
Oncogenic role of SKA2 and its ceRNA network in hepatocellular carcinoma based on a comprehensive analysis
Highlight box
Key findings
• Spindle and kinetochore associated complex subunit 2 (SKA2) is highly expressed in hepatocellular carcinoma (HCC) and is associated with tumor stage, immune infiltrating cells, and overall survival (OS) of HCC.
• SKA2 is a member of competing endogenous RNA, it is related to SPACA6P-AS/hsa-miR-378a-5p/SKA2, SNHG14/hsa-miR-378a-5p/SKA2, and SNHG15/hsa-miR-378a-5p/SKA2, which play significant roles in tumor progression.
What is known and what is new?
• Relevant studies have shown that the expression of SKA2 correlate with tumor staging, tumor grading, patient race, and TP53 mutation status in HCC.
• In this study, we constructed the competing endogenous RNA (ceRNA) network for SKA2 and further elucidated its role in HCC.
What is the implication, and what should change now?
• SKA2 plays an important role in HCC and affects tumorigenesis as a member ceRNA network. These findings might provide a theoretical foundation for further research in the field of HCC.
Introduction
According to the Global Cancer Statistics 2020 report, liver cancer is the sixth most common primary cancer worldwide and has the third highest mortality rate. In most countries, men have a higher incidence of liver cancer than women (1). In 2030, liver cancer is projected to cause over 1 million deaths globally (2). In China, hepatitis B virus (HBV) infection and aflatoxin exposure are the primary causes of liver cancer (3), HBV infection is a chronic condition that affects over 250 million people worldwide, and nearly 1 million deaths annually are caused by liver cirrhosis and liver cancer (4). Despite existing research on hepatocellular carcinoma (HCC), its therapeutic effect is not ideal. A previous study showed that the disease-free survival rates of patients who underwent resection was <70%, with a median over survival of 60.19 months. Moreover, the median time to recurrence was 20.2 months (5). Hence, its pathogenesis should be further explored. Another research has revealed that alterations or loss of function in certain genes can result in cancer development (6). Alpha-fetoprotein (AFP) is a biomarker currently used for the early diagnosis of HCC. However, the sensitivity of AFP in screening HCC ranges from 39% to 64%. Notably, in up to 20% of patients with HCC, AFP is not produced (7). Therefore, it is necessary to screen novel biomarkers for early diagnosis and to identify novel therapeutic targets for HCC by assessing changes in the gene function network related to the HCC development and progression.
The spindle and kinetochore-associated (SKA) complex is a mitotic component required for precise division of human cells and a key complex at the kinetochore microtubule interface, SKA complex and NdC80 complex participate in kinetochore-microtubule attachment during mitosis (8,9). The SKA complex is associated with the development and prognosis of many cancers, and its score may serve as a predictor of patients receiving anti-programmed cell death protein-1 (PD-1)/programmed death ligand-1 (PD-L1) therapy (10). The core structure of the SKA complex is the W-shaped coiled coil dimer, which is formed by the interaction between SKA1 (spindle and kinetochore associated complex subunit 1), SKA2 (spindle and kinetochore associated complex subunit 2), and SKA3 (spindle and kinetochore associated complex subunit 3). SKA2 and SKA3 form the skeleton of the dimerization interface and play a key role in maintaining the silence of the intermediate plate and spindle checkpoints, SKA3 affects the migration, proliferation, reproduction of HCC cells through the notch signaling pathway, SKA2 is located in chromosome 17 (9,11-13), moreover, it plays essential roles in cell cycle progression, cell proliferation and tumorigenesis. Furthermore, the SKA2 gene expression is regulated by micro-RNAs (14). It has been shown that there is a significant association between circ_0008039, miR-140-3p and SKA2 in breast cancer (15). The competing endogenous RNA (ceRNA) axis is composed of mRNA, microRNA, and lncRNA, and it can significantly affect the development of some diseases. However, it may also provide novel avenues for disease therapies (16). Several studies have revealed that SKA2 influences liver cancer proliferation and migration via the β-catenin signaling pathway (17,18). SKA2 is related to tumor stage and tumor grade in HCC. The expression of SKA2 in liver cancer is related to race. The expression level is higher in Asian HCC patients, and the expression level of SKA2 is higher in HCC patients with TP53 mutations (19,20). However, knowledge regarding the regulatory network of SKA2 in HCC is limited. Therefore, this study aimed to analyze SKA2 using HCC data from The Cancer Genome Atlas (TCGA). Moreover, the ceRNA network was evaluated via bioinformatics analysis.
