
Identifying functional cuproptosis-related long non-coding RNAs in patients with bladder cancer
Highlight box
Key findings
• The study used Pearson correlation analysis, univariate Cox regression, and least absolute shrinkage and selection operator Cox regression to analyze data from The Cancer Genome Atlas, identifying 19 cuproptosis-related genes, 14 prognostic long non-coding RNAs (lncRNAs), and constructing a seven-prognostic risk signature with good predictive power for survival rates, which identified high-risk patients with worse prognosis.
What is known and what is new?
• Cuproptosis is the process where copper binds to lipases in the tricarboxylic acid cycle, causing protein aggregation, proteotoxic stress, and cell death.
• LncRNAs are a class of non-coding transcripts over 200 nucleotides in length. lncRNAs are involved in tumor occurrence and progression.
• Creating a seven-risk signature with good predictive power for 1-, 3-, and 5-year survival rates. The high-risk group had a worse prognosis and was marked by high chemokines, T cell inhibition, and low antigen-presenting cells. The risk signature is an independent prognostic factor, with related genes primarily involved in tumorigenesis and migration.
What is the implication, and what should change now?
• Cuproptosis-related lncRNAs are central to tumorigenesis, providing a novel therapeutic target for patients with bladder cancer.
Introduction
Bladder cancer is one of the predominant urinary system tumors, with approximately 550,000 new cases and 200,000 deaths each year (1,2). Approximately 25% of the patients are initially diagnosed with muscle-invasive bladder cancer (MIBC) or metastatic bladder cancer (3). Transurethral resection of bladder tumor is the mainstay therapy for both MIBC and non-MIBC (NMIBC) (4).
Immunotherapies, including programmed cell death protein 1 (PD-1) and programmed death-ligand 1 (PD-L1) inhibitors, appear promising and have been approved for patients with metastatic urothelial carcinoma progressing after platinum-containing chemotherapy (5,6). However, immunotherapy remains ineffective for numerous patients (7). This necessitates understanding the heterogeneity of bladder cancer and establishing a marker to predict treatment response.
Copper is an essential mineral nutrient and acts as a signaling agent regulating or triggering several biological pathways in response to external stimuli (8). Copper-mediated programmed cell death, termed cuproptosis, promotes angiogenesis, tumor progression, and metastasis (9). Cuproptosis refers to copper binding to lipases in the tricarboxylic acid cycle. Their combination results in protein aggregation, proteotoxic stress, and cell death (10).
Long non-coding RNAs (lncRNAs) are a class of non-coding transcripts over 200 nucleotides in length. LncRNAs are involved in tumor occurrence and progression (11). lncRNAs, such as H19, UCA1, and MALAT1, regulate biological processes (12). However, cuproptosis-related lncRNAs have limited evidence, particularly in bladder cancer. Thus, we aimed to evaluate the prognostic implication of cuproptosis-related genes in bladder cancer and assess their correlation with tumorigenesis. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2367/rc).
Methods
Data preparation
We downloaded the transcriptome analysis dataset, clinical data, and single nucleotide variations of patients with bladder cancer from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). This dataset consists of 414 bladder cancer samples and 19 healthy samples. Incomplete clinicopathological characteristics were excluded. Data were analyzed using R (version 4.2) and the R Bioconductor software package. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identifying prognostic cuproptosis-related lncRNAs
The following 19 genes associated with copper death were screened: nuclear factor erythroid 2-related factor 2, nucleotide-binding domain, leucine-rich-containing family, pyrin domain-containing-3 (NLRP3), ATPase copper transporting beta, ATPase copper transporting alpha (ATP7A), solute carrier family 31 member 1, ferredoxin 1, lipoic acid synthase, lipoyl transferase 1, lipoyl transferase 2, dihydrolipoamide dehydrogenase, dihydrolipoamide S-acetyltransferase, pyruvate dehydrogenase E1 subunit alpha 1, pyruvate dehydrogenase E1 subunit beta, metal regulatory transcription factor 1, glutaminase, cyclin-dependent kinase inhibitor 2A, dihydrolipoamide branched chain transacylase E2, glycine cleavage system H-protein, and dihydrolipoamide S-succinyltransferase. Pearson correlation analysis assessed the correlation between cuproptosis-related gene expression and lncRNAs. The criteria for cuproptosis-related lncRNAs were P values <0.01 and |Pearson R| >0.3. A univariate Cox regression analysis analyzed the prognostic cuproptosis-related lncRNAs (P<0.05).
