
SKA1/2/3 link to poor prognosis and immune infiltration of head and neck squamous cell carcinomas
Highlight box
Key findings
• Spindle and kinetochore-associated complex 1/2/3 (SKA1/2/3) were identified as potential biological markers and therapeutic targets for head and neck squamous cell carcinoma (HNSC).
What is known and what is new?
• SKA1 expression levels are elevated in non-small cell lung cancer.
• SKA1/2/3 were related to the prognosis and immune cell infiltration of HNSC.
What is the implication, and what should change now?
• SKA1/2/3 play a role in HNSC malignancy and could be used as potential therapeutic targets for HNSC.
Introduction
Background
The head and neck squamous cell carcinomas (HNSCs) are a diverse collection of tumors that originate in the oral cavity, pharynx, and larynx and pose a severe threat to human health, contributing to a fast-rising burden of cancer morbidity and mortality (1). Smoking, heavy alcohol consumption, human papillomavirus (HPV), UV light, and betel nut chewing are all common risk factors (2). Every year, around 600,000 new cases are detected worldwide, and more than 60% of HNSC are already locally intermediate or have progressed at the time of initial diagnosis, necessitating a treatment regimen of surgery, radiation, and chemotherapy (3). On average, men are 2–4 times more likely than women to develop HNSC. Non-viral HNSC has a median age of 66 years, whereas HPV-associated oropharyngeal cancer and Epstein-Barr virus-associated nasopharyngeal cancer have median ages of 53 and 50 years, respectively (4). Survival rates for HNSC have improved moderately over the last three decades, rising from 55% in 1992–1996 to 66% in 2002–2006 in a Surveillance, Epidemiology, and End Results (SEER) registry analysis of all age groups, excluding elderly patients (aged >75 years) (5). Subgroup studies revealed that survival rates improved in all age categories. HNSC are abundantly diverse in terms of primary sites and subtypes. As a consequence, despite advancements in cancer detection and therapy, the overall 5-year survival rate for head and neck malignancies remains poor (6). This is due to the difficulties in determining the tumor’s site and the absence of substantial early clinical indications. As a result, most individuals are diagnosed with HNSC at an advanced stage. Some new genes linked to HNSC prognosis have recently been identified, including autophagy-related genes (7), ferroptosis-related genes (8), and so on. However, more prognostic and therapeutic targets for HNSC are required. As a result, identifying the molecular pathways linked with the development and progression of HNSC is critical for successful and dependable care of this form of cancer.
Rationale and knowledge gap
Spindle and kinetochore-associated complex 1/2/3 (SKA1/2/3) are three members of the SKA, which stabilize the spindle microtubules at the kinetosite during mitosis metaphase, ensuring mitosis completion (9). The SKA complex is required for the spindle microtubule’s attachment to the exciter and the maintenance of the phase plate, and hence for correct chromosomal separation during mitosis. The efficient elimination of the SKA complex has been demonstrated to result in severe chromosomal organization abnormalities (10). SKA complex dysexpression is closely linked to the prognosis of various human malignancies, including breast, cervical, liver, and lung cancers. Patients with high SKA1 expression in the liver have been shown to have a worse prognosis than those with low SKA1 expression (11). SKA1 expression levels are elevated in non-small cell lung cancer (NSCLC) and are linked to cancer progression via cell proliferation, migration, and invasion regulation (12). Patients with breast cancer who have high SKA2 expression have a poor prognosis. Interfering with SKA2 can prevent breast cancer cells from proliferating, migrating, and invading (9). SKA2 plays an important role in controlling esophageal cancer cell proliferation, migration, and invasion (13). SKA3 overexpression has been shown to increase the proliferation and migration of cervical cancer cells, as well as tumor growth (14).
Objective
The clinical significance of SKA complexes (SKA1, SKA2, and SKA3) as predictive biomarkers for HNSC has yet to be determined. We used a vast variety of public datasets and web-based technologies to investigate SKA complex expression and its link to patient prognosis, and discover multiple pathways by which the SKA complex is regulated in HNSC. Further research into the potential clinical applications of the SKA complex for cancers will contribute to a better understanding of the pathophysiology of HNSC and the identification of new prognostic biomarkers. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-862/rc).