Methods
Collection of data on the SKA2 expression
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Genomic Data Commons (GDC)-client tool was used to download HCC data on RNA-seq and miRNA-seq raw counts and clinical information from the TCGA database (http://cancergenome.nih.gov/). After removing repeated samples, 50 normal tissue samples and 373 tumor tissues samples were analyzed. Raw counts were converted to transcripts per kilobase of exon model per million mapped reads (TPM) and the expression of SKA2 was compared between normal and tumor samples.
Associations between the SKA2 expression and prognostic, tumor stage, and immune infiltrating cells
To integrate RNA-seq data with clinical information (survive days >0), the surv_cutpoint function of the survminer package in R was utilized to determine the optimal cutoff of continuous independent variables based on survival data. In addition, the association between the SKA2 expression and overall survival (OS) and tumor stage was analyzed using R software (version 4.2.0). In particular, TIMER is a database for comprehensive analysis of tumor-infiltrating immune cells, it was used to detect the association between the SKA2 expression and six types of immune cells (21).
Functional enrichment analysis
The co-expression genes of SKA2 were determined using cBioPortal v3.7.1 (https://www.cbioportal.org/), cBioPortal is an open-source software project that allows us to explore multidimensional cancer genomics data. The screening criteria with a |correlation coefficient| >0.4 and P value <0.05 were applied were applied. The identified genes were used to perform Gene Ontology (GO) analysis to determine BP (biological process), CC (cellular component), and MF (molecular function) term enrichment and perform Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis using the R package clusterProfiler (22). In addition, the TCGA data of HCC were sorted based on the SKA2 expression and divided into the SKA2 high and low expression group according to the median. The data set was analyzed using gene set enrichment analysis (GSEA) 4.3.2 software, GSEA is used to evaluate the distribution trends of genes from a predefined gene set in the gene table sorted with phenotype correlation, and thereby determine their contribution to the phenotype (23).
Identification of miRNAs
MiRWalk3.0 (http://mirwalk.umm.uniheidelberg.de/), TargetScanHuman8.0 (https://www.targetscan.org/vert_80/), and miRDB (http://mirdb.org/) were utilized to predict the miRNAs of SKA2 (24-26). The miRNAs predicted by these tools were then intersected, and the miRNAs negatively associated with the SKA2 expression were further calculated. In addition, the expression of these miRNAs was determined using HCC miRNA-seq data from TCGA. To investigate the association between these miRNAs and OS, the surv_cutpoint function of the survminer package was used to determine the high and low expression groups.
Identification of lncRNAs
Based on the miRNAs predicted above, miRWalk2.0 (miRWalk, miRanda, RNAhybrid, Targetscan) and DIANA (LncBase v2) were further utilized to predict their lncRNAs (24,27). Meanwhile, the predicted results were intersected, and the expression of these lncRNAs in both tumor and normal tissues was calculated using HCC data from TCGA.
Associations between lncRNAs and prognostic, miRNA expression, and immunity
The RNA-seq and clinical data were downloaded and integrated, and the surv_cutpoint function was used to determine the optimal cutoff value. Then, the association between the identified lncRNAs and OS was examined. Furthermore, the association between the identified lncRNAs, miRNA and SKA2 expression was evaluated.
