
Sini decoction-polysaccharide compound regulates proliferation, apoptosis, and glycolysis of liver cancer cells through PHLDA2/ANXA2
Highlight box
Key findings
• The binding of PHLDA2 and ANXA2 was predicted and verified in hepatocellular carcinoma (HCC). Sini decoction (SND)-polysaccharide compound (SNDPC) reduced the expressions of PHLDA2 and ANXA2 and suppressed the proliferation and glycolysis of HCCLM3 cells while promoting apoptosis via PHLDA2/ANXA2.
What is known and what is new?
• SND plays a critical role in the treatment of liver disease.
• The effect and mechanism of the SNDPC on HCCLM3 cells remain unclear.
What is the implication, and what should change now?
• The efficacy of SNDPC on HCCLM3 cells was assessed, and its mechanism of action was clarified, which might provide a novel option for the treatment of HCC.
• In the future, corresponding in vivo experiments are needed to further verify the efficacy and mechanism of SNDPC in the treatment of HCC.
Introduction
Primary liver cancer is the second deadliest cancer and is accountable for 9% of deaths globally (1). Intrahepatic cholangiocarcinoma and hepatocellular carcinoma (HCC) are two predominant histologic subtypes of primary liver cancer (2). It is estimated that HCC constitutes approximately 80–90% of all primary liver cancer cases (3). Risk factors that contribute to liver cancer include alcohol intake, hepatitis B virus, hepatitis C virus, nonalcoholic steatohepatitis, and exposure to aflatoxin B (4). There are numerous treatment strategies for liver cancer, including surgery, targeted drug therapy, immunotherapy, and chemoradiotherapy (5). However, due to the high invasiveness and the difficulty in the diagnosis of liver cancer, most patients are diagnosed at advanced stages.
Recent studies have indicated that traditional Chinese medicine (TCM) provides favorable therapeutic effects in primary liver cancer (6-8). Sini decoction (SND), a popular TCM formula officially recorded in the Chinese pharmacopeia, consists of three herbs—Fuzi, Zhigancao, and Ganjiang (9,10). It has been established that SND exhibits anticancer activities in treating some cancers such as gastric cancer, colorectal cancer, liver cancer, and lung cancer (11-13). Polysaccharides are considered to be a critical source of bioactive polymers primarily due to of their strong antioxidant and antitumor properties (14). Additionally, a growing body of evidence indicates that polysaccharides extracted from TCM, including Sargassum fusiforme, Lycium chinensis, and Astragalus, can exert antitumor effects in liver cancer (15-17). SND-polysaccharide compound (SNDPC) is composed of radix Aconiti carmichaeli, Glycyrrhiza, and rhizoma Zingiberis. The extracts from the three herbs have been found to possess tumor-suppressive properties (18-20). However, the effect of SNDPC on liver cancer has not been extensively studied. Therefore, we aimed to investigate the efficacy of SNDPC in primary liver cancer and discuss its underlying mechanism of affect. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1625/rc).
Methods
Isolation and purification of SNDPC
The separation and purification of the polysaccharide from radix Aconiti carmichaeli were performed as follows: radix Aconiti carmichaeli (200 g of dry powder) was degreased with acetone three times and followed by centrifugation; the precipitation was then collected and dried at 45 ℃. Deionized water was added to the licorice-defatted powder at a ratio of 1:30, and papain, accounting for 2.5% of the weight of the licorice-defatted powder, was then added. After 12 h of stirring at 60 ℃, the solution was inactivated by heating at 100 ℃ for 15 min and cooled. Next, the solution was added to 2.5% trypsin at 37 ℃ and stirred for 12 h, which was followed by inactivation at 100 ℃ for 15 min and centrifugation. The supernatant obtained was concentrated to about 50 mL on a rotary evaporator. The concentrated solution was dialyzed using a dialysis membrane with a molecular weight cutoff of 3,500 and later freeze-dried to obtain a crude polysaccharide fraction. The isolation and purification of polysaccharides of Glycyrrhiza and rhizoma Zingiberis were conducted as described above. Three polysaccharides (radix Aconiti carmichaeli, rhizoma Zingiberis, and Glycyrrhiza) were combined in ratio of 3:2:3.
