correlations with clinical pathological characteristics and prognosis using TCGA RNA-seq data. Then, the correlations between AF117829.1 expression and the TIME of CRC, including TMB, MSI, StromalScore, ImmuneScore, ESTIMATEScore, ICI levels, ICP genes expression and IPS, were clarified. Subsequently, in vitro experiments were conducted to investigate the effects of AF117829.1 expression on the proliferation, migration and invasion of CRC cells. Finally, GSEA and rescue assays were performed to explore the potential signaling pathways and target genes by which AF117829.1 regulated CRC progression.
AF117829.1 is a lncRNA gene located on chromosome 8: 89,545,424–89,757,812 reverse strand and its Ensembl ID is ENSG00000251136 (38). Its transcript ENST00000504145 (1,601bp), also called RIPK2-DT, LOC101929709 or palmitic acid regulated anti-inflammatory lncRNA (PARAIL), is divergently transcribed from gene RIPK2 (18,39). Previously, according to the TCGA and ImmPort databases and bioinformatics methods, Ma et al. (16) and Sun et al. (17) independently identified the immune-related lncRNA AF117829.1. They found that it was differentially expressed between CRC tissues and normal tissues, and could participate in constructing the immune-related lncRNA pair signature for prognostic prediction and immunological evaluation in CRC. Our results revealed that the expression of AF117829.1 was upregulated in pan-cancer, and AF117829.1 overexpression was positively associated with high TNM stage and poor prognosis in CRC, suggesting that AF117829.1 possibly played a tumor-promoting role in CRC.
Li et al. (18) found that lncRNA AF117829.1 regulated lymphocyte function in severe aplastic anaemia patients by promoting RIPK2 expression. Tanwar et al. (40) reported that lncRNA PARAIL, or named AF117829.1, regulated inflammation via interaction with RNA-binding protein ELAVL1 in monocytes and macrophages. Xu et al. (39) confirmed that LOC101929709, also named AF117829.1, promoted gastric cancer progression by aiding LIN28B to stabilize c-MYC mRNA. All these findings suggested that AF117829.1 might participate in regulating TIME. In our study, the results of ESTIMATE analysis revealed that the high-AF117829.1 expression group had a higher StromalScore than the low-AF117829.1 expression group in CRC, suggesting that CRC patients with a higher AF117829.1 expression had a higher percentage of tumor stroma. As known, the tumor stroma can promote the resistance of cancer cells to anticancer therapies, eventually leading to fatal diseases (41). Then, the relationships of AF117829.1 expression with ICI levels and ICP genes expression were investigated in CRC. The results showed that in TCGA-CRC dataset, AF117829.1 expression was significantly correlated with the infiltration levels of naive B cells, M0 macrophages, resting mast cells, T follicular helper cells, CD8 T cells, plasma cells and T regulatory cells. Furthermore, we proved that AF117829.1 expression was significantly correlated with multiple ICP genes expression in CRC. Immune response in the tumor microenvironment is essential to the cancer prognosis, and immune cells, such as lymphocytes, macrophages, dendritic cells and so on, play a key role in determining the effectiveness of cancer immunotherapy (42-44). Studies also have proven that ICP molecules have an important influence on immune cells recruitment and immunotherapy response (31,45,46). In order to study the value of AF117829.1 in predicting the immunotherapy response in CRC, we obtained the IPS scores of TCGA-CRC dataset using TCIA database, and found that the IPS scores of low-AF117829.1 expression group were higher than those of high-AF117829.1 expression group in all of the four subgroups. The existing evidence proved that a higher IPS for a cancer patient represented a better immunotherapy response (25), indicating that CRC patients with low-AF117829.1 expression could benefit more from the immunotherapy based on anti-CTLA-4 and anti-PD-1. Besides, it has been proved that CRC patients with high MSI (MSI-H) are more sensitive to immunotherapy (47), which coincides with our correlation analysis result that AF117829.1 expression was negatively correlated with MSI in COAD. Therefore, we proposed that AF117829.1 could serve as a promising immunotherapeutic predictor for CRC.