Establishment of the ceRNA network
As mentioned above, The Cytoscape software (version 3.10.2) was used to draw the ceRNA network of the above-mentioned genes, Cytoscape is an open-source software project for the integration of biomolecular interaction networks with high-throughput expression data and other molecular states into a unified conceptual framework (28).
Statistical analysis
Data were analyzed using the R software (version 4.2.0) and the unpaired t-test was used to calculate the difference between two groups of continuously distributed variables. P value <0.05 was considered statistically significant.
Results
High SKA2 expression in HCC
According to the HCC data from TCGA, the tumor tissues had a significantly higher SKA2 expression than the normal tissues (P<0.05) (Figure 1A). As shown in Figure 1B, this was further confirmed by examining 50 paired normal and tumor tissues, which showed that SKA2 was overexpressed in tumor tissues (P<0.05).

Association between SKA2 and OS, tumor stage and immunity
Data were divided into the two groups, SKA2 high group and the SKA2 low group based on the cutoff value of 26.01625 according to the surv_cutpoint function. As shown in Figure 2A, the SKA2 high expression group had a worse OS than the SKA2 low expression group (P<0.05). In addition, the SKA2 expression was upregulated with increased tumor stage. However, it decreased in patients with stage IV disease (P<0.05), as shown in Figure 2B. Moreover, a significant association was observed between SKA2 expression and tumor stage (P<0.05). According to the TIMER, there was an association between the SKA2 expression and various immune cells. In particular, SKA2 was found to be associated with B cell (correlation coefficient =0.255, P<0.05), CD8+ cell (correlation coefficient =0.203, P<0.05), CD4+ cell (correlation coefficient =0.329, P<0.05), macrophage (correlation coefficient =0.409, P<0.05), neutrophil (correlation coefficient =0.265, P<0.05), and dendritic cell (correlation coefficient =0.283, P<0.05). This information sheds light on the correlation between the SKA2 expression and the immune system (Figure 2C).

Enrichment analysis of GO, KEGG, and GSEA
According to the screening criteria (Spearman’s correlation >0.4, P<0.05), the study identified 188 co-expression genes of the SKA2 in the cBioPortal website (https://www.cbioportal.org/). The enrichment pathways of these co-expression genes were analyzed. According to the analysis results using BP GO terms, these co-expression molecules were mainly enriched in mechanisms such as nuclear division, DNA replication, and regulation of cell cycle phase transition. The CC showed that these genes were related to chromosomal region, spindle, and condensed chromosome. Based on the MF, they were enriched in ATPase activity, DNA-dependent ATPase activity, and microtubule binding (Figure 3A). The KEGG results show that the co-expression molecules were mainly enriched in cell cycle, DNA replication, p53 signaling pathway, and so on (Figure 3B). As shown in Figure 3C, the GSEA results based on SKA2 revealed that the group with high SKA2 expression were mainly enriched in cell cycle, oocyte meiosis and DNA replication.

Determination of miRNAs of SKA2
In total, 749 miRNAs of SKA2 were obtained from miRWalk 3.0 using a screening standard score >0.95. Meanwhile, 695 miRNAs were screened using TargetScan, and 78 miRNAs were obtained using miRDB. Then, 23 miRNAs were obtained from the intersection of three databases [namely, miRWalk 3.0 (http://mirwalk.umm.uni-heidelberg.de/), TargetScan (https://www.targetscan.org/vert_80/), and miRDB (https://mirdb.org/)]. The 23 miRNAs identified were hsa-miR-135a-3p, hsa-miR-19b-1-5p, hsa-miR-3160-5p, hsa-miR-3176, hsa-miR-3177-5p, hsa-miR-378a-5p, hsa-miR-3916, hsa-miR-3925-5p, hsa-miR-4330, hsa-miR-4433a-3p, hsa-miR-4524b-3p, hsa-miR-4690-5p, hsa-miR-5584-5p, hsa-miR-5588-5p, hsa-miR-595, hsa-miR-642a-3p, hsa-miR-6785-5p, Hsa-miR-6799-5p, hsa-miR-6859-5p, hsa-miR-6867-3p, hsa-miR-6867-5p, hsa-miR-6886-3p, and hsa-miR-877-5p. Furthermore, based on the downloaded data on miRNA in HCC, as shown in Figure 4A,4B, the cancer tissues had a lower hsa-miR-19b-1-5p and hsa-miR-378a-5p expression than the normal tissues. hsa-miR-19b-1-5p and hsa-miR-378a-5p were significantly associated with OS (Figure 4C,4D). Furthermore, the association between SKA2 and these two miRNAs was evaluated. As shown in Figure 4, the expression of SKA2 was negatively correlated with hsa-miR-378a-5p (P<0.05) (Figure 4F), but not with hsa-miR-19b-1-5p (Figure 4E).