Bioinformatics
A heatmap was generated with Heatmapper (http://www.heatmapper.ca/), and volcano plots were analyzed (https://hiplot.com.cn/). The analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) was implemented using the Database for Annotation, Visualization and Integrated Discovery (DAVID) online database (https://david.ncifcrf.gov/). A Venn diagram was constructed to analyze the intersection among the differential genes, more specifically, the differential genes from The Cancer Genome Atlas (TCGA)-liver cancer dataset and from GSE14520. The Multi-Experiment Matrix (MEM) database (https://biit.cs.ut.ee/mem), HCCDB (http://lifeome.net/database/hccdb), LinkedOmics database (http://www.linkedomics.org/admin.php), and Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn) were used to determine the correlation between PHLDA2 and ANXA2. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture and treatment
The human HCC cell line (HCCLM3) obtained from the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China) was cultivated in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA; Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 ℃ with 5% CO2.
Cell transfection
PHLDA2-specific protruding clustered DNA (pcDNA)-overexpression vector (Ov-PHLDA2), empty vector (Ov-NC), a specific small interfering RNA (siRNA) against PHLDA2 (siRNA-PHLDA2-1/2), and the corresponding control siRNA (siRNA-NC) were constructed by GenePharma (Shanghai, China). With the application of Lipofectamine 2000 reagent (Invitrogen), the transfection of 100 nM recombinants into HCCLM3 cells was carried out according to the manufacturer’s instructions.
Cell Counting Kit-8 (CCK-8) assay
To determine the half-maximal inhibitory concentration (IC50) value of SNDPC, HCCLM3 cells, inoculated into 96-well plates (5×104 cells/mL), were pretreated to varying concentrations (0–160 mg/mL) of SNDPC for 24 h. Subsequently, 10 µL of CCK-8 (Beyotime Biotechnology, Nantong, China) was added to each well and incubated for an additional 2 h at 37 ℃. The optical density (OD) value at 450 nm was measured using a microplate reader (Bio-Rad, Hercules, CA, USA). In addition, HCCLM3 cells, inoculated into 96-well plates (5×104 cells/mL), were pre-treated by SNDPC at IC50 for 12, 24, and 48 h.
Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay
The apoptotic level of HCCLM3 cells was estimated using apoptosis detection kits (Roche, Basel, Switzerland) as per specifications. Pretreated HCCLM3 cells (20, 40, and 80 mg/mL SNDPC) were fixed with 4% paraformaldehyde and exposed to proteinase K for 15 min. Subsequently, cells were probed to 3% H2O2 for 15 min and stained with a TUNEL kit, following which cell nuclei were counterstained using 4',6-diamidino-2-phenylindole (DAPI) for 10 min. The labeled cells were photographed under a fluorescence microscope (Olympus, Tokyo, Japan).
Measurement of glucose uptake and lactate generation
Glucose uptake and lactate generation were estimated using the Glucose Uptake Colorimetric Assay Kit (Biovision, Milpitas, CA, USA) and Lactate Colorimetric Assay Kit (Biovision) according to the manufacturer’s instructions.
Extracellular acidification rate (ECAR)
The ECAR was evaluated using Seahorse XFe 96 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA) with Seahorse XF Glycolysis Stress Test Kit according to the manufacturer’s instructions. ECAR data (mpH/min) were processed using Seahorse XF-96 Wave software.
Coimmunoprecipitation
Following the extraction of proteins with immunoprecipitation lysis buffer (20 mM of Tris-HCl, 150 mM of NaCl, and 1% Triton X-100 at pH 7.5), cell lysates were subjected to PHLDA2 antibody or ANXA2 antibody at 4 ℃ overnight before exposure to protein A/G PLUS-Agarose (sc-2003; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). Next, the precipitated proteins were resuspended and boiled. After which, it was eluded from phosphate-buffered saline (PBS)-rinsed beads. Finally, the eluates were collected via magnetic separation, and Western blotting was conducted to analyze immuno-complexes.