In our in vitro studies, we also explored the effect of AF117829.1 expression on the biological behavior of CRC cells. First, we found that AF117829.1 was highly expressed in both CRC cell lines and tissues. Furthermore, AF117829.1 overexpression could promote the proliferation, migration and invasion of CRC cells, indicating that AF117829.1 acted as an oncogene in the pathogenesis of CRC. Then, the results of GSEA showed that olfactory transduction-related signaling pathways might be targeted by AF117829.1 in CRC. Previous research showed that olfactory receptors were detected not only in normal tissues, but also in tumor tissues and cells, and olfactory transduction-related signaling pathways could affect carcinogenesis (48). Interestingly, Morita et al. (32) reported that OR7C1 high expression was correlated with higher tumorigenicity and poorer prognosis in CRC. They found that OR7C1-specific cytotoxic T lymphocyte (CTL) clone showed significantly stronger antitumor effect than that of a shared antigen-specific CTL clone, indicating that OR7C1 could be a promising target for CIC-targeting immunotherapy (32). Our rescue assays demonstrated that AF117829.1 overexpression could upregulate OR7C1 expression in CRC cells, and decreased OR7C1 expression reversed the increased proliferation, migration and invasion abilities induced by AF117829.1 overexpression. Consequently, we concluded that AF117829.1 functioned as an oncogenic lncRNA by targeting OR7C1 in CRC.
However, there are certain limitations in this study. Our results were mainly based on the analysis of public databases, and further biological experiments such as in vivo verification are needed to determine the effect of AF117829.1 on CRC. In addition, an absence of commercially available antibodies against OR7C1 used for Western blot analysis, limited the evaluation of OR7C1 protein expression levels in CRC cells and tissues.
Conclusions
Taken all together, our study demonstrated that AF117829.1 overexpression indeed promoted CRC progression and could predict a worse immunotherapy response in CRC patients. Therefore, AF117829.1 may be a potential therapeutic target for CRC patients.
Acknowledgments
The authors sincerely thank all participants in this study.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-378/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-378/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-378/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-378/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University, China (No. 2023-SR-206) and informed consent was obtained from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Saraiva MR, Rosa I, Claro I. Early-onset colorectal cancer: A review of current knowledge. World J Gastroenterol 2023;29:1289-303. [Crossref] [PubMed]
- Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229-63. [Crossref] [PubMed]
- Shin AE, Giancotti FG, Rustgi AK. Metastatic colorectal cancer: mechanisms and emerging therapeutics. Trends Pharmacol Sci 2023;44:222-36. [Crossref] [PubMed]
- Xiang M, Gao Y, Zhou Y, et al. A novel nomogram based on cell cycle-related genes for predicting overall survival in early-onset colorectal cancer. BMC Cancer 2023;23:595. [Crossref] [PubMed]
- Liu ZY, Tang JM, Yang MQ, et al. The role of LncRNA-mediated autophagy in cancer progression. Front Cell Dev Biol 2024;12:1348894. [Crossref] [PubMed]
- Huang P, Wen F, Li Y, et al. The tale of SOX2: Focusing on lncRNA regulation in cancer progression and therapy. Life Sci 2024;344:122576. [Crossref] [PubMed]
