
Optimal preservation condition for the extraction of cell-free DNA from bile of patients with biliary tract cancer
Highlight box
Key findings
• Cell-free DNA (cfDNA) tubes maintain the stability of bile cfDNA at room temperature for 10 days.
• CfDNA tubes keep bile cfDNA stable at 4 ℃ for 2 months; regular tubes cause slight loss of integrity.
• No significant difference in bile cfDNA stability between cfDNA tubes and regular tubes at −20 or −80 ℃ for 2 months.
What is known and what is new?
• Traditional methods of diagnosing biliary tract cancer (BTC) have poor specificity and sensitivity. Liquid biopsy can compensate for these deficiencies. Bile directly reflects biliary tract lesions due to its anatomical location, and its cfDNA tumor information is richer and highly consistent with tumor tissues. Bile may be superior to blood as a BTC liquid biopsy sample.
• The stability of the biological fluid is crucial for liquid biopsy, making the proper preservation of these samples absolutely essential. We delved into the effects of varying storage parameters, including temperature, duration, and the protectant, on the stability of cfDNA within bile. This rigorous exploration enabled us to ascertain the optimal conditions for bile storage, thereby laying a solid foundation for the future utilization of bile in liquid biopsy applications.
What is the implication, and what should change now?
• We have provided the optimal bile conditions for maintaining the stability of bile cfDNA in patients with BTC, which lays a theoretical foundation for the application of bile cfDNA in liquid biopsy for early diagnosis, medication guidance, therapeutic efficacy monitoring, and prognosis assessment in BTC patients.
Introduction
Biliary tract cancer (BTC), originating from the epithelium of biliary tract, is the second most common type of liver cancer after hepatocellular carcinoma, which is the third leading cause of cancer-related death globally, and accounts for 10–15% of all primary liver malignancies (1). BTC is considered a rare disease globally, but in some regions the incidence rate is relatively high, such as cholangiocarcinoma in South Korea, China and Thailand, and Gallbladder cancer in South America and Northern India (2-4). Given that BTC is usually asymptomatic at early stages, patients with BTC are often diagnosed at advanced disease stages, thereby missing the opportunity for radical surgery. Among patients who undergo surgery, more than half recover, but the overall 5-year survival rate is less than 20% (5). Therefore, early diagnosis is vital to improve the prognosis of BTC patients.
The standard first-line treatment for advanced unresectable or metastatic BTC, based on clinical therapeutic strategies, involves the addition of immune checkpoint inhibitors to gemcitabine-cisplatin chemotherapy, while FOLFOX (consisting of oxaliplatin, leucovorin, irinotecan, and fluorouracil) should be considered as the second-line treatment standard for patients with BTC undergoing preoperative therapy (6-9). Furthermore, several studies have comprehensively summarized and elaborated on the emerging neoadjuvant and targeted therapeutic approaches specifically tailored for BTC (10,11).
Medical imaging, cytological examination, and serum biomarkers are often used to diagnose BTC. However, imaging has limited sensitivity or specificity because some BTCs show no peculiar characteristics. Percutaneous transhepatic cholangiography (PTC), endoscopic retrograde cholangiopancreatography (ERCP), and biliary brush cytology have unmet sensitivity for BTC diagnosis. Serum biomarkers for BTC diagnosis, such as carbohydrate antigen 19-9 (CA19-9), are not specific biomarkers for BTCs, and they are influenced by physiological status, causing an inaccurate diagnosis in patients with BTC.
To compensate for the shortcomings of traditional detection methods, liquid biopsies have been investigated. Blood, serving as the most frequently utilized bodily fluid in liquid biopsy, encapsulates a myriad of cellular elements, notably cell-free DNA (cfDNA) derived from cancer patients, which harbors invaluable information pertaining to tumor-specific genetic and epigenetic alterations, thereby playing a pivotal role in facilitating early cancer diagnosis, monitoring the efficacy of therapeutic interventions, and elucidating the intricate mechanisms underlying drug resistance (12). In addition to plasma cfDNA, some studies have also suggested the value of bile in liquid biopsies for BTC diagnosis recently, considering that bile is secreted by liver cells, flows through bile ducts, and is concentrated in the gallbladder, keeping direct contact with the biliary tract (13-16). Bile cfDNA have shown higher concordance with tumor tissues compared to plasma cfDNA, with a sensitivity of 94.7% and a specificity of 99.9% in BTC patients, and more gene mutations could be detected in bile than in tumor tissues (17-19). Furthermore, bile cfDNA shows better performance than plasma cfDNA in detecting pancreatic duct adenocarcinoma (PDAC) (20). Therefore, bile may serve as a better source of liquid biopsies for investigating biliopancreatic cancers.
