
Exploring SSR1 as a novel diagnostic and prognostic biomarker in hepatocellular carcinoma, and its relationship with immune infiltration
Highlight box
Key findings
• Signal sequence receptor subunit 1 (SSR1) serves as a new diagnostic and potentially prognostic biomarker, showing an association with immune cell infiltration and cell proliferation in hepatocellular carcinoma (HCC).
What is known and what is new?
• Although SSR1 has undergone thorough examination in different cancer types, its importance in HCC remains largely unknown.
• Our data suggested the potential correlation between SSR1 and HCC onset and progression, rendering SSR1 a plausible target molecule for HCC therapy.
What is the implication, and what should change now?
• SSR1 emerges as a potential biomarker for HCC diagnosis and prognosis, paving the way for further exploration and potential clinical applications.
Introduction
Among primary liver cancers, hepatocellular carcinoma (HCC) stands as the deadliest malignant tumor and is a leading contributor to cancer-related fatalities globally (1). Notably, HCC exhibits the highest incidence rates in Asia, with a higher prevalence among males (2). Common etiological factors contributing to HCC include alcohol consumption, chronic hepatitis B/C virus infections, chronic liver diseases and cirrhosis resulting from non-alcoholic fatty liver disease (3-5). Despite substantial advancements in liver cancer diagnosis, surgical interventions, and the development of novel molecular targeted therapies, the treatment outcomes for liver cancer remain constrained, diagnostic accuracy remains suboptimal, and live caner patients’ 5-year overall survival (OS) rate continues to languish at a low level (5,6). Moreover, a subset of liver cancer patients has progressed to advanced stages, missing out on optimal treatment opportunities (7-10). Hence, there is an imperative need to expedite the quest for superior diagnostic markers or oncogenic factors associated with liver cancer. This endeavor is crucial for establishing the theoretical framework necessary for the development of precisely targeted therapeutic interventions.
Proteins undergoing synthesis on ribosomes bound to the endoplasmic reticulum (ER) membrane must traverse or integrate into the ER membrane. This process, referred to as ER translocon translocation, necessitates the involvement of a membrane protein complex comprised of multiple subunits localized within the ER (11). The translocon-associated protein (TRAP) complex, alternatively known as the signal sequence receptor (SSR) complex, plays a pivotal role in translocation processes (12,13). The SSR complex physically associates with the heterotrimeric protein-conducting channel Sec61 and has been chemically cross-linked to newly synthesized proteins destined for the ER lumen. This interaction aids in the translocation of proteins bearing signal peptides that may not efficiently interact directly with Sec 61 (12,14-16). The SSR complex comprises four transmembrane subunits, namely SSR1, SSR2, SSR3, and SSR4 (17). Specifically, SSR1, SSR2, and SSR3 are single-spanning membrane proteins of type I (Nlum/Ccyt) with signal peptides, while SSR4 is a multi-spanning membrane protein that traverses the membrane four times and features a prominent cytosolic domain but lacks a signal peptide (18).
SSR1 is ubiquitously present in eukaryotes, where it may involve facilitating critical factors transport in cardiac cushion development, including interferon-γ (IFN-γ) and atrial natriuretic peptide (ANP). These proteins counter inhibitory effects of transforming growth factor (TGF) on formation of mesenchymal cells in endocardial cushions (19-21). Currently, limited research exists on the involvement of SSR1 in cancer initiation and progression. One study suggests that SSR1 could function as a potential biomarker due to its significantly elevated expression in cervical, endometrial, and vulvar cancers (22). Additionally, curcumin treatment has demonstrated inhibition of colorectal cancer cell proliferation and downregulation of SSR1 expression (23). In the context of hypopharyngeal squamous cell carcinoma (HSCC), a study revealed that the long-chain noncoding RNA RP11 156L14.1, acting as competing endogenous RNA (ceRNA), may interact with miR-548a-3p. This interaction interferes with miR-548ao-3p binding to SSR1 3'-untranslated region and subsequently impacts epithelial-mesenchymal transition (EMT) (24). However, investigations into the role of SSR1 in hepatoma cell formation have been relatively scarce.
To our best knowledge, bioinformatics analysis has been extensively to decipher the molecular mechanisms underlying HCC progression (25-27). To explore clinical significance of SSR1 in HCC, we utilized a combination of bioinformatics tools. Additionally, we employed a nomogram and a forest plot to unveil the predictive utility of SSR1 in patients with liver cancer. Our findings deciphered that SSR1 was markedly upregulated in HCC and exhibited associations with factors such as age, pathologic stage, T classification, cancer status, histologic grade, and AFP levels. Notably, SSR1 gene expression emerged as an independent variable influencing HCC patient prognosis. Subsequently, we employed quantitative real-time polymerase chain reaction (qRT-PCR) to validate SSR1 in HCC tissues and cell lines. We assessed SSR1 impact on cell proliferation and migration and conducted preliminary investigations into the molecular mechanisms through which SSR1 may exert its influence on HCC. As a result, our data suggested potential correlation between SSR1 and HCC onset and progression, rendering SSR1 a plausible target molecule for HCC therapy. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-277/rc).
Methods
Data preprocessing
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Gene expression data along with clinical information from liver HCC studies were collected using The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov). Dataset included 50 normal and 374 tumor tissues, with a workflow type of transcripts per kilobase million. Missing values referred to clinical characteristics that were either unavailable or unknown. A summary of the data is presented in Table 1.