- Guo Y, Xie Y, Luo Y. The Role of Long Non-Coding RNAs in the Tumor Immune Microenvironment. Front Immunol 2022;13:851004. [Crossref] [PubMed]
- Zhang M, Wu Y, Mou J, et al. The global landscape of immune-derived lncRNA signature in colorectal cancer. Heliyon 2024;10:e25568. [Crossref] [PubMed]
- Henrich LM, Greimelmaier K, Wessolly M, et al. The Impact of Cancer-Associated Fibroblasts on the Biology and Progression of Colorectal Carcinomas. Genes (Basel) 2024;15:209. [Crossref] [PubMed]
- Kong XX, Xu JS, Hu YT, et al. Circulation immune cell landscape in canonical pathogenesis of colorectal adenocarcinoma by CyTOF analysis. iScience 2024;27:109229. [Crossref] [PubMed]
- Zhai X, Chen B, Hu H, et al. Identification of the molecular subtypes and signatures to predict the prognosis, biological functions, and therapeutic response based on the anoikis-related genes in colorectal cancer. Cancer Med 2024;13:e7315. [Crossref] [PubMed]
- Khaliq AM, Erdogan C, Kurt Z, et al. Refining colorectal cancer classification and clinical stratification through a single-cell atlas. Genome Biol 2022;23:113. [Crossref] [PubMed]
- Chen Y, Wang D, Li Y, et al. Spatiotemporal single-cell analysis decodes cellular dynamics underlying different responses to immunotherapy in colorectal cancer. Cancer Cell 2024;42:1268-1285.e7. [Crossref] [PubMed]
- Becker WR, Nevins SA, Chen DC, et al. Single-cell analyses define a continuum of cell state and composition changes in the malignant transformation of polyps to colorectal cancer. Nat Genet 2022;54:985-95. [Crossref] [PubMed]
- Liu B, Li S, Cheng Y, et al. Distinctive multicellular immunosuppressive hubs confer different intervention strategies for left- and right-sided colon cancers. Cell Rep Med 2024;5:101589. [Crossref] [PubMed]
- Ma B, Cao L, Li Y. A novel 10-gene immune-related lncRNA signature model for the prognosis of colorectal cancer. Math Biosci Eng 2021;18:9743-60. [Crossref] [PubMed]
- Sun M, Zhang T, Wang Y, et al. A Novel Signature Constructed by Immune-Related LncRNA Predicts the Immune Landscape of Colorectal Cancer. Front Genet 2021;12:695130. [Crossref] [PubMed]
- Li Y, Deng L, Pan X, et al. The Role of lncRNA AF117829.1 in the Immunological Pathogenesis of Severe Aplastic Anaemia. Oxid Med Cell Longev 2021;2021:5587921. [Crossref] [PubMed]
- Bonneville R, Krook MA, Kautto EA, et al. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis Oncol 2017;2017:PO.17.00073.
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 2015;27:450-61. [Crossref] [PubMed]
- Cheng L, Zou X, Wang J, et al. The role of CRYAB in tumor prognosis and immune infiltration: A Pan-cancer analysis. Front Surg 2023;9:1117307. [Crossref] [PubMed]
- Sun D, Wang J, Han Y, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res 2021;49:D1420-30. [Crossref] [PubMed]
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248-62. [Crossref] [PubMed]
- Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417-25. [Crossref] [PubMed]
- Shen H, Huang C, Wu J, et al. SCRIB Promotes Proliferation and Metastasis by Targeting Hippo/YAP Signalling in Colorectal Cancer. Front Cell Dev Biol 2021;9:656359. [Crossref] [PubMed]
- Ros J, Baraibar I, Saoudi N, et al. Immunotherapy for Colorectal Cancer with High Microsatellite Instability: The Ongoing Search for Biomarkers. Cancers (Basel) 2023;15:4245. [Crossref] [PubMed]
- Li DD, Tang YL, Wang X. Challenges and exploration for immunotherapies targeting cold colorectal cancer. World J Gastrointest Oncol 2023;15:55-68. [Crossref] [PubMed]