Methods
SKA1/2/3 expression and prognostic value by Gene Expression Profiling Interactive Analysis (GEPIA)
GEPIA (15) (http://gepia.cancer-pku.cn/) incorporates gene expression profile data from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) projects, allowing for data analysis and visualization. In this study, we used GEPIA to compare SKA1/2/3 expression between normal patient samples (n=44) and HNSC samples (n=518). According to the median of SKA1/2/3 expression, HNSC samples were separated into two groups (an SKA1/2/3 high expression group and an SKA1/2/3 low expression group), and Kaplan-Meier survival curves of HNSC patients were plotted. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
SKA1/2/3 expression in different subgroups by University of ALabama at Birmingham CANcer data analysis Portal (UALCAN) and Tumor-Immune System Interactions Database (TISIDB)
The UALCAN (http://ualcan.path.uab.edu/index.html) (16) is a comprehensive, interactive web resource for studying cancer data. It integrates expression profile data for all tumors in the TCGA database. It displays graphs and plots of expression profiles and patient survival data for protein-coding, microRNA (miRNA)-coding, and long intergenic non-coding RNA (lincRNA)-coding genes. The TISIDB (http://cis.hku.hk/TISIDB) (17) is an online platform that incorporates several heterogeneous data sources for tumor and immune system interaction. Both of the abovementioned resources are based on the TCGA’s genomics, transcriptomics, and clinical data for 30 cancer types. We used UALCAN and TISIDB to classify SKA1/2/3 expression values into different subgroups, including grade, stage, gender, nodal metastasis, immune subtype, and molecular subtype.
Protein-protein interaction (PPI) network by gene multiple association network integration algorithm (GeneMANIA)
GeneMANIA (http://genemania.org/) (18) investigates a vast number of publicly available biological datasets for linked genes. Protein-protein, protein-DNA, and genetic interactions, pathways, reactions, gene and protein expression data, protein domains, and phenotypic screening profiles are examples of these. SKA1/2/3 related proteins were identified by GeneMANIA. Metascape (https://metascape.org/) was used for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses.
Immune cells infiltration level with SKA1/2/3 expression
Tumor Immune Estimation Resource (TIMER; http://timer.cistrome.org/) (19) detects immune cell infiltration in tumor tissues using RNA-Seq expression profiling data. The gene module is chosen to show the relationship between SKA1/2/3 expression and immune infiltration in HNSC. The somatic copy number alteration (SCNA) module compares tumor infiltration levels between tumors with different somatic copy number changes for SKA1/2/3. Xiantao bioinformatics tool (https://www.xiantaozi.com/) is a free online analytics platform. The data of TCGA were collated and standardized for easy operation. The tool was used to compare the infiltration scores of different groups of immune cells. Gene set enrichment analysis (GSEA) was performed through the Xiantao bioinformatics tool.
SKA1/2/3 expression by Tumor Immune Single Cell Hub 2 (TISCH2)
The TISCH2 (http://tisch.comp-genomics.org/) is a tumor microenvironment (TME) single-cell RNA-sequence (scRNA-seq) database. TISCH2 provides precise cell-type annotation at the single-cell level, allowing for the investigation of TME across cancer types. TISCH2 was used to show the SKA1/2/3 expression in different cell types of HNSC in scRNA-seq.
Nomogram and validation of SKA1/2/3
RNAseq data were downloaded and collated from the TCGA database (https://portal.gdc.cancer.gov) for the TCGA-HNSC project STAR process and the data were extracted in transcripts per million (TPM) format as well as clinical data. The rms package (version 6.3-0) was applied to construct a nomogram, which predicted the survival probability of HNSC patients in different years. A calibration showed the relation between actual probability and model probability. The transcriptomes microarray datasets (GSE31056) of normal margins tissues (n=73) and tumor tissues (n=23) of oral squamous cell carcinoma were downloaded from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/). The expression values of SKA1/2/3 were extracted. Wilcoxon rank-sum test was applied to compare the expressions of SKA1/2/3 in normal and tumor tissues. The protein levels of SKA1/2/3 expressed in HNSC and normal tissues were gained from The Human Protein Atlas database (https://www.proteinatlas.org/).