Table 1
| Characteristics | Low expression of SSR1 (n=187) | High expression of SSR1 (n=187) | P |
|---|---|---|---|
| T stage (n=371) | 0.009* | ||
| T1 | 107 (28.8) | 76 (20.5) | |
| T2 | 39 (10.5) | 56 (15.1) | |
| T3 | 35 (9.4) | 45 (12.1) | |
| T4 | 4 (1.1) | 9 (2.4) | |
| N stage (n=258) | 0.62 | ||
| N0 | 128 (49.6) | 126 (48.8) | |
| N1 | 1 (0.4) | 3 (1.2) | |
| M stage (n=272) | 0.37 | ||
| M0 | 137 (50.4) | 131 (48.2) | |
| M1 | 1 (0.4) | 3 (1.1) | |
| Pathologic stage (n=350) | 0.02* | ||
| Stage I | 101 (28.9) | 72 (20.6) | |
| Stage II | 37 (10.6) | 50 (14.3) | |
| Stage III | 35 (10) | 50 (14.3) | |
| Stage IV | 2 (0.6) | 3 (0.9) | |
| Tumor status (n=355) | 0.37 | ||
| Tumor-free | 106 (29.9) | 96 (27) | |
| With tumor | 72 (20.3) | 81 (22.8) | |
| Gender (n=374) | 0.08 | ||
| Female | 52 (13.9) | 69 (18.4) | |
| Male | 135 (36.1) | 118 (31.6) | |
| Race (n=362) | 0.70 | ||
| Asian | 75 (20.7) | 85 (23.5) | |
| Black or African American | 8 (2.2) | 9 (2.5) | |
| White | 95 (26.2) | 90 (24.9) | |
| Age (n=373) | 0.03* | ||
| ≤60 years | 78 (20.9) | 99 (26.5) | |
| >60 years | 109 (29.2) | 87 (23.3) | |
| Weight (n=346) | 0.06 | ||
| ≤70 kg | 85 (24.6) | 99 (28.6) | |
| >70 kg | 92 (26.6) | 70 (20.2) | |
| Height (n=341) | 0.02* | ||
| <170 cm | 91 (26.7) | 110 (32.3) | |
| ≥170 cm | 82 (24) | 58 (17) | |
| BMI (n=337) | 0.61 | ||
| ≤25 kg/m2 | 87 (25.8) | 90 (26.7) | |
| >25 kg/m2 | 84 (24.9) | 76 (22.6) | |
| Residual tumor (n=345) | 0.62 | ||
| R0 | 172 (49.9) | 155 (44.9) | |
| R1 | 8 (2.3) | 9 (2.6) | |
| R2 | 0 (0) | 1 (0.3) | |
| Histologic grade (n=369) | 0.006* | ||
| G1 | 35 (9.5) | 20 (5.4) | |
| G2 | 97 (26.3) | 81 (22) | |
| G3 | 49 (13.3) | 75 (20.3) | |
| G4 | 4 (1.1) | 8 (2.2) | |
| Adjacent hepatic tissue inflammation (n=237) | 0.33 | ||
| None | 64 (27) | 54 (22.8) | |
| Mild | 49 (20.7) | 52 (21.9) | |
| Severe | 12 (5.1) | 6 (2.5) | |
| AFP (n=280) | 0.002* | ||
| ≤400 ng/mL | 121 (43.2) | 94 (33.6) | |
| >400 ng/mL | 22 (7.9) | 43 (15.4) | |
| Albumin (n=300) | 0.71 | ||
| <3.5 g/dL | 34 (11.3) | 35 (11.7) | |
| ≥3.5 g/dL | 122 (40.7) | 109 (36.3) | |
| Prothrombin time (n=297) | >0.99 | ||
| ≤4 s | 106 (35.7) | 102 (34.3) | |
| >4 s | 46 (15.5) | 43 (14.5) | |
| Child-Pugh grade (n=241) | 0.36 | ||
| A | 119 (49.4) | 100 (41.5) | |
| B | 9 (3.7) | 12 (5) | |
| C | 1 (0.4) | 0 (0) | |
| Fibrosis Ishak score (n=215) | 0.96 | ||
| 0 | 39 (18.1) | 36 (16.7) | |
| 1/2 | 15 (7) | 16 (7.4) | |
| 3/4 | 15 (7) | 13 (6) | |
| 5/6 | 40 (18.6) | 41 (19.1) | |
| Vascular invasion (n=318) | 0.12 | ||
| No | 115 (36.2) | 93 (29.2) | |
| Yes | 50 (15.7) | 60 (18.9) | |
| OS event (n=374) | 0.16 | ||
| Alive | 129 (34.5) | 115 (30.7) | |
| Dead | 58 (15.5) | 72 (19.3) | |
| DSS event (n=366) | 0.45 | ||
| Alive | 147 (40.2) | 140 (38.3) | |
| Dead | 36 (9.8) | 43 (11.7) | |
| PFI event (n=374) | 0.84 | ||
| Alive | 97 (25.9) | 94 (25.1) | |
| Dead | 90 (24.1) | 93 (24.9) | |
| Age (years) | 64 [52, 69] | 59 [51, 68] | 0.11 |
Data are presented as n (%) or median [IQR]. *, P<0.05. AFP, alpha-fetoprotein; BMI, body mass index; DSS, disease-specific survival; IQR, interquartile range; OS, overall survival; PFI, progression-free interval; SSR1, signal sequence receptor subunit 1.
ONCOMINE database
ONCOMINE dataset (www.oncomine.org) is an openly accessible online cancer microarray database utilized for validating SSR1 in cancer. A comparison of SSR1 transcriptional levels in liver cancer tissues and normal controls was conducted using Student’s t-test, with critical values: fold change >1.5 and P<0.05 (28,29).
Tumor IMmune Estimation Resource (TIMER)
TIMER (https://cistrome.shinyapps.io/timer/) serves as an online tool for systematically analyzing clinical impact of six immune cell types in diverse cancer types. The database distinctly illustrates the variation in SSR1 between cancer and para-cancerous tissues (30).
Differential expression analysis of SSR1 in the TCGA database
The SSR1 expression levels in normal liver tissues, HCC tumor samples, and adjacent non-tumor samples were analyzed and compared using Student’s t-test. Additionally, the SSR1 expression levels in different HCC stages including T stage, pathological stage, histological grade, gender, vascular invasion, weight, height, tumor status, and AFP level were analyzed using the ggplot2 package in R.
SSR1 protein expression analysis using the Human Protein Atlas (HPA) database
SSR1 protein expression in normal liver tissue and HCC tissue via immunohistochemical staining was determined by searching the online HPA (http://www.proteinatlas.org/) database.
Survival analysis of SSR1 in the TCGA database
Survival analysis was conducted using the Kaplan-Meier method and the Cox regression model. The Kaplan-Meier method assessed survival in patient groups with low and high SSR1 expression across various clinical subgroups. The Cox regression model analyzed the impact of multiple factors, including T staging, pathological staging, histological grading, gender, and age, on the survival of HCC patients.
Nomogram and diagnostic efficacy
A nomogram was created by combining scores of individual prognostic factor. HCC patients’ OS was analyzed with survival and rms R packages. Calibration plots were employed to evaluate predictive accuracy of the nomogram. Diagnostic and predictive capabilities of SSR1 were assessed by generating ROC curves with pROC package and calculating area under the curve (AUC). An AUC greater than 0.8 indicates satisfactory discriminative ability.
Immune infiltration analysis
TIMER tool was employed to examine the correlation between gene expression levels in TCGA dataset and immune cell presence. Gene set variation analysis (GSVA) R package and TIMER tool were used to assess SSR1 expression levels across various immune cells and their correlation with immune infiltration extent. Statistical significance was defined as P<0.05 and a correlation coefficient greater than 0.3.
Functional enrichment analysis
The ClusterProfiler package in R was used to functionally annotate SSR1-related genes according to Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Gene set enrichment analysis (GSEA)
GSEA, a computerized technique, assesses statistical significance and consistent differences in a predetermined set of genes between two biological states. Genes associated with SSR1 expression from recent research findings was identified by GSEA. Additionally, it was utilized to explore survival differences between high- and low-SSR1 groups. Analysis involved 1,000 gene set permutations. Enriched gene sets were considered significant with P<0.05 and a false discovery rate <25%.