The premise of liquid biopsy relies heavily on stable preservation of biological fluid samples, therefore, reasonable preservation of body fluid samples is particularly important in translational medical research and clinical applications. Biological fluid sample preservation requires maintenance of sample quality, stability, integrity, and function (21). The major factors affecting sample preservation include the type of container material, container pretreatment, temperature of storage, and duration of storage (22). The storage condition must maintain the applicability of clinical biospecimens, ranging from macromolecules, such as cfDNA, to variable cells and their derivatives (23-25). Blood sample preservation methods have been widely studied (26,27), however, the optimal storage conditions for bile to avoid interference with subsequent experiments remain to be explored, especially when considering that different from plasma cfDNA of short fragments, bile cfDNA contains long DNA fragments and may have some special storage requirements (19,28). In this study, we investigated effects of different storage conditions, such as temperature, duration, and nucleic acid protectant, on bile cfDNA stability and determined suitable storage conditions for bile. Our findings will provide support for future applications of bile in liquid biopsy. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-843/rc).
Methods
Study design
Temperature and time are two important factors in the storage of biomedical samples. Here, we investigated the effects of different time and temperatures on bile storage and searched for the optimal storage conditions. A total of 8 patients with BTC were included in this study, numbered from Patient #1 to Patient #8. All of the patients originate from the Third Affiliated Hospital of Naval Medical University (Eastern Hepatobiliary Surgery Hospital). Specifically, the bile of Patient #1 was collected with regular tubes, while the bile of Patients #2 through #8 were collected in tubes covered with a protective agent. After centrifugation to remove cell debris and other sediments, the bile supernatant was divided into aliquots and subjected to the bile cfDNA extraction and cfDNA quality control procedures. To test the effect of storage durations (1, 4, 7, and 10 days), bile samples (Patient #1 to Patient #7) were placed at RT prior to bile cfDNA isolation. To test the effect of storage temperature, bile samples (Patient #1, #2, #4, #6, #8) were stored at different temperatures (4, −20, and −80 ℃) for 2 months before the stability of bile cfDNA was tested (Figure 1).
Bile sample preparation
Bile samples were collected using percutaneous transhepatic cholangial drainage (PTCD) prior to surgery or during surgery, in BEAVER cfDNA tubes with protectant (Beaver, Suzhou, China) or regular sample tubes without protectant (Axygen, Siliconvalley, US). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Third Affiliated Hospital of Naval Medical University (Eastern Hepatobiliary Surgery Hospital) (No. EHBHKY2022-K-004) and informed consent was obtained from all individual participants. After shipment within 24 h, bile samples were centrifuged at 1,600 g for 10 min at 4 ℃, and the supernatant underwent centrifuge at 16,000 g for 15 min at 4 ℃. The supernatant was collected and mixed evenly, and then divided in 300 µL aliquots for subsequent experiments.
Bile storage grouping
Bile samples were separated into two groups, with or without protectant, and each group was tested for effects of storage temperature and time. In the group without protectant, bile samples were collected in regular polypropylene centrifuge tubes (Axygen), while in the group with protectant, bile samples were collected in BEAVER cfDNA tubes (Beaver). After shipment within 24 h, bile samples were pretreated for further experiments. To test the effect of storage temperature, bile samples were stored at 4, −20, and −80 ℃ for 2 months prior to bile cfDNA isolation. To test the effect of storage time, bile samples were placed at room temperature (RT) for 1, 4, 7, and 10 days prior to bile cfDNA isolation.