Constructing the prognostic cuproptosis-associated lncRNA risk signature
First, cuproptosis-associated lncRNAs were utilized to conduct a predictive risk signature. Second, the multivariate Cox regression analysis and the least absolute shrinkage and selection operator (LASSO) Cox regression (R package “glmnet”) were utilized to establish a risk signature in patients with bladder cancer (13). Finally, based on the predictive signature, the risk score of every patient was calculated as follows:
The datasets were divided into low- and high-risk groups based on the median risk scores. Kaplan-Meier survival analysis, time-dependent receiver operating characteristic (ROC) curve analysis, and Cox relative risk recurrence study were utilized to determine the sensitivity and validity of the predictive signature.
Constructing and validating the individualized prognostic signature
A nomogram can calculate the survival rate of patients with tumors and has excellent clinical application value (14). A nomogram signature was established to predict the 1-, 3-, and 5-year overall survival (OS) of patients with bladder cancer. The prognostic nomogram was completed using the R package “rms” and “Hmisc”, which includes the risk score, age, grade, sex, and stage.
Principal component analysis (PCA) and gene set enrichment analysis (GSEA)
GSEA was utilized to identify the biological processes and molecular functions between the low- and high-risk groups (15). The PCA was conducted to confirm our assessments using the R package “limma” and “scatterplot3d” (16).
Immune cell infiltration and tumor mutation burden (TMB) analysis
The “GSEABase”, “GSVA”, and “CIBERSORT” were utilized to assess immune infiltration in patients with bladder cancer. Perl language and R package “maftools” were utilized to calculate the TMB.
Statistical analyses
Pearson correlation analysis was utilized to screen the cuproptosis-related lncRNAs. Wilcox-test or Student’s t-test was conducted to analyze cuproptosis-related lncRNA expressions between cancer and healthy tissues. Additionally, univariate and multivariate Cox regression analyses were utilized to evaluate the independence of the cuproptosis-related lncRNA signature. P values <0.05 indicated statistical significance. R software 4.2.0 was utilized for all statistical analyses.
Results
Identifying cuproptosis-related lncRNAs
Figure 1 illustrates the search strategy. The data included 414 bladder cancer samples and 19 healthy samples. Clinicopathological features included the age, sex, grade, stage, and tumor, node, and metastasis stage. A total of 14,049 lncRNAs were identified from TCGA dataset for bladder cancer. Cuproptosis-related lncRNA was defined as one whose expression was correlated with one or more of the 19 cuproptosis-related genes (|Pearson R| >0.4 and P<0.001). We identified 480 cuproptosis-related lncRNAs (Figure 2A). Combined with the prognostic information, univariate Cox regression was implemented to screen the prognostic cuproptosis-related lncRNAs (P<0.05). Finally, we identified 14 prognostic cuproptosis-related lncRNAs (Figure 2B).
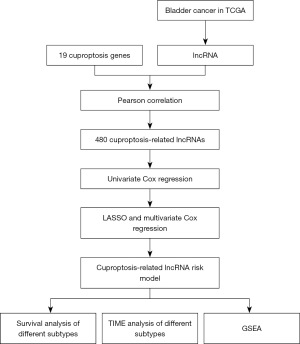
Constructing the cuproptosis-related lncRNA prognostic signature
We constructed a prognostic signature to determine the predictive value of prognostic cuproptosis-related lncRNAs. Samples in the TCGA database were randomly assigned to the training and validation groups. The 14 prognostic cuproptosis-related lncRNAs were screened by the LASSO regression signature. Ten cuproptosis-related lncRNAs, including AC124069.1, AC092794.1, GRK5-IT1, AC005519.1, AC253576.2, AC02132.1, AC103746.1, AL031429.2, AC093535.1, and ZNF436-AS1, were generated using the “glmnet” package of R software. Of them, seven were introduced into the multi-Cox proportional risk signature (Table S1, Figure 2C,2D). A seven-lncRNA signature was established by a multivariate Cox proportional signature (Figure 2E). The risk score was calculated as follows:
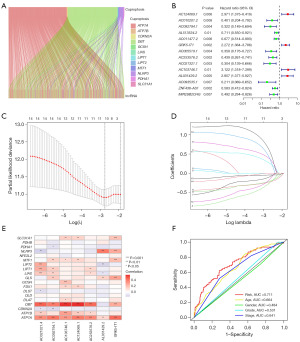
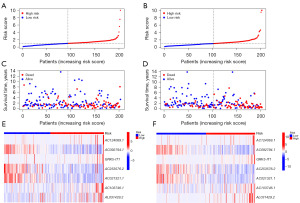
In TCGA dataset, the time-dependent ROC curve demonstrated an accuracy of 0.711, 0.673, and 0.684 for the 1-, 3-, and 5-year survival rates, respectively (Figures 2F,3A). Patients in the high-risk group demonstrated worse survival than patients in the low-risk group, both in the TCGA dataset and in the training or test groups (Figure 3B-3D). Figure 4 illustrates that a high-risk score was correlated with a worse prognosis.