Cell culture and transfection
THLE-2, LO2 and seven HCC cell lines (QGY-7703, BEL-7404, Hep3B, MHCC97L, SMMC-7721, SK-hep-1 and HepG2) were purchased from the Cell Resource Center, Chinese Academy of Science Committee (Shanghai, China). Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum (FBS) (GIBCO, Grand Island, USA), 100 µg/mL streptomycin, and 100 U/mL penicillin was used to culture these cells in a humidified incubator containing 5% (v/v) of CO2, at 37 ℃. SSR1 siRNA duplexes (5'-AGAAAACAAGGGUUUUGGCAA-3') were sourced from RiboBio company (Guangzhou, China). Transfection of QGY-7703 and SMMC-7721 with 100 nM siRNA was conducted using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA). Confirmation of RNA interference was performed through qRT-PCR 72 hours later.
qRT-PCR
TRIzol reagent (Invitrogen, Carlsbad, CA) was employed for RNA extraction from liver cancer, adjacent normal tissues, and HCC cells. GoScriptTM Reverse Transcription System (Promega) was utilized to synthesize complementary DNA (cDNA). qRT-PCR was carried out with SYBR Green (Promega, Madison, USA) in Roche LightCycler 96 (Roche Applied Science, Penzberg, Germany), with each reaction performed in triplicate wells. Specific primers: 5'-CTGCTTCTCTTACTCGTGTTCC-3' (F) and 5'-TCTTCTTCTACCTCGGCTTCAT-3' (R) for SSR1, 5'-CGAGAGCTACACGTTCACGG-3' (F) and 5'-GGGTGTCGAGGGAAAAATAGG-3' (R) for E-cadherin, 5'-TCAGGCGTCTGTAGAGGCTT-3' (F) and 5'-ATGCACATCCTTCGATAAGACTG-3' (R) for N-cadherin, 5'-GACGCCATCAACACCGAGTT-3' (F) and 5'-CTTTGTCGTTGGTTAGCTGGT-3' (R) for Vimentin, and 5'-GAGCGAGATCCCTCCAAAT-3' (F) and 5'-GCTGTTGCATACTTCTCATG-3' (R) for GAPDH. The relative expression of SSR1 was calculated using the 2−ΔΔCt method.
Cell Counting Kit-8 (CCK-8) assay
SMMC-7721 and QGY-7703 liver cancer cells were transfected with SSR1-siRNA and negative control siRNAs for 48 hours. Afterward, 10 µL of CCK8 solution (CCK-8, Dojindo, Japan) was added per well 2 hours before the end of each 37 ℃ incubation cycle every 24 hours. Viable cell counts were determined daily measuring 450 nm absorbance with a plate reader (BITELX800, BiTek, Boston, USA).
5-Ethynyl-2'-deoxyuridine (EdU) cell proliferation assay
The EdU cell proliferation detection kit was obtained from RiboBio Biotechnology Co., Ltd. (Guangzhou, China). Briefly, the cells were fixed in 4% paraformaldehyde and permeabilized with 0.3% Triton X-100. Afterwards, cells were incubated with 50 µM EdU for 2 hours followed by counterstaining the cell nuclei with Hoechst 33342. EdU-positive nuclei were detected using a fluorescence microscope (Leica, Wetzlar, Germany). The cell proliferation rate was calculated based on the proportion of nucleated cells incorporating EdU.
Wound healing
About 4.0×105 cells were cultured to attain >80% confluence. Following this, pipette tip (200 µL) was employed to longitudinally scrape the center of the well’s bottom. Detached cells were washed away before being placed in serum-free media. Images were captured at 0 and 48 hours post-wounding.
Transwell migration assay
The migration ability of the cells was evaluated using 24-well Transwell plates (Corning; Corning Inc., Corning, NY, USA). The lower chamber was filled with 800 µL of medium containing 10% FBS, while the upper chamber received 200 µL of serum-free medium with 6×104 transfected cells. After 48 hours, the non-migrated cells in the upper chamber were discarded, and the cells that had migrated to the lower chamber were fixed with crystal violet. The stained cells were then counted under a light microscope.
Statistical analysis
Box plots were employed to assess SSR1 levels in HCC patients. Relationship between clinical characteristics and SSR1 expression in HCC was investigated using Wilcoxon signed-rank test and logistic regression. Kaplan-Meier with log-rank test P value was utilized to compare OS rates between high and low SSR1 level groups. SSR1 diagnostic value was determined through a receiver operating characteristic (ROC) curve, with the area under the ROC curve indicating diagnostic efficacy. Univariate Cox analysis tested for potential prognostic factors, while multivariate Cox analysis confirmed SSR1 on survival alongside other clinical variables. A nomogram integrating SSR1 expression and clinical factors was created to predict HCC OS. R statistical software (version 3.5.3) or SPSS software (version 24.0) was employed for all statistical analyses, with significance defined as P<0.05.
Results
Baseline characteristics
Patients (n=374) exhibiting necessary clinical features were obtained from TCGA. Clinical characteristics are outlined in Table 1. Within 374 subjects, male were 253 (67.6%), and female were 121 (32.4%). Among them, 177 patients (47.3%) were aged 60 years or younger, while 196 patients (52.4%) were older than 60 years. Regarding HCC stage, 173 patients were classified as stage I (46.3%), 87 as stage II (23.3%), 85 as stage III (22.7%), and 5 as stage IV (1.3%). Cancer status comprised 202 tumor-free patients (56.9%) and 153 patients with tumors (43.1%).
High SSR1 expression in HCC
First, ONCOMINE was used to search for SSR1 expression levels. SSR1 was shown to be more overexpressed in HCC as compared to adjacent liver tissue (Roessler Liver, Figure 1A,1B). On the TIMER, SSR1 was overexpressed in HCC tissue (Figure 1C). Downloaded TCGA data is then used for further verification [liver hepatocellular carcinoma (LICH), TCGA]. SSR1 level in HCC tissue was markedly higher (P<0.001) than normal ones (Figure 1D). Findings were confirmed in HCC samples and matched normal liver tissue (P<0.001) (Figure 1E). Additionally, SSR1 protein was also highly expressed in HCC tissues compared to normal liver tissues as revealed by analysis of HPA database (Figure 1F).