- Sharma P, Goswami S, Raychaudhuri D, et al. Immune checkpoint therapy-current perspectives and future directions. Cell 2023;186:1652-69. [Crossref] [PubMed]
- Choi Y, Seok SH, Yoon HY, et al. Advancing cancer immunotherapy through siRNA-based gene silencing for immune checkpoint blockade. Adv Drug Deliv Rev 2024;209:115306. [Crossref] [PubMed]
- Morita R, Hirohashi Y, Torigoe T, et al. Olfactory Receptor Family 7 Subfamily C Member 1 Is a Novel Marker of Colon Cancer-Initiating Cells and Is a Potent Target of Immunotherapy. Clin Cancer Res 2016;22:3298-309. [Crossref] [PubMed]
- Shakhpazyan NK, Mikhaleva LM, Bedzhanyan AL, et al. Long Non-Coding RNAs in Colorectal Cancer: Navigating the Intersections of Immunity, Intercellular Communication, and Therapeutic Potential. Biomedicines 2023;11:2411. [Crossref] [PubMed]
- Marima R, Basera A, Miya T, et al. Exosomal long non-coding RNAs in cancer: Interplay, modulation, and therapeutic avenues. Noncoding RNA Res 2024;9:887-900. [Crossref] [PubMed]
- Liang ZX, Liu HS, Wang FW, et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis 2019;10:829. [Crossref] [PubMed]
- Chen F, Chen J, Yang L, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol 2019;21:498-510. [Crossref] [PubMed]
- Nappi F. Non-Coding RNA-Targeted Therapy: A State-of-the-Art Review. Int J Mol Sci 2024;25:3630. [Crossref] [PubMed]
- Martin FJ, Amode MR, Aneja A, et al. Ensembl 2023. Nucleic Acids Res 2023;51:D933-41. [Crossref] [PubMed]
- Xu TP, Yu T, Xie MY, et al. LOC101929709 promotes gastric cancer progression by aiding LIN28B to stabilize c-MYC mRNA. Gastric Cancer 2023;26:169-86. [Crossref] [PubMed]
- Tanwar VS, Reddy MA, Das S, et al. Palmitic Acid-Induced Long Noncoding RNA PARAIL Regulates Inflammation via Interaction With RNA-Binding Protein ELAVL1 in Monocytes and Macrophages. Arterioscler Thromb Vasc Biol 2023;43:1157-75. [Crossref] [PubMed]
- Liu L, Xu L, Wu D, et al. Impact of tumour stroma-immune interactions on survival prognosis and response to neoadjuvant chemotherapy in bladder cancer. EBioMedicine 2024;104:105152. [Crossref] [PubMed]
- Laskowski TJ, Biederstädt A, Rezvani K. Natural killer cells in antitumour adoptive cell immunotherapy. Nat Rev Cancer 2022;22:557-75. [Crossref] [PubMed]
- Oliveira G, Wu CJ. Dynamics and specificities of T cells in cancer immunotherapy. Nat Rev Cancer 2023;23:295-316. [Crossref] [PubMed]
- Park J, Hsueh PC, Li Z, et al. Microenvironment-driven metabolic adaptations guiding CD8(+) T cell anti-tumor immunity. Immunity 2023;56:32-42. [Crossref] [PubMed]
- Qu C, Cui H, Xiao S, et al. The landscape of immune checkpoint-related long non-coding RNAs core regulatory circuitry reveals implications for immunoregulation and immunotherapy responses. Commun Biol 2024;7:327. [Crossref] [PubMed]
- Santry LA, van Vloten JP, AuYeung AWK, et al. Recombinant Newcastle disease viruses expressing immunological checkpoint inhibitors induce a pro-inflammatory state and enhance tumor-specific immune responses in two murine models of cancer. Front Microbiol 2024;15:1325558. [Crossref] [PubMed]
- Fu X, Huang J, Zhu J, et al. Prognosis and immunotherapy efficacy in dMMR&MSS colorectal cancer patients and an MSI status predicting model. Int J Cancer 2024;155:766-75. [Crossref] [PubMed]
- Lee HJ, Ku CR, Cho A, et al. Acetate-Mediated Odorant Receptor OR51E2 Activation Results in Calcitonin Secretion in Parafollicular C-Cells: A Novel Diagnostic Target of Human Medullary Thyroid Cancer. Biomedicines 2023;11:1688. [Crossref] [PubMed]