Determination of lncRNAs of miRNAs
DIANA (LncBase v2) was used to obtain 2317 lncRNAs related to hsa-miR-378a-5p, with a threshold >0.9. At least three algorithms from miRWalk, miRanda, RNAhybrid, and Targetscan were taken as the standard, and data from miRWalk2.0 were used to predicted 57 lncRNAs. Meanwhile, 26 lncRNAs were obtained from the intersection of the two databases. As shown in Figure 5, 12 lncRNAs were significantly highly expressed in the tumor tissues (P<0.05). These lncRNAs were AC005154.6, EXTL3-AS1, GABPB1-AS1, KCNQ1OT1, MALAT1, MUC19, NEAT1, RAMP2-AS1, SNHG14, SNHG15, SPACA6P-AS, TPTEP1.
Survival assessment of lncRNAs
The association between prognosis and 12 lncRNAs was analyzed. Figure 6 shows that 9 of the 12 genes were associated with OS (P<0.05). These genes were EXTL3-AS1, GABPB1-AS1, NEAT1, RAMP2-AS1, SNHG14, SNHG15, SPACA6P-AS, TPTEP1, and AC005154.6.

Analysis of the association between SKA2, miRNAs and lncRNAs
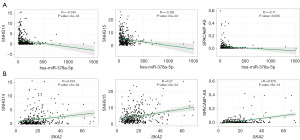
The association between SKA2 and miRNA as well as lncRNAs was analyzed. As shown in Figure 7A, SNHG14, SNHG15, and SPCA6P-AS were significantly negatively associated with hsa-378a-5p (P<0.05). In addition, these three lncRNAs were positively associated with SKA2 (P<0.05), as depicted in Figure 7B.
Establishment of ceRNA network
As shown in Figure 8, the SPACA6P-AS/hsa-miR-378a-5p/SKA2, SNHG14/hsa-miR-378a-5p/SKA2, and SNHG15/hsa-miR-378a-5p/SKA2 regulatory axes were identified.

Discussion
SKA2 is a protein-coding gene that plays a crucial role in the normal metabolism of the human body. Moreover, it is associated with various cellular processes such as cell cycle, tumorigenesis, and mental illnesses (14). For example, overexpression of SKA2 promotes the invasion and metastasis of breast cancer cells (15,29,30). SKA2 is highly expressed in gastric cancer and influences the tumor growth (31). In addition, it is associated with psychiatric disorders (32-35). However, its role in HCC is not completely elucidated. The current study aims to comprehensively investigate the role of SKA2 in HCC.
In this study, using the TCGA datasets, SKA2 was found to be overexpressed in the HCC tissues. This result is consistent with that of previous studies (17,18). Based on our findings, a high SKA2 expression could be a predictor of worse prognosis. This finding is supported by the study of Wang et al., which showed that a high SKA2 expression is associated with negative prognostic outcomes in breast cancer (29). In addition, Yu et al. found that the SKA family is associated with immune infiltrating cells (19). Our study revealed that the SKA2 expression was related to tumor stage and immune infiltrating cells.