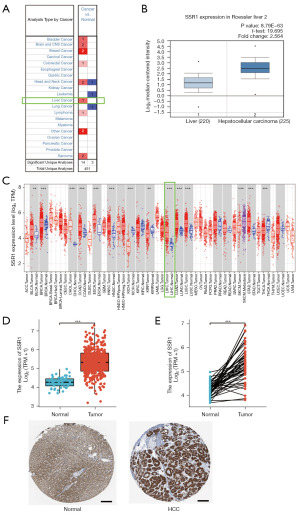
Correlation between SSR1 and clinical features
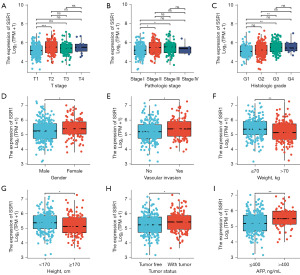
Table 2 summarizes association observed between SSR1 and clinical features in HCC patients. Elevated SSR1 showed significant correlations with age (P=0.03), T stage (P=0.001), histologic grade (P=0.001), pathologic stage (P=0.002), cancer status (P=5.51e−04) and AFP levels (P=0.002). Additionally, high SSR1 expression was linked with histologic grade, T stage, gender, vascular invasion, weight, height, tumor status, pathologic stage and AFP levels (P<0.05), as illustrated in Figure 2.
Table 2
| Characteristics | Total, N | OR (95% CI) | P value |
|---|---|---|---|
| T stage (T2 & T3 & T4 vs. T1) | 371 | 1.985 (1.316–3.009) | 0.001* |
| N stage (N1 vs. N0) | 258 | 3.048 (0.384–62.060) | 0.34 |
| M stage (M1 vs. M0) | 272 | 3.137 (0.396–63.865) | 0.33 |
| Pathologic stage (stage II & stage III & stage IV vs. stage I) | 350 | 1.953 (1.279–2.995) | 0.002* |
| Tumor status (with tumor vs. tumor-free) | 355 | 1.242 (0.816–1.894) | 0.31 |
| Gender (female vs. male) | 374 | 1.518 (0.983–2.356) | 0.06 |
| Age (>60 vs. ≤60 years) | 373 | 0.629 (0.417–0.946) | 0.03* |
| BMI (>25 vs. ≤25 kg/m2) | 337 | 0.875 (0.570–1.342) | 0.54 |
| Residual tumor (R1 & R2 vs. R0) | 345 | 1.387 (0.534–3.718) | 0.50 |
| Histologic grade (G3 & G4 vs. G1 & G2) | 369 | 2.047 (1.334–3.162) | 0.001* |
| Adjacent hepatic tissue inflammation (mild & severe vs. none) | 237 | 1.127 (0.676–1.880) | 0.65 |
| AFP (>400 vs. ≤400 ng/mL) | 280 | 2.516 (1.422–4.557) | 0.002* |
| Child-Pugh grade (B & C vs. A) | 241 | 1.428 (0.592–3.517) | 0.43 |
| Vascular invasion (yes vs. no) | 318 | 1.484 (0.934–2.367) | 0.10 |
| Fibrosis Ishak score (3/4&5/6 vs. 0&1/2) | 215 | 1.020 (0.597–1.742) | 0.94 |
| Albumin (≥3.5 vs. <3.5 g/dL) | 300 | 0.868 (0.506–1.488) | 0.61 |
| Prothrombin time (>4 vs. ≤4 s) | 297 | 0.971 (0.590–1.597) | 0.91 |
| Race (Black or African American & White vs. Asian) | 362 | 0.848 (0.559–1.284) | 0.44 |
| Weight (>70 vs. ≤70 kg) | 346 | 0.653 (0.426–0.998) | 0.050 |
| Height (≥170 vs. <170 cm) | 341 | 0.585 (0.377–0.903) | 0.02* |
*, P<0.05. AFP, alpha-fetoprotein; BMI, body mass index; CI, confidence interval; OR, odds ratio; SSR1, signal sequence receptor subunit 1.
SSR1 is an independent OS risk factor
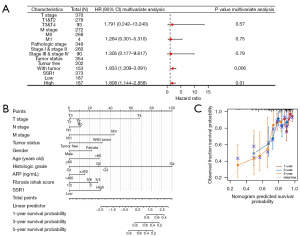
Kaplan-Meier survival analysis revealed that elevated SSR1 was linked to poorer prognosis (P=0.01), as depicted in Figure 3A. Subgroup analysis demonstrated a significant association between high SSR1 and poor prognosis in T1–3 stages (P=0.04), T3–4 stage (P=0.049), pathologic stage I–IV (P=0.03), pathologic stage I–III (P=0.03), histologic grade G1–3 (P=0.003), histologic grade G1 (P=0.007), male (P=0.001), and age >60 years old (P=0.04), as illustrated in Figure 3B-3I. Univariate Cox analysis demonstrated a significant correlation between high SSR1 and poor OS [hazard ratio (HR) =1.554, 95% CI: 1.098–2.201, P=0.01]. Furthermore, SSR1 was an independent OS risk factor in HCC patients (HR =1.808, 95% CI: 1.144–2.858, P=0.01), as presented in Table 3 and Figure 4A. Consequently, nomogram was developed to predict OS by combining SSR1 with clinical variables (Figure 4B,4C).
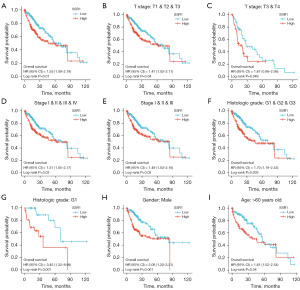
Table 3
| Characteristics | Total, N | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |||
| T stage | 371 | |||||
| T1 & T2 | 278 | Reference | ||||
| T3 & T4 | 93 | 2.598 (1.826–3.697) | <0.001* | 1.791 (0.242–13.240) | 0.57 | |
| N stage | 258 | |||||
| N0 | 254 | Reference | – | – | ||
| N1 | 4 | 2.029 (0.497–8.281) | 0.32 | – | – | |
| M stage | 272 | |||||
| M0 | 268 | Reference | ||||
| M1 | 4 | 4.077 (1.281–12.973) | 0.02* | 1.264 (0.301–5.310) | 0.75 | |
| Pathologic stage | 350 | |||||
| Stage I & stage II | 260 | Reference | ||||
| Stage III & stage IV | 90 | 2.504 (1.727–3.631) | <0.001* | 1.305 (0.177–9.617) | 0.79 | |
| Tumor status | 355 | |||||
| Tumor-free | 202 | Reference | ||||
| With tumor | 153 | 2.317 (1.590–3.376) | <0.001* | 1.933 (1.209–3.091) | 0.006* | |
| Gender | 374 | |||||
| Male | 253 | Reference | – | – | ||
| Female | 121 | 1.261 (0.885–1.796) | 0.20 | – | – | |
| Race | 362 | |||||
| Asian & Black or African American | 177 | Reference | – | – | ||
| White | 185 | 1.265 (0.881–1.816) | 0.20 | – | – | |
| Age | 373 | |||||
| ≤60 years | 177 | Reference | – | – | ||
| >60 years | 196 | 1.205 (0.850–1.708) | 0.30 | – | – | |
| Residual tumor | 345 | |||||
| R0 | 327 | Reference | – | – | ||
| R1 & R2 | 18 | 1.604 (0.812–3.169) | 0.17 | – | – | |
| BMI | 337 | |||||
| ≤25 kg/m2 | 177 | Reference | – | – | ||