Bile cfDNA extraction
Bile cfDNA extraction was performed in line with our previously established method (29). Briefly, bile samples were centrifuged at 16,000 g for 5 min at 4 ℃ after thawing. Then, the supernatant was added to 240 µL Buffer ACL (939017, Qiagen, Shanghai, China) and 1.68 µL Carrier RNA (1017647, Qiagen) and mixed, and 30 µL proteinase K (19133, Qiagen) was added. The mixture was incubated at 60 ℃ for 30 min, and then 540 µL Buffer ACB (1069275, Qiagen) was added and vortexed, followed by placing at it −20 ℃ for 5 min. Then, we added the mixture to the QIAquick column (28115, Qiagen) and incubated it for 5 min at RT. After incubation, it was centrifuged at 6,000 rpm for 1 min, and we discarded the flow-through. We filled the column with 500 µL buffer PE (19065, Qiagen) and centrifuged at 6,000 rpm for 1 min to wash the column twice. Then, it was centrifuged at 13,000 rpm for 1 min to remove any remaining liquid in the column. We added 30 µL of elution buffer (19086, Qiagen) into the column and incubated for 3 min at RT, then centrifuged at 13,000 rpm for 1 min to elute and store cfDNA.
Concentration measurement, purity test, and fragment distribution detection of bile cfDNA
A Qubit 3.0 fluorometer (Thermo Fisher, Massachusetts, USA), NanoDrop 2000 (Thermo Fisher), and Agilent 2100 bioanalyzer (Agilent Technologies, California, USA) were used to detect the cfDNA concentration, purity, and fragment distribution of the bile cfDNA, respectively.
Statistical analysis
Statistical analyses were performed with Graphpad Prism 7, and unpaired Student’s t-tests or Mann-Whitney tests were used for comparison of quantitative variables between groups. All the hypothesis tests were 2 sided, with P<0.05 considered to be statistically significant.
Results
Bile cfDNA is less stable at RT
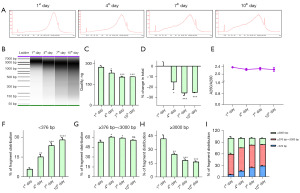
We first detected whether bile could be stored at RT for a short period. Bile samples that were collected in tubes without protectant were placed at RT for 1, 4, 7, and 10 days before bile cfDNA isolation. We previously showed that long fragments of DNA are predominant in bile (19), which was confirmed in bile cfDNA on day 1 (Figure 2A). However, cfDNA fragments became shorter with placing bile samples at RT, as indicated by the more and more flattened peaks (Figure 2A) and the dispersion of DNA bands (Figure 2B). We also detected bile cfDNA in bile samples stored at RT for 2 months without protectant; cfDNA showed dispersed bands without enrichment of long DNA fragments (Figure S1A,S1B), indicating that cfDNA continued losing integrity when bile samples were stored at RT without protectant. The amount of extracted cfDNA per sample decreased over time; after 7 days at RT without protectant, approximately 25% of cfDNA in bile was lost (Figure 2C,2D), and 55% was lost after 2 months at RT (Figure S1C,S1D), but the purity of extracted cfDNA was not affected (Figure 2E). In addition, the fragment size analysis showed a significant increase in short cfDNA fragments (<376 bp) and decrease in long cfDNA fragments (>3,000 bp) during 10 days at RT (Figure 2F-2I), and the change was even greater during 2 months (Figure S1E-S1H). These results indicated that when bile samples were collected without protectant and stored at RT, bile cfDNA degraded with time.
Protectant protects bile cfDNA from degradation at RT
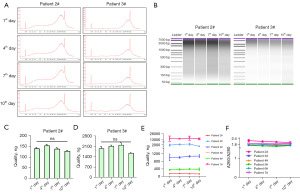
We then tested whether bile cfDNA was stable in bile samples collected in tubes covered with protectant for storage up to 10 days at RT (Figure 3, Figure S2). In contrast to the storage at RT without protectant, the bile cfDNA fragment size distribution of bile stored with cfDNA tubes showed quite similar patterns across 10 days (Figure 3A,3B, Figure S2A,S2B), and there was no significant decrease in cfDNA amounts in each sample regardless of cfDNA contents (Figure 3C-3E, Figure S2C-S2F). The cfDNA purity was also stable during 10 days (Figure 3F). Taken together, tubes covered with protectant were beneficial to bile cfDNA stability, and bile samples could be stored with protectant for 10 days without worrying about cfDNA degradation, which is of great convenience for bile sample shipment.