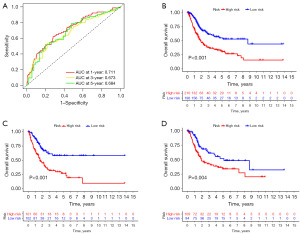
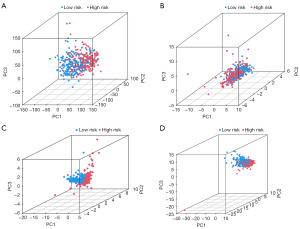
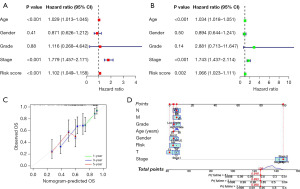
To establish the validity of the seven cuproptosis-related lncRNAs involved in signature construction, we conducted PCA analysis. We explored whether these seven cuproptosis-related lncRNAs could distinguish the differences between the high- and low-risk groups in TCGA dataset, cuproptosis genes, cuproptosis-related lncRNAs, and risk signatures. The seven cuproptosis-related lncRNAs differentiated between the low- and high-risk groups (Figure 5A-5D). Univariate and multivariate Cox regression analyses suggested the prognostic signature value as an independent prognostic factor for bladder cancer. The predictive signature constructed by the seven cuproptosis-related lncRNAs was an independent prognostic character (Figure 6A,6B).
Establishing an individualized prognostic signature
We considered the clinical application of the risk scores of cuproptosis-related lncRNAs in predicting OS in patients with bladder cancer. Subsequently, we constructed an individualized prognostic signature using the nomogram signature (Figure 6C,6D). The signature demonstrated good predictive power to predict the 1-, 3-, and 5-year OS.
Functional enrichment analysis and immune infiltrate analysis
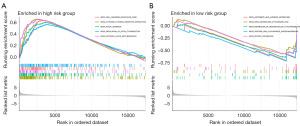
We performed a GSEA analysis to explore the biological processes and signaling pathways. Pathways involved in cytokine regulation, tumorigenesis, and migration, such as the focal adhesion pathway and cytokine-cytokine receptor interaction pathway, were enriched in the high-risk group (Figure 7A,7B). The immune cell infiltration analysis demonstrated that the high-risk group expressed high chemokines and T cell inhibition and low antigen-presenting cells (Figure 8A). We analyzed the correlation between tumor-infiltrating immune cells and risk scores using the “CIBERSORT” R package. Monocytes, endothelial cells, and cluster of differentiation 8 (CD8)+ T cells were positively correlated with the risk score (Figure 8B). Thus, patients with a high content of these cells and high-risk score demonstrated worse prognosis.
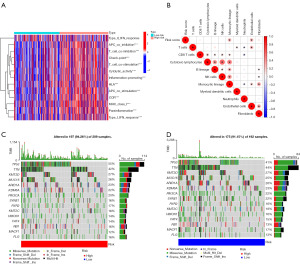
According to the genetic change analysis, 15 genes, including TP53, TTN, KMT2D, and so on, were significantly different between the high- and low-risk score subgroups (Figure 8C,8D).
Discussion
Bladder cancer is the predominant cancer among urinary malignancies; it is one of the most prevalent cancers worldwide (17). Multidisciplinary treatment, including surgery, chemotherapy, and immunotherapy, has considerably improved survival in patients with bladder cancer; nonetheless, the prognosis remains unsatisfactory (18). Therefore, clinicians should identify the prognostic and molecular features specific to bladder cancer to improve patient outcomes.