| >25 kg/m2 | 160 | 0.798 (0.550–1.158) | 0.24 | – | – | |
| Histologic grade | 369 | |||||
| G1 | 55 | Reference | – | – | ||
| G2 | 178 | 1.162 (0.686–1.969) | 0.58 | – | – | |
| G3 | 124 | 1.185 (0.683–2.057) | 0.55 | – | – | |
| G4 | 12 | 1.681 (0.621–4.549) | 0.31 | – | – | |
| Adjacent hepatic tissue inflammation | 237 | |||||
| None | 118 | Reference | – | – | ||
| Mild | 101 | 1.204 (0.723–2.007) | 0.48 | – | – | |
| Severe | 18 | 1.144 (0.447–2.930) | 0.78 | – | – | |
| AFP | 280 | |||||
| ≤400 ng/mL | 215 | Reference | – | – | ||
| >400 ng/mL | 65 | 1.075 (0.658–1.759) | 0.77 | – | – | |
| Child-Pugh grade | 241 | |||||
| A | 215 | Reference | – | – | ||
| B & C | 22 | 1.643 (0.811–3.330) | 0.17 | – | – | |
| Fibrosis Ishak score | 214 | |||||
| 0 | 75 | Reference | – | – | ||
| 1/2 | 31 | 0.935 (0.437–2.002) | 0.86 | – | – | |
| 3/4 | 28 | 0.698 (0.288–1.695) | 0.43 | – | – | |
| 5/6 | 81 | 0.737 (0.410–1.325) | 0.31 | – | – | |
| Vascular invasion | 318 | |||||
| No | 208 | Reference | – | – | ||
| Yes | 110 | 1.344 (0.887–2.035) | 0.16 | – | – | |
| SSR1 | 374 | |||||
| Low | 187 | Reference | ||||
| High | 187 | 1.554 (1.098–2.201) | 0.01* | 1.808 (1.144–2.858) | 0.01* | |
*, P<0.05. HR, hazard ratio; CI, confidence interval; BMI, body mass index; AFP, alpha-fetoprotein; SSR1, signal sequence receptor subunit 1.
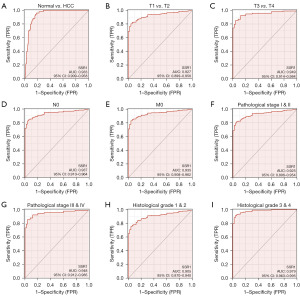
SSR1 diagnostic value in HCC
AUC was 0.933, indicating a good diagnostic value, as depicted in Figure 5A. Subgroup analysis further highlighted diagnostic value of SSR1 in various HCC features, with AUC of 0.927 for T1–2 stage, 0.949 for T3–4 stage, 0.937 for N0, 0.935 for M0, 0.925 for pathological stage I–II, 0.948 for pathological stage III–IV, 0.905 for histological grade 1–2, and 0.979 for histological grade 3–4 (Figure 5B-5I).
SSR1-related genes in HCC
To anticipate the functional enrichment details of genes interacting with SSR1, we conducted GO&KEGG enrichment analysis. The results revealed that genes associated with SSR1 were engaged in numerous biological processes (BPs) and cellular components (CCs), encompassing digestion, integrator complex, and catenin complex, as illustrated in Table 4.
Table 4
| Ontology | ID | Description | Gene ratio | Bg ratio | P value | P.adjust | q-value |
|---|---|---|---|---|---|---|---|
| BP | GO:0007586 | Digestion | 8/127 | 139/18,670 | 4.91e−06 | 0.008 | 0.008 |
| CC | GO:0032039 | Integrator complex | 3/137 | 28/19,717 | 9.47e−04 | 0.09 | 0.080 |
| CC | GO:0016342 | Catenin complex | 3/137 | 29/19,717 | 0.001 | 0.09 | 0.080 |
Gene sets with NOM P value <0.05 and FDR q-value <0.25 were considered as significantly enriched. SSR1, signal sequence receptor subunit 1; GO, Gene Ontology; CC, cellular component; BP, biological process; NOM, nominal; FDR, false discovery rate.
GSEA analysis
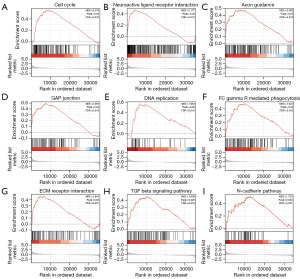
Utilizing the low- and high-SSR1 expression datasets, we conducted GSEA to pinpoint signaling pathways activated in HCC. GSEA revealed a substantial contrast in MSigDB collection enrichment as outlined in Table 5. Gene sets associated with “cell cycle”, “neuroactive ligand receptor interaction”, “axon guidance”, “gap junction”, “DNA replication”, “gamma R mediated phagocytosis”, “ECM receptor interaction”, “TGF-β signaling pathway”, and” N-cadherin pathway” in cancer exhibited distinct enrichment in high SSR1 level phenotype, as depicted in Figure 6.
Table 5
| Gene set name | Size | ES | NES | P.adjust | q-value |
|---|---|---|---|---|---|
| KEGG_CELL_CYCLE | 124 | 0.55593464 | 2.37597553 | 0.02227843 | 0.01531513 |
| KEGG_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | 270 | 0.46818856 | 2.17666659 | 0.02227843 | 0.01531513 |
| KEGG_AXON_GUIDANCE | 129 | 0.47694547 | 2.05459499 | 0.02227843 | 0.01531513 |
| KEGG_GAP_JUNCTION | 90 | 0.4916483 | 1.98700116 | 0.02227843 | 0.01531513 |
| KEGG_DNA_REPLICATION | 36 | 0.57337191 | 1.96390079 | 0.02227843 | 0.01531513 |
| KEGG_FC_GAMMA_R_MEDIATED_PHAGOCYTOSIS | 96 | 0.47317723 | 1.94104323 | 0.02227843 | 0.01531513 |
| KEGG_ECM_RECEPTOR_INTERACTION | 83 | 0.46085435 | 1.84094866 | 0.03080098 | 0.02117388 |
| KEGG_TGF_BETA_SIGNALING_PATHWAY | 85 | 0.45887259 | 1.83880859 | 0.02562905 | 0.01761848 |
| PID_NCADHERIN_PATHWAY | 33 | 0.521961501 | 1.72023064 | 0.03885773 | 0.02734905 |
Gene sets with P value <0.05 and q-value <0.05 were considered as significantly enriched. SSR1, signal sequence receptor subunit 1; GSEA, gene set enrichment analysis; ES, enrichment score; NES, normalized enrichment score; KEGG, Kyoto Encyclopedia of Genes and Genomes; PID, Pathway Interaction Database.