Statistical analysis
First, normality and homogeneity of variance were assessed, which verified that the data were normally distributed and there was homogeneity of variance. The t-test was used for comparisons between the two groups. For comparisons among multiple groups, one- or two-way analysis of variance (ANOVA) was performed, followed by Tukey’s multiple comparisons test for post hoc analysis. A P value <0.05 was considered statistically significant.
Results
The expression and prognostic value of SKA1/2/3 in HNSC
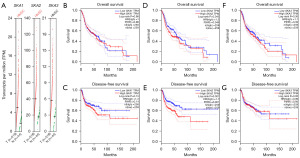
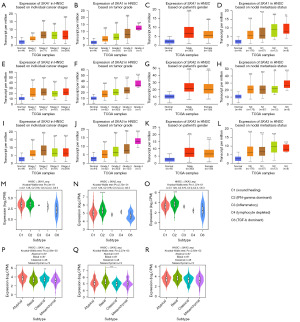
SKA1 and SKA2 were significantly highly expressed in HNSC, whereas SKA3 was up-regulated in HNSC, without significance (Figure 1A). The expression of SKA1 had no influence on the overall survival (OS) and disease-free survival (DFS) of HNSC patients (Figure 1B,1C). High expression of SKA2 was significantly related to poor OS and poor DFS of HNSC patients (Figure 1D,1E). There was no correlation between SKA3 expression and survival of HNSC patients (Figure 1F,1G). SKA1/2/3 was up-regulated in different stages, grades, gender, and nodal metastasis compared to the normal tissues (Figure 2A-2L). There was no obviously difference of SKA1 expression in different immune subtypes (Figure 2M). Up-regulated SKA2/3 were observed in interferon (IFN)-gamma dominant subtype compared to wound healing subtype (Figure 2N,2O). Molecular subtypes of HNSC included atypical, basal, classical, and mesenchymal subtypes. The expression of SKA1 was down-regulated in the classical subtype compared to that in the atypical subtype (Figure 2P). The expression of SKA2 was lower in the classical subtype than in the basal subtype (Figure 2Q). There was no obviously difference of SKA3 expression in different molecular subtypes (Figure 2R). The correlation between SKA1/2/3 and clinical information of HNSC patients is shown in Tables S1-S3.
PPI network and GO/KEGG analysis of SKA1/2/3
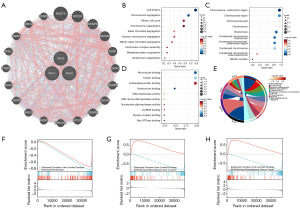
SKA1/2/3-related proteins were identified by GeneMANIA. A PPI network was constructed, as shown in Figure 3A. Regarding biological process, SKA1/2/3 and the related proteins were enriched in cell division, chromosome segregation, mitotic cell cycle, and so on (Figure 3B). Regarding cellular component, they were involved in chromosome, centromeric region, and so on (Figure 3C). Regarding molecular function, they were associated with microtubule binding, cytoskeletal protein binding, etc. (Figure 3D). As for molecular function, they were related to oocyte meiosis, apoptosis, and so on (Figure 3E). GSEA showed that SKA1/2/3 were related to formation of cornified envelope and retinoblastoma gene cancer (Figure 3F-3H).