RNA extraction and quantitative real-time polymerase chain reaction (PCR)
The reversed transcription of RNA isolated from HCCLM3 cells with TRIzol reagent (Invitrogen) into complement DNC (cDNA) was implemented using a cDNA Synthesis Kit (Takara Bio, Kusatsu, Japan) according to manufacturer’s instructions. The templates were amplified using real-time quantitative PCR with the SYBR Premix Ex Taq II kit (Accurate Biotechnology, China). The relative gene expression was estimated utilizing the comparative cycle threshold (Ct) (2−ΔΔCt) method.
Western blotting
The protein concentration of sample HCCLM3 cells was determined with radioimmunoprecipitation (RIPA) buffer (Bio-Rad) and quantified using a BCA Protein Assay Kit (Beyotime, Shanghai, China). Before transfer onto polyvinylidene fluoride membranes (Millipore, Burlington, MA, USA), proteins (20 µg/lane) were loaded onto 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels. After the membranes were incubated overnight, they were sealed with 5% bovine serum albumin for 2 h and with primary antibodies against Bcl-2 (ab196495; Abcam, Cambridge, UK), Bax (ab32503; Abcam), cleaved caspase 3 (25128-1-AP; Proteintech, Rosemont, IL, USA), caspase 3 (82202-1-RR; Proteintech), LDHA [#2012; Cell Signaling Technology (CST), Danvers, MA, USA], HK2 (22029-1-AP; Proteintech), PKM2 (#3198; CST), PHLDA2 (14661-1-AP; Proteintech), SPINK1 (13477-1-AP; Proteintech), ANXA2 (#8235; CST), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; #2118; CST) at 4 ℃. Subsequently, the membranes were exposed to horseradish peroxidase-labeled secondary antibodies for 1 h. Protein bands were visualized using an enhanced chemiluminescence (ECL) detection system (Amersham Pharmacia Biotech, Piscataway, NJ, USA) according to the manufacturer’s instructions. Protein density was analyzed using ImageJ software (NIH, Bethesda, MD, USA).
Statistical analysis
Statistical analysis was performed using the SPSS 18.0 software package (IBM Corp., Armonk, NY, USA). Data were expressed as the mean ± standard deviation (SD). To examine the differences between the two groups, a t-test was employed. For the comparisons among multiple groups, one-way analysis of variance (ANOVA) followed by the Bonferroni post hoc test was used. A P value less than 0.05 was considered statistically significant.
Results
SNDPC inhibited the proliferation and induced the apoptosis of HCC cells
To investigate the efficacy of SNDPC in HCC, we detected the effect of SNDPC on cell proliferative ability and cell apoptotic level in HCC. Our results suggested that SNDPC enhanced the cell inhibition rate of HCCLM3 cells, and the control group was 0 mg/mL SNDPC on HCCLM3 cells (Figure 1A) in concentration-dependent fashion. The IC50 value was 38.2 mg/mL. Thus, we chose 40 mg/mL of SNDPC for the subsequent assays. Our results showed that 40 mg/mL of SNDPC markedly suppressed the proliferation of HCCLM3 cells compared with the control cells (Figure 1B). Additionally, SNDPC pretreatment increased the cell apoptosis rate relative to that of the control cells (Figure 1C,1D). Meanwhile, SNDPC resulted in reduced Bcl-2 expression and elevated Bax and cleaved caspase 3 expression in HCCLM3 cells (Figure 1E).
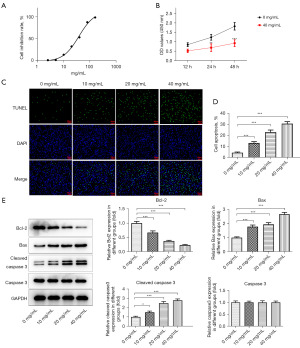
SNDPC suppressed the glycolysis of HCCLM3 cells
The impact of SNDPC on HCCLM3 cell glycolysis was investigated. SNDPC reduced the amount of glucose uptake and lactate levels in a concentration-dependent manner (Figure 2A,2B) and decreased ECAR, and the control group was 0 mg/mL SNDPC on HCCLM3 cells (Figure 2C). Moreover, results from Western blotting showed that expression of LDHA, HK2, and PKM2 was remarkably decreased after treatment with 10–40 mg/mL of SNDPC (Figure 2D).