SSR1 expression and immune infiltration
We assessed correlation between SSR1 and quantified immune cell infiltration level using single sample GSEA (ssGSEA), employing Spearman correlation. SSR1 exhibited a negative correlation with cytotoxic cells, dendritic cells, and plasmacytoid dendritic cells, while showing a positive correlation with Th2, follicular helper T, and T helper cells, as illustrated in Figure 7 and detailed in Table 6 (P<0.001).
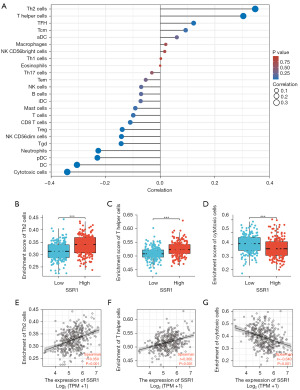
Table 6
| Molecular | Cells | Pearson | Spearman | |||
|---|---|---|---|---|---|---|
| Correlation coefficient | P value | Correlation coefficient | P value | |||
| SSR1 | aDC | 0.075 | 0.15 | 0.061 | 0.24 | |
| SSR1 | B cells | −0.049 | 0.34 | −0.070 | 0.17 | |
| SSR1 | CD8 T cells | −0.109 | 0.04 | −0.110 | 0.03 | |
| SSR1 | Cytotoxic cells | −0.328 | <0.001 | −0.341 | <0.001 | |
| SSR1 | DC | −0.293 | <0.001 | −0.305 | <0.001 | |
| SSR1 | Eosinophils | 0.006 | 0.91 | −0.000 | 0.99 | |
| SSR1 | iDC | −0.051 | 0.33 | −0.072 | 0.17 | |
| SSR1 | Macrophages | 0.051 | 0.33 | 0.020 | 0.70 | |
| SSR1 | Mast cells | −0.104 | 0.044 | −0.092 | 0.08 | |
| SSR1 | Neutrophils | −0.199 | <0.001 | −0.227 | <0.001 | |
| SSR1 | NK CD56bright cells | 0.018 | 0.73 | 0.016 | 0.76 | |
| SSR1 | NK CD56dim cells | −0.116 | 0.03 | −0.142 | 0.006 | |
| SSR1 | NK cells | −0.029 | 0.58 | −0.070 | 0.18 | |
| SSR1 | pDC | −0.253 | <0.001 | −0.231 | <0.001 | |
| SSR1 | T cells | −0.073 | 0.16 | −0.099 | 0.056 | |
| SSR1 | T helper cells | 0.341 | <0.001 | 0.304 | <0.001 | |
| SSR1 | Tcm | 0.104 | 0.044 | 0.093 | 0.07 | |
| SSR1 | Tem | −0.021 | 0.69 | −0.053 | 0.30 | |
| SSR1 | Tfh | 0.128 | 0.01 | 0.122 | 0.02 | |
| SSR1 | Tgd | −0.100 | 0.054 | −0.143 | 0.005 | |
| SSR1 | Th1 cells | −0.010 | 0.85 | 0.002 | 0.97 | |
| SSR1 | Th17 cells | −0.052 | 0.31 | −0.031 | 0.55 | |
| SSR1 | Th2 cells | 0.359 | <0.001 | 0.348 | <0.001 | |
| SSR1 | Treg | −0.168 | 0.001 | −0.140 | 0.007 | |
SSR1, signal sequence receptor subunit 1; HCC, hepatocellular carcinoma; aDC, activated dendritic cell; DC, dendritic cell; iDC, immature dendritic cell; NK, natural killer; pDC, plasmacytoid dendritic cell; Tcm, central memory T cell; Tem, effector memory T cell; Tfh, follicular helper T cell; Tgd, γ/δ T cell; Th, T helper cell; Treg, regulatory T cell.
SSR1 in HCC tissues and cell lines
We assessed SSR1 expression in seven liver cancer cell lines, along with two normal hepatic cell lines. A higher level of SSR1 in liver cancer cell lines was observed comparing to normal ones (Figure 8A). Notably, SSR1 in the SMMC-7721 and QGY-7703 liver cancer cell lines was notably higher than in other liver cancer cell lines. Consequently, SMMC-7721 and QGY-7703 were selected for subsequent experiments.
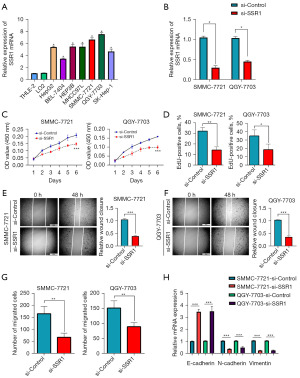
SSR1 promotes HCC proliferation/migration
To investigate SSR1 impact on cell proliferation/migration, we transfected distinct siRNAs into the SMMC-7721 and QGY-7703 cell lines. Knockdown efficacy was validated through qRT-PCR (Figure 8B), revealing a significant reduction in SSR1 expression post-si-SSR1 transfection. SSR1 in cell proliferation was assessed using CCK-8 and EdU cell proliferation assay, demonstrating a marked inhibition of proliferation in SMMC-7721 and QGY-7703 cells following SSR1 knockdown (Figure 8C,8D). Wound-healing and Transwell migration assays were conducted to evaluate SSR1 impact on cell migration, indicating a substantial decrease in migration ability for SMMC-7721 and QGY-7703 cells upon SSR1 knockdown (Figure 8E-8G). To elucidate the underlying mechanism, relevant signaling pathway proteins including N-cadherin, Vimentin, and E-cadherin were determined by qRT-PCR. SSR1 knockdown significantly attenuated EMT (Figure 8H).
Discussion
Protein translocation refers to the movement of proteins between cellular compartments. After translation, proteins have to be transported from the ribosome across the ER, from where they form part of other cell organelles, localized to the membrane or are secreted (31). Translocation to the ER occurs via the translocon, a protein conducting channel created by Sec61 and related proteins such as the SSR, also known as the TRAP complex (31,32). SSR complex, located on the ER membrane and is associated with channel protein Sec61 as a heterotetramer, is critical in defining the orientation of membrane proteins that lack strong hydrophobic signal sequences and require help translocating and localizing to the ER membrane (32). SSR consists of four subunits, namely SSR1–4 (33).
The expression and functional implications of the SSR1 gene in various diseases have recently gained attention. A study demonstrates that reduced SSR1 expression inhibits pre-insulin translocation, leading to a significant decrease in insulin secretion in type 2 diabetes patients (17). Additionally, SSR1 overexpression in glioma has been linked to the regulation of the JAK/STAT signaling pathway (34). In the context of HSCC, SSR1 expression is significantly elevated in tumor tissues/cells (24). However, SSR1 in HCC prognosis remains unclear. This study analyzed SSR1 expression profiles in various human solid tumors using ONCOMINE/TIMER databases. Higher SSR1 expression was observed in HCC tumor tissues compared to normal ones. This finding was validated in resected HCC tissues and cell lines.