The correlation between SKA1/2/3 expression and immune cells infiltration
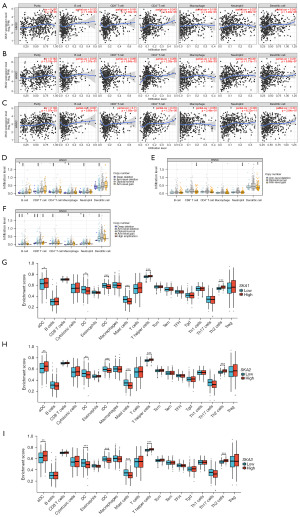
SKA1 expression was obviously positively correlated to the purity, B cell infiltration, CD8+ T cell infiltration, CD4+ T cell infiltration, macrophage infiltration, neutrophil infiltration, and dendritic cell (DC) infiltration (Figure 4A). SKA2 expression was significantly positively correlated with infiltration of CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and DC (Figure 4B). SKA3 expression was significantly positively correlated with CD8+ T cell, CD4+ T cell, neutrophil, and DC (Figure 4C). Copy number mutations, especially arm-level deletion, of SKA1 could significantly limit immune infiltration of B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and DC (Figure 4D). Copy number mutations, especially arm-level gain, of SKA2 could significantly reduce immune infiltration of CD8+ T cell, CD4+ T cell, neutrophil, and DC (Figure 4E). Copy number mutations, especially arm-level deletion, of SKA1 could significantly limit immune infiltration of B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and DC (Figure 4F). HNSC samples were divided into two parts according to the median of SKA1/2/3. In the SKA1/2/3 high expression group, the enrichment scores of active DCs (aDCs), T helper cells, and Th2 cells were obviously higher than that in SKA1/2/3 low expression group, and the enrichment scores of DC, immature DC (iDC), and mast cells were significantly lower than that in SKA1/2/3 low expression group (Figure 4G-4I).
The SKA1/2/3 expression in scRNA-seq of HNSC
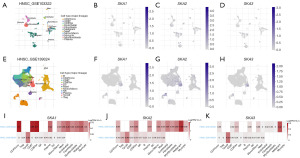
Two HNSC datasets (GSE103322 and GSE139324) were obtained from the TISCH2 database. GSE103322 contained 5,902 single cells from 18 patients with oral cavity tumors. The cell annotation showed 11 kinds of cells which were identified by scRNA-seq (Figure 5A). The SKA1/2/3 expression distributions in different kinds of cells are displayed in Figure 5B-5D. GSE139324 included 130,721 single cells from 26 patients with HNSC. The cell annotation showed 11 kinds of cells which were identified by scRNA-seq (Figure 5E). The SKA1/2/3 expression distributions in different kinds of cells are displayed in Figure 5F-5H. The expression value of SKA1/2/3 was high in malignant cells (Figure 5I-5K).
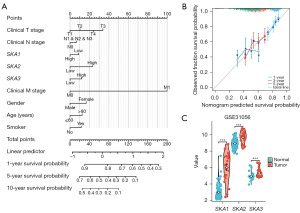
Nomogram and validation of SKA1/2/3
The nomogram included clinical information and SKA1/2/3, containing T stage, N stage, M stage, gender, age, and smoking history (Figure 6A). Each indicator corresponded to different scores. The total score indicated the probability of 1-, 5-, and 10-year survival. The predicted curve coincided better with the ideal curve, indicating that the nomogram demonstrated good predictability (Figure 6B). The expressions of SKA1/2/3 were higher in tumor tissues than that in normal tissues in GSE31056 (Figure 6C).
Discussion
The majority of patients with HNSC present with locally advanced disease that necessitates multimodality treatment. Despite that this therapy is intended to be curative, a considerable proportion of these patients will present localized regional failure and/or distant metastases (20). These patients have a dismal prognosis. Due to the high risk of local disease recurrence after initial treatment, the limited treatment options for metastatic disease, and the spatiotemporal genetic heterogeneity of solid tumors associated with poor prognosis due to decreased treatment response and high rates of tumor recurrence, there is a strong need to identify new biomarkers for HNCC (21).
SKA1, SKA2, and SKA3 shape an intricate system at the kinetochore-microtubule interface, and concurrent depletion of SKA1 and SKA3 leads to a breakdown of chromosomal aggregation and cell death (11). The SKA1/2/3 complex is necessary for correct chromosomal segregation during mitosis by stabilizing spindle microtubule attachment to kinetochores and maintaining the phase plate. Depletion of the SKA complex causes the production of unstable kinetochore-microtubule structures as well as severe chromosomal alignment problems. SKA mutations cause spindle checkpoint abnormalities, a common occurrence in cancer, and play an important role in cell cycle regulation and carcinogenesis (21). We found that the biological function of SKA1/2/3 and related genes was related to formation of cornified envelope and retinoblastoma gene cancer. It was indicated that SKA1/2/3 played an important role in the biological process.