Protectant protects bile cfDNA for 2 months
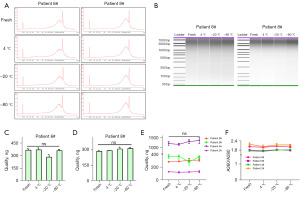
Biomedical samples are usually stored at low temperature for long periods of time, therefore, we also tested the effects of low storage temperature (4, −20, and −80 ℃) for a long storage period (2 months). Given that protectant is beneficial to bile cfDNA stability, bile samples were collected with cfDNA tubes, and then divided into aliquots for the following experiments. Compared to fresh bile samples, the fragment distributions of bile cfDNA of bile samples stored at 4, −20, and −80 ℃ for 2 months displayed similar patterns (Figure 4A,4B, Figure S3A,S3B). Neither bile cfDNA contents in each sample nor cfDNA purity showed significant fluctuation during storage for 2 months (Figure 4C-4F, Figure S3C,S3D). In addition, bile samples without protectant contained cfDNA similar to fresh bile samples (Figure S4A-S4D); however, cfDNA showed slightly decreased integrity in bile samples stored at 4 ℃ for 2 months without protectant (Figure S4E-S4H).
In conclusion, for short-term storage of bile samples at RT (within 10 days), tubes with protectant are recommended to protect bile cfDNA from degradation and provide a convenient method to ship bile samples. For long-term storage (such as 2 months), bile samples collected in tubes with protectant could be stored at 4 ℃ without affecting bile cfDNA integrity, while bile samples collected in tubes without protectant should be stored below −20 ℃ to maintain cfDNA integrity.
Discussion
Biological samples need to be stored properly; otherwise, the subsequent analysis would be inaccurate. In this study, we investigated the storage conditions for bile samples to preserve bile cfDNA. Bile is a unique body fluid secreted by the liver cells; it flows within the biliary tree, is stored in the gallbladder, and is discharged into the duodenum to facilitate digestion (30). Bile is exposed to the biliary epithelium all the time, which makes it an ideal biofluid for liquid biopsy in patients with biliary tract malignancies (31). In patients with obstructive jaundice, the accumulated bile is usually drained to prevent liver failure; however, it may contain a relatively high amount of tumor cell-originated molecules that may be used as biomarkers or therapeutic targets for hepatobiliary diseases (32-34). Bile can be defined as a complex alkaline aqueous medium that contains most of the solutes found in blood and micelles formed by bile salts, electrolytes, phospholipids, cholesterol, bile pigments, and proteins (13,35). Therefore, to better seek the potential of bile as a new source of liquid biopsy, the proper storage conditions of bile need to be explored.
In addition to cells, cfDNA is also found in bile, and the sensitivity of cfDNA for the diagnosis of gallbladder cancer is higher than that of cytology (36). We have previously found that bile cfDNA shows high concordance with tumor tissue biopsies (19). Moreover, bile cfDNA outperforms plasma cfDNA in detecting BTCs (18), thus suggesting that bile cfDNA may be a powerful tool for liquid biopsy in bile. The exploration of bile cfDNA represents a major advance that not only elevates the sensitivity and specificity of BTCs diagnosis but also enables the identification of potentially mutated, druggable genes for targeted therapy, based on its rich tumor-specific information, while simultaneously facilitating the assessment of patient prognosis and the monitoring of treatment response (12,15,17-19). Despite its immense potential in BTC research, the detection sensitivity and accuracy of bile cfDNA may be compromised by factors such as patient status, bile composition, processing methods, and storage conditions, while the origin, release mechanisms, and functional mechanisms of bile cfDNA remain elusive. To overcome the mentioned drawbacks, there is an urgent need to optimize and standardize bile processing and storage to reduce interferences and ensure accurate detection results, while also delving deeper into the mechanistic role of bile cfDNA in BTC to provide a theoretical basis for clinical application, potentially improving diagnostic accuracy when combined with clinical serum markers. With the profound comprehension of bile cfDNA mechanisms, coupled with the prevalent adoption of advanced genetic testing technologies and the relentless progress in medical care, bile cfDNA is poised to occupy a pivotal position in BTC diagnosis, treatment strategy formulation, and prognostic assessment (14,37,38). Its anticipated expansion into broader medical areas will require continuous scrutiny, relentless refinement, and adaptation to ensure its sustained, robust, and healthy development within this dynamic field. In this study, we collected bile samples in protectant-free regular tubes and with cfDNA tubes with protectant to test the effect of storage conditions on bile cfDNA stability.