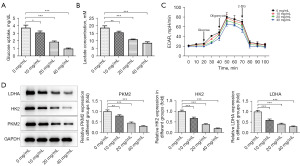
Identification of difference-related genes affected by SNDPC in HCC
To determine how SNDPC affects HCC progression, we identified the genes associated with HCC that were affected by SNDPC. The data were displayed as heatmaps and volcano plots (Figure 3A,3B). The GO enrichment and pathway analysis identified the upregulated and downregulated genes related to critical physiological processes, molecular functions, cellular components, and signaling pathways (Figure 3C-3F). To further pinpoint the aberrantly expressed and prognostic genes, we intersected the obtained differential genes with the differential genes for HCC in the TCGA dataset and those in GSE14520, obtained a total of 4 highly expressed genes and 3 lowly expressed genes. The control group was HCCLM3 cells before treatment with SNDPC. Our data revealed that the four highly expressed genes are AKR1C3, FDPS, SPINK1, and PHLDA2; the three lowly expressed genes are DNAJC12, SERPINE1, and IGFBP3. According to the analysis of the TCGA database GSE14520 database, PHLDA2 and SPINK1 have expression prognostic value (Figure 3G,3H). In addition, through Western blotting, we further discovered that SNDPC reduced PHLDA2 and SPINK1 expression, with SNDPC exerting a greater effect on PHLDA2 level, the control group was 0 mg/mL SNDPC on HCCLM3 cells (Figure 3I).
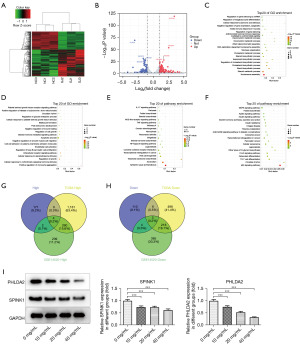
SNDPC regulated the expression of ANXA2 by PHLDA2
PHLDA2 was predicted to be associated with ANXA2 according to the MEM database. The LinkedOmics database predicted that PHLDA2 could be positively correlated with ANXA2. In addition, the HCCDB database also showed that PHLDA2 could be associated with ANXA2. GEPIA showed that PHLDA2 could be positively correlated with ANXA2 in HCC, with a correlation coefficient of 0.47. Therefore, this study further explored whether PHLDA2 could participate in liver cancer through ANXA2. The control group was HCCLM3 cells before treatment with SNDPC (Figure 4A-4D). Results from Western blotting showed that SNDPC reduced ANXA2 expression (Figure 4E). Coimmunoprecipitation assay further confirmed the combination of PHLDA2 and ANXA2 in HCCLM3 cells (Figure 4F). To investigate the role of PHLDA2 in ANXA2 expression within HCCLM3 cells treated with SNDPC, we overexpressed PHLDA2 (the transfection efficiency is shown in Figure 4G) and found that PHLDA2 overexpression reversed the effects of SNDPC on ANXA2 expression, and the control group was 0mg/mL SNDPC on HCCLM3 cells (Figure 4H).
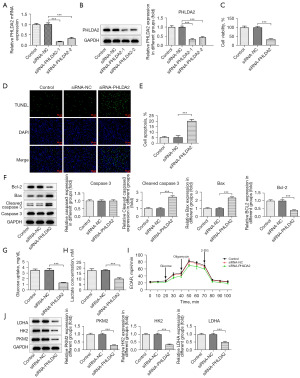
PHLDA2 silencing suppressed the proliferation and glycolysis of HCCLM3 cells and promoted cell apoptosis
To clarify the biological roles of PHLDA2 in HCCLM3 cells treated with SNDPC, we silenced PHLDA2. The transfection efficiency is shown in Figure 5A,5B. It was noted that siRNA-PHLDA2-1 had better transfection efficiency. Thus, siRNA-PHLDA2-1 (labeled as siRNA-PHLDA2) was adopted for follow-up assays. Relative to the siRNA-NC group, the PHLDA2-depleted group exhibited remarkably diminished proliferation of HCCLM3 cells (Figure 5C). As shown in Figure 5D,5E, the apoptosis rate in cells transfected with siRNA-PHLDA2 was increased, which was in line the finding that Bcl-2 expression was reduced while Bax and cleaved caspase 3 expression was elevated by PHLDA2 silencing (Figure 5F). In addition, knockdown of PHLDA2 reduced glucose uptake, lactate concentration, and ECAR (Figure 5G-5I). Finally, LDHA, HK2, and PKM2 expressions were remarkably reduced after PHLDA2 silencing (Figure 5J).