Next, GO and KEGG enrichment analysis of the co-expression genes were performed. Results showed that several genes were mainly enriched in several processes including DNA replication, chromosomal region, ATPase activity, and cell cycle. The GSEA analysis revealed that a high SKA2 expression was involved in cell cycle and DNA replication, which is consistent with the results of GO and KEGG enrichment analysis. Ding et al. have reported that the SKA family is associated with prognostic value and immune infiltration in breast cancer (36). Yu found that SKA complex is related to prognosis in gliomas (37).
MicroRNA is a single-stranded non-coding small RNAs, which mainly acts in post-transcriptional regulation and plays different roles in various pathological processes. It can be oncogenes or tumor suppressor factors (38). Recent literature has shown that miR-29a-3p affects the metastasis of HCC by targeting LOX, LOXL2, and VEGFA (39), Moreover, miR-125b expression is downregulated in HCC, and miR-125b has anti-metastatic properties (40), Zou et al.’s emphasized that LIX1L drives hepatocarcinogenesis and tumor progression via miR-21-3p (41). Our study performed a comprehensive screening and analysis, results showed that hsa-miR-378a-5p was related to SKA2. Furthermore, the hsa-miR-378a-5p expression is low in tumors and is associated with prognosis. Previous studies have revealed that hsa-miR-378a-5p is significantly associated with the prognostic outcome of digestive system cancer (42) and sarcoma (43).
LncRNA is a type of RNA molecule that is >200 nucleotides long. They play an essential role in various biological processes, including tumor metastasis, cancer development, DNA damage, tumor immune microenvironment, and metabolic reprogramming (44-49). It has been shown that lncRNA HERH-1 and HERH-4 are involved in regulating cell cycle progression in advanced HCC (50). We predicted lncRNAs (SNHG14, SNHG15 and SPACA6P-AS) about hsa-miR-378a-5p, and they are overexpressed in HCC and associated with OS. According to RNALocate (51), SNHG14 is located in the insoluble cytoplasm, nuclear, cytosol, and membrane in HCC cell lines. A high SNHG14 expression promotes cell proliferation and colony formation and inhibits cell apoptosis in the HCC cells (52). In addition, SNHG14 is negatively associated with hsa-miR-378a-5p. SNHG14 is overexpressed in colorectal cancer, leading to enhanced cell proliferation and invasion (53). Furthermore, SNHG14 promotes cell proliferation and angiogenesis via PTEN signaling in HCC (54). According to Du et al. research, SNHG15 is associated with breast cancer (55), and lncRNA SNHG15 contributes to oncogenesis by targeting miR-506-5p (56). Finally, SPACA6P-AS act as a ceRNA, which promotes HCC oncogenicity (57).
Previous studies have revealed that ceRNA plays a significant role in various types of tumors. For instance, GAS6-AS1/miR-24-3p/GIMAP6 regulatory axes affect the progression of lung adenocarcinoma (58). Cao et al. has revealed that TMEM220-AS1 affects the proliferation and metastasis of liver cancer via the miR-484/MAG1 axis (59). In addition, lncRNA SNHG14 enhances HCC growth by acting as a molecular sponge of miR-876-5p to regulate SSR2 expression (60). Our study investigated the SPACA6P-AS/hsa-miR-378a-5p/SKA2, SNHG14/hsa-miR-378a-5p/SKA2, and SNHG15/hsa-miR-378a-5p/SKA2 regulatory axes HCC. Results showed that they may influence tumor growth, metastasis, and OS.
Conclusions
SKA2 is significantly overexpressed in HCC and is associated with OS, tumor stage, and immune infiltrating cells. Furthermore, SKA2 is a member of ceRNA that affects tumorigenesis. These findings might provide a theoretical foundation for further research in the field of HCC.