The storage temperature and storage time have been found to significantly influence bile cell viability (39). CfDNA mainly originates from cell apoptosis and necrosis, which means that only a small proportion of cfDNA is real tumor cell-derived (40). The cfDNA tubes are designed to prevent cell lysis and to protect cfDNA from nuclease in blood samples, thereby protecting the integrity of the cfDNA population. We tested bile storage at RT for up to 10 days, and we found that bile cfDNA was stable when bile samples were collected with cfDNA tubes and placed at RT for 10 days (Figure 3), but bile cfDNA degraded at RT in tubes without protectant (Figure 2). Hence, bile should be collected with cfDNA tubes for further investigation of bile cfDNA, in which scenario the shipment of bile samples at RT is also acceptable.
Storage temperature is another vital factor for bile sample preservation. We investigated the effects of storage at low temperatures for a long period (2 months) on bile cfDNA stability. For bile samples collected with cfDNA tubes, storage at 4 ℃ for 2 months did not affect bile cfDNA stability, and lower temperature (−20 or −80 ℃) preservation seems unnecessary (Figure 4). As for bile collected in tubes without protectant, low temperature storage seems to be a better choice for bile cfDNA preservation. Although it has been suggested to store bile samples at 4 ℃ rather than at RT (39), we observed slightly decreased integrity of bile cfDNA even in bile samples stored at 4 ℃ for 2 months with regular tubes. However, when frozen at −20 or −80 ℃ for 2 months, bile cfDNA with regular tubes did not exhibit significant degradation or loss of integrity (Figure S4). Taken together, these results indicate that if collected with cfDNA tubes, bile samples could be stored at 4 ℃ without affecting bile cfDNA for further research, however, if collected in tubes without protectant, bile samples should be stored frozen, such as −20 or −80 ℃ for long-term preservation.
There are several limitations in this study. First, a larger sample size is needed to address the proper storage conditions more clearly. Bile samples included in this study were collected using different methods such as PTCD, ERCP, or during surgery, and they differed substantially in physiochemical properties. A larger sample size might clarify different storage conditions for different types of bile samples. Second, we estimated cfDNA contents and integrity under different storage conditions, but it remains to be explored whether the tumor-related genetic information carried by the bile cfDNA is affected by different storage conditions. Third, we do not have whole-genome sequencing data of bile cfDNA, so we cannot assess whether the mutation, copy number landscape and structural variation spectra under different storage conditions affect its consistency with tumor tissue, whether the efficiency of genetic testing of bile is different between different storage conditions. Fourth, we have no clinical data such as drug information, progression-free survival, overall survival, immunohistochemistry of cellular-mesenchymal epithelial transition factor (c-MET), epidermal growth factor receptor (EGFR), human epidermal growth factor receptor 2 (HER2), and serum carcinoembryonic antigen (CEA) and CA19-9, so we cannot provide references for targeted therapy and prognostic evaluation. Finally, bile contains various kinds of solutes that may be valuable in diagnosis or therapeutic target development, so the proper storage conditions need to be investigated for different constituents.
Conclusions
In this study, we explored the optimal storage conditions for the extraction of cfDNA from bile samples. The cfDNA tubes and the regular sample tubes without protectant inside were used to collect bile samples of patients with BTC, and the effects of storage temperature and storage time for the preservation of bile samples were evaluated. It was found that bile cfDNA degradated with time if bile samples were collected with regular tubes and stored at RT. CfDNA tubes prevented degradation of bile cfDNA and were convenient to transport at RT for 10 days. Bile samples collected with cfDNA tubes can be stored at 4 ℃ for 2 months without affecting the stability of bile cfDNA. When collecting with regular tubes, bile samples remained stable at −20 or −80 ℃. These results suggested that for bile cfDNA research, bile samples should be collected with cfDNA tubes for short-term shipment at RT, and bile samples could be stored at 4 ℃ in cell-free tubes, or frozen at −20 ℃ with regular tubes. We proposed the optimal storage conditions of bile samples for the extraction of cfDNA from patients with BTC, providing insights into the feasibility of bile cfDNA in liquid biopsy.