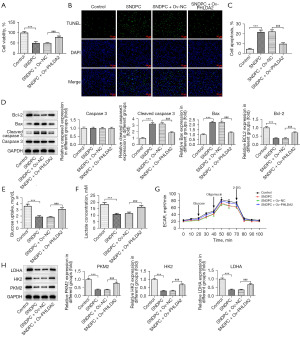
SNDPC suppressed the proliferation and glycolysis HCCLM3 cells and promoted apoptosis by PHLDA2/ANXA2
Cell viability was decreased and increased by SNDPC and PHLDA2 overexpression, respectively (Figure 6A). Moreover, SNDPC induced HCCLM3 cell apoptosis while PHLDA2 overexpression inhibited the cell apoptosis that was accelerated by SNDPC (Figure 6B,6C). In addition, PHLDA2 overexpression reduced Bcl-2 expression and promoted Bax and cleaved caspase 3 expression (Figure 6D). Finally, PHLDA2 overexpression reversed the effect of SNDPC on glucose uptake, lactate concentration, ECAR, and levels of LDHA, HK2, and PKM2 (Figure 6E-6H).
Discussion
HCC is a major contributor to deaths resulting from cancers globally (21). A previous study showed that the alterations in tumor microenvironment stress and genes could modulate glycolysis-related transporters and enzymes (22). It has been established that aerobic glycolysis is a pivotal regulator in HCC growth, metastasis, and drug resistance (23). In addition, the apoptosis process is associated with the cell-autonomous removal of superfluous, infected, or damaged cells, thereby constituting the most prominent defense mechanism against the occurrence of HCC (24). Therefore, effective glycolysis- and apoptosis-targeted therapies may represent a novel treatment strategy for HCC. This study was intended to ascertain the efficacy of SNDPC in HCC treatment and investigate whether SNDPC can affect cell proliferation, glycolysis, and apoptosis by targeting PHLDA2/ANXA2.
TCM is widely used clinically for the treatment of different types of cancers such as HCC and gastric carcinoma (25,26). SNDPC is composed of three polysaccharides from radix Aconiti carmichaeli, Glycyrrhiza, and rhizoma Zingiberis. Yao et al. found that aconitine is a main component in radix Aconiti Lateralis Preparata, exhibits antitumor effects in HCC by enhancing the ability of the immunocyte to kill tumor cells (27). Wang et al. reported that licorice could reduce the effects of radiation therapy, chemotherapy, or other adverse effects caused by cancer treatments (12). Furthermore, Ansari et al. demonstrated that the anticancer activity of the methanolic extract of Zingiber officinale rhizome (ZOME) in cervical cancer and breast cancer (28). In this study, we found that SNDPC could reduce cell proliferative capability and suppress glycolysis but facilitate cell apoptosis in HCC, which indicates that SNDPC might have a potential anticancer effect in HCC.
To clarify the mechanism of SNDPC function on HCC cells, we selected several related genes from bioinformatic databases. PHLDA2 was ultimately chosen, as it was highly expressed in HCC and had good prognostic value. PHLDA2, the apoptosis-related gene, is a PH domain-containing protein with phosphoinositide-binding capacity (29). It has been reported that PHLDA2 has close relation with poor prognosis in patients with glioma, HCC, or renal cell carcinoma (30). However, the functional role that PHLDA2 exerts in HCC has not yet been elucidated. In this study, we silenced PHLDA2 in HCCLM3 cells and found that PHLDA2 deficiency significantly suppressed the proliferative capability of HCCLM3 cells, suppressed glycolysis, and facilitated cell apoptosis. Additionally, PHLDA2 overexpression reversed the effects of SNDPC on HCC cells, which indicates that SNDPC might regulate HCC by targeting PHLDA2.