Considered to be one of the most significant inhibitory checkpoints, programmed death protein 1 (PD-1) and its programmed death ligands 1 (PD-L1) and 2 (PD-L2) are directly engaged in cancer immunomodulation, leading to the inactivation of cell-mediated anti-tumor immune responses (22). Immunotherapies using PD1/PD-L1 inhibitors have been authorized for treatment in advanced HNSC. Immunotherapies are dependent on PD-L1 expression in cancer cells; if this expression is low in these precancerous lesions, immunotherapy will not work as well as it should. When detecting PD-L1 expression in premalignant squamous lesions using various immunohistochemistry assays, the rate of variation varies greatly, and a meta-analysis (23) report verified the significant study heterogeneity degree. For these included studies, the combined estimate of PD-L1 positivity was 48%. Precancerous lesions had higher PD-L1 positivity than normal mucosa, but lower positivity than squamous cell carcinomas. Tumor tissue can be collected in clinical practice by several methods such as fine-needle aspiration, core biopsies, and surgical specimens. These methods often involve distinct anatomical regions. Additionally, there are variations in PD-L1 expression between main and metastatic samples, with the latter typically exhibiting higher levels of PD-L1 expression. These variations may contribute to the clonal evolution of tumors and complicate the process of choosing the best samples for PD-L1 testing (24). Prior to receiving treatment with PD-L1 inhibitors, patients may have received several regimens of traditional chemotherapy and radiotherapy. PD-L1 expression may be impacted by these medicines. It is therefore recommended to re-biopsy and re-evaluate PD-L1 expression levels in cases of relapse after chemotherapy and/or radiotherapy.
In the present study, SKA1 was found to be highly expressed in different stages and grades of HNSC. But the expression of SKA1 was not linked to the prognosis of HNSC patients. The expression of SKA1 was down-regulated in the classical subtype compared to that in the atypical subtype. From the scRNA-seq analysis, SKA1 was found to be highly expressed in malignant cells of HNSC. SKA1 had been reported to be related to many cancers. By using immunohistochemistry analysis, 42 cases of salivary gland cystic carcinoma and 20 cases of normal tissue adjacent to the tumor were shown to express the SKA1 gene. The positive SKA1 staining rates in salivary gland cystic carcinoma were 78.6%, which was significantly higher than the rates in healthy salivary glands (25). The histological features of salivary gland cystic carcinoma that were progressed and solid were favorably linked with SKA1 expression. In T-24 bladder cancer cells, SKA1 knockdown drastically decreased the expression of CDK4 and cyclin D1, which was helpful in preventing cell proliferation (26). Prostate-specific SKA1 up-regulation induced spontaneous cancer in transgenic mice models (27).
Up-regulated SKA2 expression was identified in HNSC. In different immune subtypes of HNSC, SKA2 expression was higher in the IFN-gamma dominant subtype compared to that in the wound healing subtype. Our findings indicated that SKA2 expression was significantly positively correlated with infiltration of CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and DC. Moreover, highly expressed SKA2 was considerably positively related to the poor OS and DFS of HNSC patients. Breast cancer tissues exhibit increased SKA2 expression, and SKA2 deletion can considerably lessen breast cancer cell invasion and migration (28). Patients with strong SKA2 expression in their adrenocortical carcinoma have a bad prognosis (29). SKA2 immunohistochemical staining and messenger RNA (mRNA) levels in hepatocellular carcinoma (HCC) tissues were both considerably higher than in normal tissues. SKA2 deletion drastically reduced HCC cell growth and invasion, but SKA2 overexpression markedly increased cell proliferation and invasion (30).