Acknowledgments
We appreciate the support of the physicians and pathologists, and participation of the patients in this study. We would like to thank Chao Sun for his assistance in collecting patients’ clinical information. Chao Sun is a former employee of 3D Medicines Inc.
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-843/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-843/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-843/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-843/coif). B.N., X.X., S.C., Z.L., W.Z., D.Z. come from 3D Medicines Inc. (a profitable company). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Banales JM, Marin JJG, Lamarca A, et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol 2020;17:557-88. [Crossref] [PubMed]
- Bridgewater JA, Goodman KA, Kalyan A, et al. Biliary Tract Cancer: Epidemiology, Radiotherapy, and Molecular Profiling. Am Soc Clin Oncol Educ Book 2016;35:e194-203. [Crossref] [PubMed]
- Torre LA, Siegel RL, Islami F, et al. Worldwide Burden of and Trends in Mortality From Gallbladder and Other Biliary Tract Cancers. Clin Gastroenterol Hepatol 2018;16:427-37. [Crossref] [PubMed]
- Valle JW, Borbath I, Khan SA, et al. Biliary cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v28-37. [Crossref] [PubMed]
- Valle J, Wasan H, Palmer DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 2010;362:1273-81. [Crossref] [PubMed]
- Guven DC, Sahin TK, Erul E, et al. The association between albumin levels and survival in patients treated with immune checkpoint inhibitors: A systematic review and meta-analysis. Front Mol Biosci 2022;9:1039121. [Crossref] [PubMed]
- Lamarca A, Palmer DH, Wasan HS, et al. Second-line FOLFOX chemotherapy versus active symptom control for advanced biliary tract cancer (ABC-06): a phase 3, open-label, randomised, controlled trial. Lancet Oncol 2021;22:690-701. [Crossref] [PubMed]
- Rizzo A, Ricci AD, Tober N, et al. Second-line Treatment in Advanced Biliary Tract Cancer: Today and Tomorrow. Anticancer Res 2020;40:3013-30. [Crossref] [PubMed]
- Rizzo A, Brandi G. Neoadjuvant therapy for cholangiocarcinoma: A comprehensive literature review. Cancer Treat Res Commun 2021;27:100354. [Crossref] [PubMed]
- De Lorenzo S, Garajova I, Stefanini B, et al. Targeted therapies for gallbladder cancer: an overview of agents in preclinical and clinical development. Expert Opin Investig Drugs 2021;30:759-72. [Crossref] [PubMed]
- Rizzo A, Ricci AD, Tavolari S, et al. Circulating Tumor DNA in Biliary Tract Cancer: Current Evidence and Future Perspectives. Cancer Genomics Proteomics 2020;17:441-52. [Crossref] [PubMed]
- Boyer JL. Bile formation and secretion. Compr Physiol 2013;3:1035-78. [Crossref] [PubMed]
- Arechederra M, Rullán M, Amat I, et al. Next-generation sequencing of bile cell-free DNA for the early detection of patients with malignant biliary strictures. Gut 2022;71:1141-51. [Crossref] [PubMed]
- Miura Y, Ohyama H, Mikata R, et al. The efficacy of bile liquid biopsy in the diagnosis and treatment of biliary tract cancer. J Hepatobiliary Pancreat Sci 2024;31:329-38. [Crossref] [PubMed]
- Kearney JF, Weighill D, Yeh JJ. Promise of bile circulating tumor DNA in biliary tract cancers. Cancer 2023;129:1643-5. [Crossref] [PubMed]
- Li Z, Liu Y, Fu J, et al. Bile is a reliable and valuable source to study cfDNA in biliary tract cancers. Front Oncol 2022;12:961939. [Crossref] [PubMed]
- Gou Q, Zhang CZ, Sun ZH, et al. Cell-free DNA from bile outperformed plasma as a potential alternative to tissue biopsy in biliary tract cancer. ESMO Open 2021;6:100275. [Crossref] [PubMed]