By using the MEM, HCCDB, LinkedOmics, and GEPIA databases, we confirmed that PHLDA2 was positively associated with ANXA2 in HCC. ANXA2 has been confirmed to facilitate cancer advancement and therapeutic resistance in patients with HCC (31). It has been reported that S100A10 triggers the mTOR pathway via the interaction with ANXA2 to facilitate tumor glycolysis in gastric cancer (32). In our study, SNDPC reduced ANXA2 expression while PHLDA2 overexpression increased it, which suggests that SNDPC might affect HCC cellular biological processes by mediating PHLDA2/ANXA2.
Conclusions
Our findings suggest that SNDPC suppresses cell proliferation and glycolysis while promoting cell apoptosis in HCC. These protective effects might depend on the regulation of PHLDA2/ANXA2 pathway and could represent a novel, fundamental insight into developing therapeutic strategies for HCC. In future research directions and follow-up studies, we will use several other liver cancer cell lines to further validate the results of a wider range of HCC phenotypes to address the limitations of the current study or explore other relevant aspects of the role of SNDPC in cancer treatment.
Acknowledgments
Funding: This study was funded by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1625/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1625/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1625/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1625/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Orcutt ST, Anaya DA. Liver Resection and Surgical Strategies for Management of Primary Liver Cancer. Cancer Control 2018;25:1073274817744621. [Crossref] [PubMed]
- Gao YX, Yang TW, Yin JM, et al. Progress and prospects of biomarkers in primary liver cancer Int J Oncol 2020;57:54-66. (Review). [Crossref] [PubMed]
- Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol 2017;34:153-9. [Crossref] [PubMed]
- Piñero F, Dirchwolf M, Pessôa MG. Biomarkers in Hepatocellular Carcinoma: Diagnosis, Prognosis and Treatment Response Assessment. Cells 2020;9:1370. [Crossref] [PubMed]
- Jiří T, Igor K. Mba. Hepatocellular carcinoma future treatment options. Klin Onkol 2020;33:26-9. [PubMed]
- Li JJ, Liang Q, Sun GC. Traditional Chinese medicine for prevention and treatment of hepatocellular carcinoma: A focus on epithelial-mesenchymal transition. J Integr Med 2021;19:469-77. [Crossref] [PubMed]
- Liu C, Yang S, Wang K, et al. Alkaloids from Traditional Chinese Medicine against hepatocellular carcinoma. Biomed Pharmacother 2019;120:109543. [Crossref] [PubMed]
- Liu X, Li M, Wang X, et al. Effects of adjuvant traditional Chinese medicine therapy on long-term survival in patients with hepatocellular carcinoma. Phytomedicine 2019;62:152930. [Crossref] [PubMed]
- Wu J, Yuan D, Yang M, et al. Sini decoction as an adjuvant therapy for angina pectoris: a systematic review of randomized controlled trials. J Tradit Chin Med 2017;37:12-22. [Crossref] [PubMed]
- Ding X, Zhang Y, Pan P, et al. Multiple mitochondria-targeted components screened from Sini decoction improved cardiac energetics and mitochondrial dysfunction to attenuate doxorubicin-induced cardiomyopathy. Theranostics 2023;13:510-30. [Crossref] [PubMed]
- Pan B, Wang Y, Wu C, et al. A Mechanism of Action Study on Danggui Sini Decoction to Discover Its Therapeutic Effect on Gastric Cancer. Front Pharmacol 2021;11:592903. [Crossref] [PubMed]
- Wang KL, Yu YC, Hsia SM. Perspectives on the Role of Isoliquiritigenin in Cancer. Cancers (Basel) 2021;13:115. [Crossref] [PubMed]
- Hao L, Li S, Chen G, et al. Study on the mechanism of quercetin in Sini Decoction Plus Ginseng Soup to inhibit liver cancer and HBV virus replication through CDK1. Chem Biol Drug Des 2024;103:e14567. [Crossref] [PubMed]