Cuproptosis induces proteotoxic stress and finally leads to cell death. Patients with tumors report a significantly higher content of copper ions in the serum and solid tumor tissues than healthy people (19,20). The increased content can promote the proliferation and metastasis of tumor cells and angiogenesis (20). Wang et al. reported that blocking Cu2+ transport can cause oxidative stress, reduce cellular ATP levels, and activate AMP-activated protein kinase, thereby reducing adipogenesis and inhibiting tumor cell proliferation (21). Safi et al. demonstrated that in hormone-sensitive and castration-resistant disease signatures, disulfiram alone did not significantly affect the growth of prostate cancer cells. In contrast, disulfiram combined with copper significantly inhibited the growth of prostate cancer cells (22). Therefore, understanding the biological process of copper drooping in tumor cells may facilitate the identification of novel therapeutic targets.
Abnormal lncRNA expression promotes the occurrence and development of bladder cancer (23,24). Bladder cancer-associated transcript 2 promotes lymphangiogenesis and lymphatic metastasis in bladder cancer (25). The copper metabolism MURR1 domain family is involved in cholangiocarcinogenesis through the EX41-miR-340-COMMD6 competitive endogenous RNA (ceRNA) network (26). However, researchers have not investigated cuproptosis-related lncRNAs in bladder cancer. In this study, we constructed a prognostic risk signature with good predictive power (1-, 3-, and 5-year survival rates of 0.711, 0.673, and 0.684, respectively) based on cuproptosis-related lncRNAs. Cox regression analysis demonstrated the risk signature as an independent factor.
We utilized LASSO regression and multivariate Cox regression analysis to examine seven lncRNAs (AC124069.1, AC092794.1, GRK5-IT1, AC253576.2, AC021321.1, AC103746.1, and AL031429.2). The cuproptosis genes co-expressed by three lncRNAs are DBT, ATP7A, and NLRP3, which are also associated with prognosis. We utilized these seven cuproptosis-related lncRNAs to construct the risk score signature and individualized prognostic signature. Risk scores can accurately predict patient outcomes without considering the traditional clinical risk markers or molecular factors. The differences in age, stage, sex, and grade did not affect the accuracy of the risk score signature in predicting prognosis. The individualized prognostic signature incorporated a cuproptosis-related lncRNA prognostic risk signature and clinicopathological parameters. This in turn could facilitate predicting the prognosis of individual patients.
GSEA suggested that the high-risk group was primarily enriched in cytokine regulation, tumorigenesis, and migration. Similarly, immune cell infiltration in the high-risk group demonstrated high expressions of chemokines, monocytes, endothelial cells, and CD8+ T cells. Therefore, cuproptosis-related lncRNAs may be involved in regulating bladder cancer progression through these immune cells and chemokines. The lymphocyte/monocyte ratio affected the prognosis of bladder cancer (27). Additionally, the genetic alteration analysis suggested that the high-risk group demonstrated higher mutation frequencies. This finding explains the mechanism underlying poor prognosis in the high-risk group. Immunotherapy may be more effective in patients with high TMB because of their relatively high number of neoantigens (28,29). Furthermore, immunotherapy is more beneficial for patients in the high-risk group with high TMB. A limitation of this study is the lack of external validation for lncRNA expression data from other sources. Researchers should validate the role of these lncRNAs in bladder cancer in the future.