- Shen N, Zhang D, Yin L, et al. Bile cell free DNA as a novel and powerful liquid biopsy for detecting somatic variants in biliary tract cancer. Oncol Rep 2019;42:549-60. [Crossref] [PubMed]
- Driescher C, Fuchs K, Haeberle L, et al. Bile-Based Cell-Free DNA Analysis Is a Reliable Diagnostic Tool in Pancreatobiliary Cancer. Cancers (Basel) 2020;13:39. [Crossref] [PubMed]
- Muller R, Betsou F, Barnes MG, et al. Preservation of Biospecimens at Ambient Temperature: Special Focus on Nucleic Acids and Opportunities for the Biobanking Community. Biopreserv Biobank 2016;14:89-98. [Crossref] [PubMed]
- Subramanian KS. Storage and preservation of blood and urine for trace element analysis. A review. Biol Trace Elem Res 1995;49:187-210. [Crossref] [PubMed]
- Betsou F, Lehmann S, Ashton G, et al. Standard preanalytical coding for biospecimens: defining the sample PREanalytical code. Cancer Epidemiol Biomarkers Prev 2010;19:1004-11. [Crossref] [PubMed]
- Lehmann S, Guadagni F, Moore H, et al. Standard preanalytical coding for biospecimens: review and implementation of the Sample PREanalytical Code (SPREC). Biopreserv Biobank 2012;10:366-74. [Crossref] [PubMed]
- Palmirotta R, Ludovici G, De Marchis ML, et al. Preanalytical Procedures for DNA Studies: The Experience of the Interinstitutional Multidisciplinary BioBank (BioBIM). Biopreserv Biobank 2011;9:35-45. [Crossref] [PubMed]
- Sakkestad ST, Skavland J, Hanevik K. Whole blood preservation methods alter chemokine receptor detection in mass cytometry experiments. J Immunol Methods 2020;476:112673. [Crossref] [PubMed]
- Kanamoto M, Shimada M, Utsunomiya T, et al. Impact of a new refrigerator for the preservation of whole blood. J Surg Res 2012;173:180-4. [Crossref] [PubMed]
- Cristiano S, Leal A, Phallen J, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385-9. [Crossref] [PubMed]
- Shen N, Zhu B, Zhang W, et al. Comprehensive Evaluation and Application of a Novel Method to Isolate Cell-Free DNA Derived From Bile of Biliary Tract Cancer Patients. Front Oncol 2022;12:891917. [Crossref] [PubMed]
- Yao Y, Jiao D, Li Z, et al. Roles of Bile-Derived Exosomes in Hepatobiliary Disease. Biomed Res Int 2021;2021:8743409. [Crossref] [PubMed]
- Shen J, Wang W, Wu J, et al. Comparative proteomic profiling of human bile reveals SSP411 as a novel biomarker of cholangiocarcinoma. PLoS One 2012;7:e47476. [Crossref] [PubMed]
- Gradilone SA, O'Hara SP, Masyuk TV, et al. MicroRNAs and benign biliary tract diseases. Semin Liver Dis 2015;35:26-35. [Crossref] [PubMed]
- Marin JJ, Bujanda L, Banales JM. MicroRNAs and cholestatic liver diseases. Curr Opin Gastroenterol 2014;30:303-9. [Crossref] [PubMed]
- Gradilone SA. Extracellular vesicles as therapeutic carriers of microRNAs for cholangiocarcinoma. Hepatology 2017;65:404-6. [Crossref] [PubMed]
- Bévalot F, Cartiser N, Bottinelli C, et al. State of the art in bile analysis in forensic toxicology. Forensic Sci Int 2016;259:133-54. [Crossref] [PubMed]
- Kinugasa H, Nouso K, Ako S, et al. Liquid biopsy of bile for the molecular diagnosis of gallbladder cancer. Cancer Biol Ther 2018;19:934-8. [Crossref] [PubMed]
- Javle M, Bekaii-Saab T, Jain A, et al. Biliary cancer: Utility of next-generation sequencing for clinical management. Cancer 2016;122:3838-47. [Crossref] [PubMed]
- Li M, Zhang Z, Li X, et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet 2014;46:872-6. [Crossref] [PubMed]
- Xia Y, Gao Y, Wang B, et al. Optimizing the Method of Cell Separation from Bile of Patients with Cholangiocarcinoma for Flow Cytometry. Gastroenterol Res Pract 2019;2019:5436961. [Crossref] [PubMed]
- Razavi P, Li BT, Brown DN, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med 2019;25:1928-37. [Crossref] [PubMed]