- Zeng P, Li J, Chen Y, et al. The structures and biological functions of polysaccharides from traditional Chinese herbs. Prog Mol Biol Transl Sci 2019;163:423-44. [Crossref] [PubMed]
- Cui B, Chen Y, Liu S, et al. Antitumour activity of Lycium chinensis polysaccharides in liver cancer rats. Int J Biol Macromol 2012;51:314-8. [Crossref] [PubMed]
- Fan S, Zhang J, Nie W, et al. Antitumor effects of polysaccharide from Sargassum fusiforme against human hepatocellular carcinoma HepG2 cells. Food Chem Toxicol 2017;102:53-62. [Crossref] [PubMed]
- Ji H, Lou X, Jiao J, et al. Preliminary Structural Characterization of Selenium Nanoparticle Composites Modified by Astragalus Polysaccharide and the Cytotoxicity Mechanism on Liver Cancer Cells. Molecules 2023;28:1561. [Crossref] [PubMed]
- de Lima RMT, Dos Reis AC, de Menezes APM, et al. Protective and therapeutic potential of ginger (Zingiber officinale) extract and [6]-gingerol in cancer: A comprehensive review. Phytother Res 2018;32:1885-907. [Crossref] [PubMed]
- Wang KL, Yu YC, Chen HY, et al. Recent Advances in Glycyrrhiza glabra (Licorice)-Containing Herbs Alleviating Radiotherapy- and Chemotherapy-Induced Adverse Reactions in Cancer Treatment. Metabolites 2022;12:535. [Crossref] [PubMed]
- Wu JJ, Guo ZZ, Zhu YF, et al. A systematic review of pharmacokinetic studies on herbal drug Fuzi: Implications for Fuzi as personalized medicine. Phytomedicine 2018;44:187-203. [Crossref] [PubMed]
- Gilles H, Garbutt T, Landrum J. Hepatocellular Carcinoma. Crit Care Nurs Clin North Am 2022;34:289-301. [Crossref] [PubMed]
- Kooshan Z, Cárdenas-Piedra L, Clements J, et al. Glycolysis, the sweet appetite of the tumor microenvironment. Cancer Lett 2024;600:217156. [Crossref] [PubMed]
- Feng J, Li J, Wu L, et al. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res 2020;39:126. [Crossref] [PubMed]
- Hu Y, Chen D, Hong M, et al. Apoptosis, Pyroptosis, and Ferroptosis Conspiringly Induce Immunosuppressive Hepatocellular Carcinoma Microenvironment and γδ T-Cell Imbalance. Front Immunol 2022;13:845974. [Crossref] [PubMed]
- Li Z, Feiyue Z, Gaofeng L. Traditional Chinese medicine and lung cancer--From theory to practice. Biomed Pharmacother 2021;137:111381. [Crossref] [PubMed]
- Kim KC, Yook JH, Eisenbraun J, et al. Quality of life, immunomodulation and safety of adjuvant mistletoe treatment in patients with gastric carcinoma - a randomized, controlled pilot study. BMC Complement Altern Med 2012;12:172. [Crossref] [PubMed]
- Yao F, Jiang GR, Liang GQ, et al. The antitumor effect of the combination of aconitine and crude monkshood polysaccharide on hepatocellular carcinoma. Pak J Pharm Sci 2021;34:971-9. [PubMed]
- Ansari JA, Ahmad MK, Khan AR, et al. Anticancer and Antioxidant activity of Zingiber officinale Roscoe rhizome. Indian J Exp Biol 2016;54:767-73. [PubMed]
- Ma Z, Lou S, Jiang Z. PHLDA2 regulates EMT and autophagy in colorectal cancer via the PI3K/AKT signaling pathway. Aging (Albany NY) 2020;12:7985-8000. [Crossref] [PubMed]
- Idichi T, Seki N, Kurahara H, et al. Molecular pathogenesis of pancreatic ductal adenocarcinoma: Impact of passenger strand of pre-miR-148a on gene regulation. Cancer Sci 2018;109:2013-26. [Crossref] [PubMed]
- Qiu LW, Liu YF, Cao XQ, et al. Annexin A2 promotion of hepatocellular carcinoma tumorigenesis via the immune microenvironment. World J Gastroenterol 2020;26:2126-37. [Crossref] [PubMed]
- Li Y, Li XY, Li LX, et al. S100A10 Accelerates Aerobic Glycolysis and Malignant Growth by Activating mTOR-Signaling Pathway in Gastric Cancer. Front Cell Dev Biol 2020;8:559486. [Crossref] [PubMed]
(English Language Editor: J. Gray)