Acknowledgments
Funding: This work was supported by
Footnote
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-833/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-833/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Villanueva A. Hepatocellular Carcinoma. N Engl J Med 2019;380:1450-62. [Crossref] [PubMed]
- Chimed T, Sandagdorj T, Znaor A, et al. Cancer incidence and cancer control in Mongolia: Results from the National Cancer Registry 2008-12. Int J Cancer 2017;140:302-9. [Crossref] [PubMed]
- Novotny LA, Evans JG, Su L, et al. Review of Lambda Interferons in Hepatitis B Virus Infection: Outcomes and Therapeutic Strategies. Viruses 2021;13:1090. [Crossref] [PubMed]
- Kluger MD, Salceda JA, Laurent A, et al. Liver resection for hepatocellular carcinoma in 313 Western patients: tumor biology and underlying liver rather than tumor size drive prognosis. J Hepatol 2015;62:1131-40. [Crossref] [PubMed]
- Haigis KM, Cichowski K, Elledge SJ. Tissue-specificity in cancer: The rule, not the exception. Science 2019;363:1150-1. [Crossref] [PubMed]
- Charrière B, Maulat C, Suc B, et al. Contribution of alpha-fetoprotein in liver transplantation for hepatocellular carcinoma. World J Hepatol 2016;8:881-90. [Crossref] [PubMed]
- Polley S, Müschenborn H, Terbeck M, et al. Stable kinetochore-microtubule attachment requires loop-dependent Ndc80-Ndc80 binding. EMBO J 2023;42:e112504. [Crossref] [PubMed]
- Jeyaprakash AA, Santamaria A, Jayachandran U, et al. Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol Cell 2012;46:274-86. [Crossref] [PubMed]
- Li Z, Huang L, Li J, et al. Immunological role and prognostic value of the SKA family in pan-cancer analysis. Front Immunol 2023;14:1012999. [Crossref] [PubMed]
- Hanisch A, Silljé HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J 2006;25:5504-15. [Crossref] [PubMed]
- Guimaraes GJ, Deluca JG. Connecting with Ska, a key complex at the kinetochore-microtubule interface. EMBO J 2009;28:1375-7. [Crossref] [PubMed]
- Bai S, Chen W, Zheng M, et al. Spindle and kinetochore-associated complex subunit 3 (SKA3) promotes stem cell-like properties of hepatocellular carcinoma cells through activating Notch signaling pathway. Ann Transl Med 2021;9:1361. [Crossref] [PubMed]
- Xie M, Bu Y. SKA2/FAM33A: A novel gene implicated in cell cycle, tumorigenesis, and psychiatric disorders. Genes Dis 2018;6:25-30. [Crossref] [PubMed]
- Dou D, Ren X, Han M, et al. Circ_0008039 supports breast cancer cell proliferation, migration, invasion, and glycolysis by regulating the miR-140-3p/SKA2 axis. Mol Oncol 2021;15:697-709. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Wang D, Suo YJ, Gong L, et al. SKA2 promotes proliferation and invasion of hepatocellular carcinoma cells via activating the β-catenin signaling pathway. Neoplasma 2020;67:743-50. [Crossref] [PubMed]
- Jiang J, Xu B, Zheng Y, Guo X, Chen F. Spindle and kinetochore-associated protein 2 facilitates the proliferation and invasion of hepatocellular carcinoma via the regulation of Wnt/β-catenin signaling. Exp Cell Res 2020;395:112181. [Crossref] [PubMed]
- Yu DC, Chen XY, Li X, et al. Transcript levels of spindle and kinetochore-associated complex 1/3 as prognostic biomarkers correlated with immune infiltrates in hepatocellular carcinoma. Sci Rep 2021;11:11165. [Crossref] [PubMed]