Conclusions
In summary, we comprehensively evaluated the correlation between cuproptosis-related genes and tumorigenesis in patients with bladder cancer. Cuproptosis-related lncRNAs may be central to tumorigenesis, providing a novel therapeutic target for patients with bladder cancer. We constructed a risk score signature and individualized predictive signature based on cuproptosis-related lncRNAs to predict the survival of these patients.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2367/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2367/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2367/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lenis AT, Lec PM, Chamie K, et al. Bladder Cancer: A Review. JAMA 2020;324:1980-91. [Crossref] [PubMed]
- Bladder cancer: diagnosis and management of bladder cancer: © NICE (2015) Bladder cancer: diagnosis and management of bladder cancer. BJU Int 2017;120:755-65. [Crossref] [PubMed]
- Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J Clin 2020;70:404-23. [Crossref] [PubMed]
- Grayson M. Bladder cancer. Nature 2017;551:S33. [Crossref] [PubMed]
- Pfail JL, Katims AB, Alerasool P, et al. Immunotherapy in non-muscle-invasive bladder cancer: current status and future directions. World J Urol 2021;39:1319-29. [Crossref] [PubMed]
- Jones G, Cleves A, Wilt TJ, et al. Intravesical gemcitabine for non-muscle invasive bladder cancer. Cochrane Database Syst Rev 2012;1:CD009294. [Crossref] [PubMed]
- Wu Z, Liu J, Dai R, et al. Current status and future perspectives of immunotherapy in bladder cancer treatment. Sci China Life Sci 2021;64:512-33. [Crossref] [PubMed]
- Jiang Y, Huo Z, Qi X, et al. Copper-induced tumor cell death mechanisms and antitumor theragnostic applications of copper complexes. Nanomedicine (Lond) 2022;17:303-24. [Crossref] [PubMed]
- Li SR, Bu LL, Cai L. Cuproptosis: lipoylated TCA cycle proteins-mediated novel cell death pathway. Signal Transduct Target Ther 2022;7:158. [Crossref] [PubMed]
- Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022;375:1254-61. [Crossref] [PubMed]
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651-69. [Crossref] [PubMed]
- Cao Y, Tian T, Li W, et al. Long non-coding RNA in bladder cancer. Clin Chim Acta 2020;503:113-21. [Crossref] [PubMed]
- Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010;33:1-22. [Crossref] [PubMed]
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364-70. [Crossref] [PubMed]
- Chen L, Zhang YH, Wang S, et al. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One 2017;12:e0184129. [Crossref] [PubMed]
- Ni M, Wang H, Wang M, et al. Comparative Efficacy of Chinese Herbal Injections Combined with Paclitaxel Plus Cisplatin for Non-Small-Cell Lung Cancer: A Multidimensional Bayesian Network Meta-Analysis. Evid Based Complement Alternat Med 2020;2020:1824536. [Crossref] [PubMed]
- Alifrangis C, McGovern U, Freeman A, et al. Molecular and histopathology directed therapy for advanced bladder cancer. Nat Rev Urol 2019;16:465-83. [Crossref] [PubMed]
- Lopez-Beltran A, Cimadamore A, Blanca A, et al. Immune Checkpoint Inhibitors for the Treatment of Bladder Cancer. Cancers (Basel) 2021;13:131. [Crossref] [PubMed]
- Rostkowska-Nadolska B, Pośpiech L, Bochnia M. Content of trace elements in serum of patients with carcinoma of the larynx. Arch Immunol Ther Exp (Warsz) 1999;47:321-5. [PubMed]
- Garber K. Cancer's copper connections. Science 2015;349:129. [Crossref] [PubMed]
- Wang J, Luo C, Shan C, et al. Inhibition of human copper trafficking by a small molecule significantly attenuates cancer cell proliferation. Nat Chem 2015;7:968-79. [Crossref] [PubMed]
- Safi R, Nelson ER, Chitneni SK, et al. Copper signaling axis as a target for prostate cancer therapeutics. Cancer Res 2014;74:5819-31. [Crossref] [PubMed]
- Zhang Y, Chen X, Lin J, et al. Biological functions and clinical significance of long noncoding RNAs in bladder cancer. Cell Death Discov 2021;7:278. [Crossref] [PubMed]
- Wu Y, Zhang L, He S, et al. Identification of immune-related LncRNA for predicting prognosis and immunotherapeutic response in bladder cancer. Aging (Albany NY) 2020;12:23306-25. [Crossref] [PubMed]
- He W, Zhong G, Jiang N, et al. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J Clin Invest 2018;128:861-75. [Crossref] [PubMed]
- Yang M, Huang W, Sun Y, et al. Prognosis and modulation mechanisms of COMMD6 in human tumours based on expression profiling and comprehensive bioinformatics analysis. Br J Cancer 2019;121:699-709. [Crossref] [PubMed]
- Shi H, Wang K, Yuan J, et al. A high monocyte-to-lymphocyte ratio predicts poor prognosis in patients with radical cystectomy for bladder cancer. Transl Cancer Res 2020;9:5255-67. [Crossref] [PubMed]
- Zheng Y, Yao M, Yang Y. Higher Tumor Mutation Burden Was a Predictor for Better Outcome for NSCLC Patients Treated with PD-1 Antibodies: A Systematic Review and Meta-analysis. SLAS Technol 2021;26:605-14. [Crossref] [PubMed]
- Deng H, Zhao Y, Cai X, et al. PD-L1 expression and Tumor mutation burden as Pathological response biomarkers of Neoadjuvant immunotherapy for Early-stage Non-small cell lung cancer: A systematic review and meta-analysis. Crit Rev Oncol Hematol 2022;170:103582. [Crossref] [PubMed]