Up-regulated SKA3 expression was identified in HNSC. Copy number mutations, especially arm-level deletion, of SKA3 could significantly limit immune cells infiltration. The downregulation of SKA3 significantly reduced tumor growth and invasion of HCC both in vivo and in vitro, and SKA3 was discovered to have an impact on tumor development through the cell cycle and P53 signaling pathway (31). SKA3 functions as an oncogene in bladder cancer and contributes to M2 macrophage polarization (32).
The expressions of SKA1/2/3 were validated in GSE31056. The nomogram included SKA1/2/3, T stage, N stage, M stage, gender, age, and smoking history. The predicted curve coincided better with the ideal curve, indicating that the nomogram demonstrated good predictability of 1-, 5-, and 10-year survival.
SKA1/2/3 were over-expressed in HNSC and were associated with the immune cells’ infiltration level. SKA2 influenced the survival of HNSC patients. These findings indicated that SKA1/2/3 could be used as biological markers and therapeutic targets for HNSC. This research could help researchers better understand molecular mechanisms that under of HNSC and aid in the development of SKA-mediated HNSC treatments. More research is required to corroborate our results and enhance SKA’s clinical applicability as a biomarker of disease progression or treatment target for HNSC.
Conclusions
In HNSC, SKA1/2/3 were highly expressed and related with immune cell infiltration. SKA2 affected HNSC patients’ survival. SKA1/2/3 could serve as potential biological markers and therapeutic targets for HNSC. This study could assist researchers better understand the molecular pathways that underlie HNSC.
Acknowledgments
The authors thank the free online platforms (UALCAN, GEPIA, Sangerbox, Xiantao bioinformatics tool, TIMER, cBioPortal, TISCH2) for data analysis and visualization.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-862/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-862/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-862/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Zheng B, Huang Z, Liu W, et al. A real-world evaluation of tislelizumab in patients with head and neck cancer. Transl Cancer Res 2024;13:808-18. [Crossref] [PubMed]
- Ganci F, Pulito C, Valsoni S, et al. PI3K Inhibitors Curtail MYC-Dependent Mutant p53 Gain-of-Function in Head and Neck Squamous Cell Carcinoma. Clin Cancer Res 2020;26:2956-71. [Crossref] [PubMed]
- Windon MJ, D'Souza G, Rettig EM, et al. Increasing prevalence of human papillomavirus-positive oropharyngeal cancers among older adults. Cancer 2018;124:2993-9. [Crossref] [PubMed]
- Pulte D, Brenner H. Changes in survival in head and neck cancers in the late 20th and early 21st century: a period analysis. Oncologist 2010;15:994-1001. [Crossref] [PubMed]
- Wei M, Zhi J, Li L, et al. Predicting therapeutic responses in head and neck squamous cell carcinoma from TP53 mutation detected by cell-free DNA. Transl Cancer Res 2023;12:3604-17. [Crossref] [PubMed]
- Zhang C, Cai Q, Ke J. Poor Prognosis of Oral Squamous Cell Carcinoma Correlates With ITGA6. Int Dent J 2023;73:178-85. [Crossref] [PubMed]
- Yang L, Chen Z, Liu Y, et al. Immunization Combined with Ferroptosis Related Genes to Construct a New Prognostic Model for Head and Neck Squamous Cell Carcinoma. Cancers (Basel) 2022;14:4099. [Crossref] [PubMed]
- Lange KI, Suleman A, Srayko M. Kinetochore Recruitment of the Spindle and Kinetochore-Associated (Ska) Complex Is Regulated by Centrosomal PP2A in Caenorhabditis elegans. Genetics 2019;212:509-22. [Crossref] [PubMed]
- Wilson BE, Shen Q, Cescon DW, et al. Exploring immune interactions in triple negative breast cancer: IL-1β inhibition and its therapeutic potential. Front Genet 2023;14:1086163. [Crossref] [PubMed]