- Song GQ, He TL, Ji KJ, et al. SKA1/2/3 is a biomarker of poor prognosis in human hepatocellular carcinoma. Front Oncol 2022;12:1038925. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Sticht C, De La Torre C, Parveen A, et al. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One 2018;13:e0206239. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005. [Crossref] [PubMed]
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 2020;48:D127-31. [Crossref] [PubMed]
- Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res 2016;44:D231-8. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Wang Y, Zhang C, Mai L, et al. PRR11 and SKA2 gene pair is overexpressed and regulated by p53 in breast cancer. BMB Rep 2019;52:157-62. [Crossref] [PubMed]
- Ren Z, Yang T, Zhang P, et al. SKA2 mediates invasion and metastasis in human breast cancer via EMT. Mol Med Rep 2019;19:515-23. [PubMed]
- Su H, Ren F, Jiang H, et al. Upregulation of microRNA-520a-3p inhibits the proliferation, migration and invasion via spindle and kinetochore associated 2 in gastric cancer. Oncol Lett 2019;18:3323-30. [Crossref] [PubMed]
- Sadeh N, Wolf EJ, Logue MW, et al. Epigenetic variation at ska2 predicts suicide phenotypes and internalizing psychopathology. Depress Anxiety 2016;33:308-15. [Crossref] [PubMed]
- Pandey GN, Rizavi HS, Zhang H, et al. The Expression of the Suicide-Associated Gene SKA2 Is Decreased in the Prefrontal Cortex of Suicide Victims but Not of Nonsuicidal Patients. Int J Neuropsychopharmacol 2016;19:pyw015. [Crossref] [PubMed]
- Boks MP, Rutten BP, Geuze E, et al. SKA2 Methylation is Involved in Cortisol Stress Reactivity and Predicts the Development of Post-Traumatic Stress Disorder (PTSD) After Military Deployment. Neuropsychopharmacology 2016;41:1350-6. [Crossref] [PubMed]
- Kalla C, Goltser-Dubner T, Pevzner D, et al. Resting mononuclear cell NR3C1 and SKA2 expression levels predict blunted cortisol reactivity to combat training stress among elite army cadets exposed to childhood adversity. Mol Psychiatry 2021;26:6680-7. [Crossref] [PubMed]
- Ding J, He X, Wang J, et al. Integrative analysis of prognostic value and immune infiltration of spindle and kinetochore-associated family members in breast cancer. Bioengineered 2021;12:10905-23. [Crossref] [PubMed]
- Yu S. Overexpression of SKA Complex Is Associated With Poor Prognosis in Gliomas. Front Neurol 2022;12:755681. [Crossref] [PubMed]
- Dong H, Lei J, Ding L, et al. MicroRNA: function, detection, and bioanalysis. Chem Rev 2013;113:6207-33. [Crossref] [PubMed]
- Yang YL, Tsai MC, Chang YH, et al. MIR29A Impedes Metastatic Behaviors in Hepatocellular Carcinoma via Targeting LOX, LOXL2, and VEGFA. Int J Mol Sci 2021;22:6001. [Crossref] [PubMed]
- Kim HS, Kim JS, Park NR, et al. Exosomal miR-125b Exerts Anti-Metastatic Properties and Predicts Early Metastasis of Hepatocellular Carcinoma. Front Oncol 2021;11:637247. [Crossref] [PubMed]
- Zou J, Zhu X, Xiang D, et al. LIX1-like protein promotes liver cancer progression via miR-21-3p-mediated inhibition of fructose-1,6-bisphosphatase. Acta Pharm Sin B 2021;11:1578-91. [Crossref] [PubMed]
- Chen Z, Shen Z, Zhang Z, et al. RNA-Associated Co-expression Network Identifies Novel Biomarkers for Digestive System Cancer. Front Genet 2021;12:659788. [Crossref] [PubMed]
- Shi D, Mu S, Pu F, et al. Development of a Novel Immune Infiltration-Related ceRNA Network and Prognostic Model for Sarcoma. Front Cell Dev Biol 2021;9:652300. [Crossref] [PubMed]