SSR1 clinical significance in HCC was comprehensively elucidated in this study. High SSR1 level was correlated with various clinical factors such as age, pathologic stage, T classification, cancer status, histologic grade, and AFP levels. Further analysis identified SSR1 as an independent prognostic factor for HCC patients OS. Constructed forest plot highlighted the relevance of SSR1 expression and tumor status to HCC poor prognosis. Kaplan-Meier survival further supported the conclusion that SSR1 overexpression is indicative of a poor prognosis. Multiple independent datasets subjected to ROC analysis demonstrated the upper-middle diagnostic ability of SSR1. A predictive nomogram for HCC based on SSR1 expression and clinical factors was developed using the TCGA dataset, offering a tool for predicting individual patient mortality.
Previous reports have linked SSR family molecules, particularly SSR2, to the tumorigenesis of HCC. SSR2 overexpression in HCC tumor tissues and its involvement in modulating EMT signaling pathways have been reported (33). Molecular processes and functions of SSR1 in HCC remain elusive. Consequently, we conducted GO and GSEA to explore genes with similar patterns to SSR1 in HCC. The GO analysis indicated that heightened SSR1 expression primarily participated in cellular components like the integrator and catenin complex. Moreover, GSEA results revealed an association between the high SSR1 expression phenotype and pathways involving the cell cycle, DNA replication, and the N-cadherin pathway in HCC. Notably, both N-cadherin and catenin are molecules linked to EMT signaling pathway (35,36). Recently, SEC61G (Sec61), a component of the SEC61 complex, was identified as overexpressed in kidney cancer tissues, predicting a poor prognosis. Knocking down Sec61G significantly downregulated N-cadherin and -catenin (37). Given the reported physical connection between the SEC61 complex and SSR complex using biochemical methods and chemical crosslinking during transit into ER lumen (15,38), we hypothesize that SSR1 may be related to EMT signaling pathway. Following the successful establishment of an SSR1 knockdown cell model, we observed decreased proliferation and migration abilities of hepatoma cells. Subsequently, we conducted qRT-PCR to evaluate the expression levels of EMT pathway-related molecules. The results indicated that upregulated SSR1 in HCC contributes to progression and metastasis by modulating EMT signaling pathway.
TME exerts significant influence on epigenetics, immune evasion, tumor differentiation and metastasis. Generally, TME is an intricate milieu consisting of diverse cell types and various chemicals released by stromal, immune and tumor cells (39). Among these components, tumor-infiltrating lymphocytes (TILs) have emerged as pivotal contributors to the development, occurrence, and treatment of HCC (40). TILs enhance the immunosuppressive microenvironment, facilitating immune escape by establishing intricate intercellular interaction networks that promote tumor progression (41). Our findings indicated an elevated presence of Th2 cells and T helper cells in the SSR1-overexpressed group. T helper cells play a crucial role in the immune system (42). These cells differentiate into two main subtypes, Th1 and Th2 cells. Th2 cells regulate eosinophils, basophils, mast cells, and B cells orchestrating immune responses, primarily humoral immunity (43). A study comparing Th1 and Th2 cytokines in hepatitis C virus (HCV)-related HCC patients with normal controls revealed significantly reduced Th1 cytokines and increased Th2 cytokine, suggesting that Th1/Th2 imbalance diminishes anti-tumor immunity, potentially playing a role in the development of HCV-related HCC (44). CD8+ T-cells eliminate cells expressing abnormal surface phenotypes due to intrinsic changes, including cancer cells, infected cells (especially viruses), and injured cells (45). Cohort analysis involving 446 HCC cases demonstrated that a high density of CD8+ cells in tumor area correlated positively with improved OS, disease-free survival, and lower recurrence rates (46). In line with these findings, our research revealed a reduced number of cytotoxic cells in HCC tumor tissues with high SSR1 expression, indicating suppressed adaptive immunity in these tumor tissues, thereby facilitating the proliferation and development of liver cancer cells. In summary, our research underscores the close relationship between SSR1 expression levels and the tumor microenvironment in liver cancer, prompting the need for further in-depth investigations.
Nonetheless, our study faces certain limitations. Firstly, it relies on a retrospective cohort analysis, necessitating prospective studies with larger sample sizes to validate our findings. Secondly, the relationship between SSR1 and immune infiltration requires further validation in animal models of HCC. Lastly, despite demonstrating SSR1’s influence on EMT, the molecular mechanisms underlying SSR1’s impact on the prognosis of HCC patients need additional validation.
Conclusions
In conclusion, our research suggests that SSR1 may be essential in HCC progression by regulating EMT pathway. SSR1 emerges as a potential biomarker for HCC diagnosis and prognosis, paving the way for further exploration and potential clinical applications.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-277/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-277/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-277/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-277/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69:363-85. [Crossref] [PubMed]
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917. [Crossref] [PubMed]
- Rinella ME. Nonalcoholic fatty liver disease: a systematic review. JAMA 2015;313:2263-73. [Crossref] [PubMed]
- Batey RG, Burns T, Benson RJ, et al. Alcohol consumption and the risk of cirrhosis. Med J Aust 1992;156:413-6. [Crossref] [PubMed]
- Balogh J, Victor D 3rd, Asham EH, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma 2016;3:41-53. [Crossref] [PubMed]
- Zhao W, Liu C, Wu Y, et al. Transarterial chemoembolization (TACE)-hepatic arterial infusion chemotherapy (HAIC) combined with PD-1 inhibitors plus lenvatinib as a preoperative conversion therapy for nonmetastatic advanced hepatocellular carcinoma: a single center experience. Transl Cancer Res 2024;13:2315-31. [Crossref] [PubMed]