- Chen Y, Xu X, Wang Y, et al. Hypoxia-induced SKA3 promoted cholangiocarcinoma progression and chemoresistance by enhancing fatty acid synthesis via the regulation of PAR-dependent HIF-1a deubiquitylation. J Exp Clin Cancer Res 2023;42:265. [Crossref] [PubMed]
- Chen C, Guo Q, Song Y, et al. SKA1/2/3 serves as a biomarker for poor prognosis in human lung adenocarcinoma. Transl Lung Cancer Res 2020;9:218-31. [Crossref] [PubMed]
- Chen J, Yang HM, Zhou HC, et al. PRR11 and SKA2 promote the proliferation, migration and invasion of esophageal carcinoma cells. Oncol Lett 2020;20:639-46. [Crossref] [PubMed]
- Hu DD, Chen HL, Lou LM, et al. SKA3 promotes lung adenocarcinoma metastasis through the EGFR-PI3K-Akt axis. Biosci Rep 2020;40:BSR20194335. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Chandrashekar DS, Karthikeyan SK, Korla PK, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022;25:18-27. [Crossref] [PubMed]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 2019;35:4200-2. [Crossref] [PubMed]
- Franz M, Rodriguez H, Lopes C, et al. GeneMANIA update 2018. Nucleic Acids Res 2018;46:W60-4. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Brandt A, Thiele B, Schultheiß C, et al. Circulating Tumor DNA in Head and Neck Squamous Cell Carcinoma. Cancers (Basel) 2023;15:2051. [Crossref] [PubMed]
- Wilson HL, D'Agostino RB Jr, Meegalla N, et al. The Prognostic and Therapeutic Value of the Mutational Profile of Blood and Tumor Tissue in Head and Neck Squamous Cell Carcinoma. Oncologist 2021;26:e279-89. [Crossref] [PubMed]
- Marletta S, Fusco N, Munari E, et al. Atlas of PD-L1 for Pathologists: Indications, Scores, Diagnostic Platforms and Reporting Systems. J Pers Med 2022;12:1073. [Crossref] [PubMed]
- Girolami I, Pantanowitz L, Munari E, et al. Prevalence of PD-L1 expression in head and neck squamous precancerous lesions: a systematic review and meta-analysis. Head Neck 2020;42:3018-30. [Crossref] [PubMed]
- Paolino G, Pantanowitz L, Barresi V, et al. PD-L1 evaluation in head and neck squamous cell carcinoma: Insights regarding specimens, heterogeneity and therapy. Pathol Res Pract 2021;226:153605. [Crossref] [PubMed]
- Zhao L, Jiang L, Du P, et al. Expression of SKA1 and MMP-9 in primary salivary adenoid cystic carcinoma: Correlation with tumor progression and patient prognosis. Acta Otolaryngol 2016;136:575-9. [Crossref] [PubMed]
- Tian F, Xing X, Xu F, et al. Downregulation of SKA1 Gene Expression Inhibits Cell Growth in Human Bladder Cancer. Cancer Biother Radiopharm 2015;30:271-7. [Crossref] [PubMed]
- Li J, Xuan JW, Khatamianfar V, et al. SKA1 over-expression promotes centriole over-duplication, centrosome amplification and prostate tumourigenesis. J Pathol 2014;234:178-89. [Crossref] [PubMed]
- Ren Z, Yang T, Zhang P, et al. SKA2 mediates invasion and metastasis in human breast cancer via EMT. Mol Med Rep 2019;19:515-23. [PubMed]
- Yu S, Ma J. Spindle and Kinetochore-Associated Complex Is Associated With Poor Prognosis in Adrenocortical Carcinoma. J Surg Res 2022;277:50-9. [Crossref] [PubMed]
- Wang D, Suo YJ, Gong L, et al. SKA2 promotes proliferation and invasion of hepatocellular carcinoma cells via activating the β-catenin signaling pathway. Neoplasma 2020;67:743-50. [Crossref] [PubMed]
- Hou Y, Wang Z, Huang S, et al. SKA3 Promotes tumor growth by regulating CDK2/P53 phosphorylation in hepatocellular carcinoma. Cell Death Dis 2019;10:929. [Crossref] [PubMed]
- Wang C, Liu S, Zhang X, et al. SKA3 is a prognostic biomarker and associated with immune infiltration in bladder cancer. Hereditas 2022;159:20. [Crossref] [PubMed]