- Liu QL, Zhang Z, Wei X, et al. Noncoding RNAs in tumor metastasis: molecular and clinical perspectives. Cell Mol Life Sci 2021;78:6823-50. [Crossref] [PubMed]
- Xu T, Xie M, Jing X, et al. Crosstalk between Environmental Inflammatory Stimuli and Non-Coding RNA in Cancer Occurrence and Development. Cancers (Basel) 2021;13:4436. [Crossref] [PubMed]
- Ghafouri-Fard S, Azimi T, Hussen BM, et al. Non-coding RNA Activated by DNA Damage: Review of Its Roles in the Carcinogenesis. Front Cell Dev Biol 2021;9:714787. [Crossref] [PubMed]
- Chang L, Li J, Ding J, et al. Roles of long noncoding RNAs on tumor immune escape by regulating immune cells differentiation and function. Am J Cancer Res 2021;11:2369-85. [PubMed]
- Varier KM, Dhandapani H, Liu W, et al. An immunotherapeutic approach to decipher the role of long non-coding RNAs in cancer progression, resistance and epigenetic regulation of immune cells. J Exp Clin Cancer Res 2021;40:242. [Crossref] [PubMed]
- Sellitto A, Pecoraro G, Giurato G, et al. Regulation of Metabolic Reprogramming by Long Non-Coding RNAs in Cancer. Cancers (Basel) 2021;13:3485. [Crossref] [PubMed]
- Liu T, Shi Q, Yang L, et al. Long non-coding RNAs HERH-1 and HERH-4 facilitate cyclin A2 expression and accelerate cell cycle progression in advanced hepatocellular carcinoma. BMC Cancer 2021;21:957. [Crossref] [PubMed]
- Cui T, Dou Y, Tan P, et al. RNALocate v2.0: an updated resource for RNA subcellular localization with increased coverage and annotation. Nucleic Acids Res 2022;50:D333-9. [Crossref] [PubMed]
- Ning L, Cui T, Zheng B, et al. MNDR v3.0: mammal ncRNA-disease repository with increased coverage and annotation. Nucleic Acids Res 2021;49:D160-4. [Crossref] [PubMed]
- Wang X, Yang P, Zhang D, et al. LncRNA SNHG14 promotes cell proliferation and invasion in colorectal cancer through modulating miR-519b-3p/DDX5 axis. J Cancer 2021;12:4958-70. [Crossref] [PubMed]
- Zhang H, Xu HB, Kurban E, et al. LncRNA SNHG14 promotes hepatocellular carcinoma progression via H3K27 acetylation activated PABPC1 by PTEN signaling. Cell Death Dis 2020;11:646. [Crossref] [PubMed]
- Du J, Zhong H, Ma B. Targeting a novel LncRNA SNHG15/miR-451/c-Myc signaling cascade is effective to hamper the pathogenesis of breast cancer (BC) in vitro and in vivo. Cancer Cell Int 2021;21:186. [Crossref] [PubMed]
- Chen Z, Zhong T, Li T, et al. LncRNA SNHG15 modulates gastric cancer tumorigenesis by impairing miR-506-5p expression. Biosci Rep 2021;41:BSR20204177. [Crossref] [PubMed]
- Di Palo A, Siniscalchi C, Mosca N, et al. A Novel ceRNA Regulatory Network Involving the Long Non-Coding Antisense RNA SPACA6P-AS, miR-125a and its mRNA Targets in Hepatocarcinoma Cells. Int J Mol Sci 2020;21:5068. [Crossref] [PubMed]
- Wang Y, Ma M, Li C, et al. GAS6-AS1 Overexpression Increases GIMAP6 Expression and Inhibits Lung Adenocarcinoma Progression by Sponging miR-24-3p. Front Oncol 2021;11:645771. [Crossref] [PubMed]
- Cao C, Li J, Li G, et al. Long Non-coding RNA TMEM220-AS1 Suppressed Hepatocellular Carcinoma by Regulating the miR-484/MAGI1 Axis as a Competing Endogenous RNA. Front Cell Dev Biol 2021;9:681529. [Crossref] [PubMed]
- Liao Z, Zhang H, Su C, et al. Long noncoding RNA SNHG14 promotes hepatocellular carcinoma progression by regulating miR-876-5p/SSR2 axis. J Exp Clin Cancer Res 2021;40:36. [Crossref] [PubMed]