- Huang Y, Wang H, Lian Y, et al. Upregulation of kinesin family member 4A enhanced cell proliferation via activation of Akt signaling and predicted a poor prognosis in hepatocellular carcinoma. Cell Death Dis 2018;9:141. [Crossref] [PubMed]
- Riehle KJ, Vasudevan SA, Bondoc A, et al. Surgical management of liver tumors. Pediatr Blood Cancer 2024; Epub ahead of print. [Crossref] [PubMed]
- Li Z, Duan D, Li L, et al. Tumor-associated macrophages in anti-PD-1/PD-L1 immunotherapy for hepatocellular carcinoma: recent research progress. Front Pharmacol 2024;15:1382256. [Crossref] [PubMed]
- Ding DY, Jiang SY, Zu YX, et al. Collagen in hepatocellular carcinoma: A novel biomarker and therapeutic target. Hepatol Commun 2024;8:e0489. [Crossref] [PubMed]
- Shen K, Arslan S, Akopian D, et al. Activated GTPase movement on an RNA scaffold drives co-translational protein targeting. Nature 2012;492:271-5. [Crossref] [PubMed]
- Hartmann E, Görlich D, Kostka S, et al. A tetrameric complex of membrane proteins in the endoplasmic reticulum. Eur J Biochem 1993;214:375-81. [Crossref] [PubMed]
- Wiedmann M, Kurzchalia TV, Hartmann E, et al. A signal sequence receptor in the endoplasmic reticulum membrane. Nature 1987;328:830-3. [Crossref] [PubMed]
- Snapp EL, Reinhart GA, Bogert BA, et al. The organization of engaged and quiescent translocons in the endoplasmic reticulum of mammalian cells. J Cell Biol 2004;164:997-1007. [Crossref] [PubMed]
- Conti BJ, Devaraneni PK, Yang Z, et al. Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events. Mol Cell 2015;58:269-83. [Crossref] [PubMed]
- Wiedmann M, Goerlich D, Hartmann E, et al. Photocrosslinking demonstrates proximity of a 34 kDa membrane protein to different portions of preprolactin during translocation through the endoplasmic reticulum. FEBS Lett 1989;257:263-8. [Crossref] [PubMed]
- Kriegler T, Kiburg G, Hessa T. Translocon-Associated Protein Complex (TRAP) is Crucial for Co-Translational Translocation of Pre-Proinsulin. J Mol Biol 2020;432:166694. [Crossref] [PubMed]
- Bañó-Polo M, Martínez-Garay CA, Grau B, et al. Membrane insertion and topology of the translocon-associated protein (TRAP) gamma subunit. Biochim Biophys Acta Biomembr 2017;1859:903-9. [Crossref] [PubMed]
- Hartmann E, Prehn S. The N-terminal region of the alpha-subunit of the TRAP complex has a conserved cluster of negative charges. FEBS Lett 1994;349:324-6. [Crossref] [PubMed]
- Li P, Wang D, Lucas J, et al. Atrial Natriuretic Peptide Inhibits Transforming Growth Factor β–Induced Smad Signaling and Myofibroblast Transformation in Mouse Cardiac Fibroblasts. Circ Res 2008;102:185-92. [Crossref] [PubMed]
- Mesbah K, Camus A, Babinet C, et al. Mutation in the Trapalpha/Ssr1 gene, encoding translocon-associated protein alpha, results in outflow tract morphogenetic defects. Mol Cell Biol 2006;26:7760-71. [Crossref] [PubMed]
- Liu Y, Yi Y, Wu W, et al. Bioinformatics prediction and analysis of hub genes and pathways of three types of gynecological cancer. Oncol Lett 2019;18:617-28. [Crossref] [PubMed]
- Van Erk MJ, Teuling E, Staal YC, et al. Time- and dose-dependent effects of curcumin on gene expression in human colon cancer cells. J Carcinog 2004;3:8. [Crossref] [PubMed]
- Yan J, Wang ZH, Yan Y, et al. RP11-156L14.1 regulates SSR1 expression by competitively binding to miR-548ao-3p in hypopharyngeal squamous cell carcinoma. Oncol Rep 2020;44:2080-92. [Crossref] [PubMed]
- Zhou Y, Zheng S, Guo Q, et al. Upregulation of RACGAP1 is correlated with poor prognosis and immune infiltration in hepatocellular carcinoma. Transl Cancer Res 2024;13:847-63. [Crossref] [PubMed]
- Wang XQ, Li LL, Lou P, et al. Noval ceRNA axis-mediated high expression of TOP2A correlates with poor prognosis and tumor immune infiltration of hepatocellular carcinoma. Transl Cancer Res 2023;12:3486-502. [Crossref] [PubMed]
- Yu X, Feng B, Wu J, et al. A novel anoikis-related gene signature can predict the prognosis of hepatocarcinoma patients. Transl Cancer Res 2024;13:1834-47. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 2007;450:663-9. [Crossref] [PubMed]
- Sommer N, Junne T, Kalies KU, et al. TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochim Biophys Acta 2013;1833:3104-11. [Crossref] [PubMed]
- Hong X, Luo H, Zhu G, et al. SSR2 overexpression associates with tumorigenesis and metastasis of Hepatocellular Carcinoma through modulating EMT. J Cancer 2020;11:5578-87. [Crossref] [PubMed]
- Li J, Zhao Z, Wang X, et al. PBX2-Mediated circTLK1 Activates JAK/STAT Signaling to Promote Gliomagenesis via miR-452-5p/SSR1 Axis. Front Genet 2021;12:698831. [Crossref] [PubMed]
- Mège RM, Ishiyama N. Integration of Cadherin Adhesion and Cytoskeleton at Adherens Junctions. Cold Spring Harb Perspect Biol 2017;9:a028738. [Crossref] [PubMed]
- Brüser L, Bogdan S. Adherens Junctions on the Move-Membrane Trafficking of E-Cadherin. Cold Spring Harb Perspect Biol 2017;9:a029140. [Crossref] [PubMed]
- Meng H, Jiang X, Wang J, et al. SEC61G is upregulated and required for tumor progression in human kidney cancer. Mol Med Rep 2021;23:427. [Crossref] [PubMed]
- Dejgaard K, Theberge JF, Heath-Engel H, et al. Organization of the Sec61 translocon, studied by high resolution native electrophoresis. J Proteome Res 2010;9:1763-71. [Crossref] [PubMed]
- Labani-Motlagh A, Ashja-Mahdavi M, Loskog A. The Tumor Microenvironment: A Milieu Hindering and Obstructing Antitumor Immune Responses. Front Immunol 2020;11:940. [Crossref] [PubMed]
- Zheng X, Jin W, Wang S, et al. Progression on the Roles and Mechanisms of Tumor-Infiltrating T Lymphocytes in Patients With Hepatocellular Carcinoma. Front Immunol 2021;12:729705. [Crossref] [PubMed]
- Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene 2008;27:5904-12. [Crossref] [PubMed]
- Luckheeram RV, Zhou R, Verma AD, et al. CD4+T cells: differentiation and functions. Clin Dev Immunol 2012;2012:925135. [Crossref] [PubMed]
- Belizário JE, Brandão W, Rossato C, et al. Thymic and Postthymic Regulation of Naïve CD4(+) T-Cell Lineage Fates in Humans and Mice Models. Mediators Inflamm 2016;2016:9523628. [Crossref] [PubMed]
- Sakaguchi E, Kayano K, Segawa M, et al. Th1/Th2 imbalance in HCV-related liver cirrhosis. Nihon Rinsho 2001;59:1259-63. [PubMed]
- Andersen MH, Schrama D, Thor Straten P, et al. Cytotoxic T cells. J Invest Dermatol 2006;126:32-41. [Crossref] [PubMed]
- Sun C, Xu J, Song J, et al. The predictive value of centre tumour CD8+ T cells in patients with hepatocellular carcinoma: comparison with Immunoscore. Oncotarget 2015;6:35602-15. [Crossref] [PubMed]


