
Elucidating the molecular and immune interplay between head and neck squamous cell carcinoma and diffuse large B-cell lymphoma through bioinformatics and machine learning
Highlight box
Key findings
• Utilizing bioinformatics approaches combined with machine learning algorithms, we have identified potential biomarkers for head and neck squamous cell carcinoma (HNSCC) and diffuse large B-cell lymphoma (DLBCL), namely ACACB, MMP8, PAX5, and TNFAIP6.
• The expression profiles of these identified genes exhibit a complex relationship with the efficacy of immunotherapeutic interventions, notably CTLA-4 and PD-1/PD-L1 checkpoints. Capitalizing on these findings, we have engineered an innovative diagnostic paradigm: the nomogram model.
What is known and what is new?
• Existing research has utilized machine learning to investigate biomarkers within individual tumor types.
• This study extends this approach by exploring shared potential targets between HNSCC and DLBCL, delineating commonalities that may serve as pivotal points of intervention.
What is the implication, and what should change now?
• Building upon the pivotal genes identified within both conditions, this research enhances not only the precision of early disease diagnosis but also facilitates the prediction of patient treatment responses and survival outcomes.
• It is anticipated that medical professionals will leverage these molecular markers to carry out clinical trials, with the objective of discovering novel therapeutic avenues or refining current treatment protocols.
Introduction
Head and neck squamous cell carcinoma (HNSCC) ranks among the most prevalent malignant cancers globally, typically manifesting in the oral squamous epithelium, oropharynx, larynx, and hypopharynx (1). Notably, pharyngeal cancer is increasingly linked to human papillomavirus (HPV) infection (2), while nasopharyngeal cancer is closely connected to Epstein-Barr virus (EBV) (3). Due to the high propensity for lymphatic metastasis in HNSCC, lymph node involvement is a common clinical feature, complicating treatment and significantly impacting prognosis negatively (4-7). Diffuse large B-cell lymphoma (DLBCL), the most prevalent subtype of non-Hodgkin lymphoma, accounts for approximately 40% of all non-Hodgkin lymphoma cases (8). It is a heterogeneous tumor noted for its invasive behavior, complex origins, and high mortality (9). R-CHOP therapy is effective in treating about 60% of patients with DLBCL (10). However, many patients develop resistance to this treatment or experience a recurrence, leading to poor outcomes, especially in EBV-positive DLBCL cases (11). Accumulating evidence confirms that both HNSCC and DLBCL share a set of pathological features that contribute to their aggressive behavior, including disruption of normal tissue structures, invasion of surrounding tissues, and metastasis through lymphatic and vascular networks (12-15). At the molecular level, similarities in abnormal signaling pathways, dysregulation of the cell cycle, and resistance to apoptosis all contribute to the malignant phenotype of these cancers (16-18). Although these cancers are primarily of squamous and lymphocytic origin, emerging research suggests that they may share a key cancer survival strategy known as immune evasion (19-22).
Immune evasion is a complex and multifaceted process by which cancer cells develop the capability to elude detection and destruction by the immune system (23,24). This mechanism is increasingly acknowledged as an indicator of cancer progression, with significant implications for prognosis and treatment responses across various cancer types (23,25). The intricate interplay between cancer cells and the immune system involves a myriad of molecular and cellular interactions. In HNSCC and DLBCL, the tumor microenvironment plays a pivotal role in fostering immune evasion (26,27). Within this environment, various cellular components such as tumor-associated macrophages, myeloid-derived suppressor cells, and regulatory T cells infiltrate the tumors, establishing an immunosuppressive milieu that supports tumor growth and dissemination (28-30). Furthermore, genetic and epigenetic alterations in cancer cells contribute to immune evasion (31,32). For instance, mutations in tumor antigens can diminish immune cell recognition, upregulation of checkpoint molecules like programmed cell death protein 1/programmed death-ligand 1 (PD-1/PD-L1) can induce T cell exhaustion, and the secretion of immunosuppressive cytokines can create a local environment unfavorable to immune effector cells (33-35).
In both HNSCC and DLBCL, the intricate interplay of genetic mutations, epigenetic alterations, and environmental factors underscores the need for reliable biomarkers. These biomarkers are crucial for early detection, accurate prognosis, and the customization of treatment options. Although traditional methods of discovering biomarkers have provided significant insights into these diseases, they often fall short in addressing the complexity and dynamics of cancer genomes. Consequently, this study leverages the strengths of bioinformatics tools and machine learning (ML) strategies to systematically identify novel biomarkers for HNSCC and DLBCL. This approach is expected to unearth new biological insights into these tumors, enhance the effectiveness and personalization of treatments for these formidable diseases, and bridge the gap between basic research and clinical application. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1064/rc).
Methods
Data collection and data processing
The GSE83632 dataset, consisting of 87 normal and 76 DLBCL samples (36,37) treated with the GPL5175 platform {Affymetrix Human Exon 1.0 ST Array [transcript (gene) version]}, was downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov). Additionally, transcriptome expression profile and survival dataset for HNSCC, which include 44 normal samples and 502 tumor samples, were obtained from The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga). Data organization and initial processing were conducted with R (version 4.3.2), which involved converting probe IDs to gene symbols based on the platform’s annotation files and removing empty probes. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). An overview of the study procedures is provided in Figure 1, detailing each step of the research process.
Identification of differentially expressed genes (DEGs) from HNSCC dataset
The expression data in the HNSCC dataset were corrected with log2, and duplicate genes were removed. DEGs between tumor and normal samples were screened using the criteria P value <0.05 and |log2 fold change (FC)| >1.5 with the limma package.
Screening of DLBCL related modules
The weighted gene co-expression network analysis (WGCNA) was performed to identify co-expressed gene modules from the DLBCL dataset with WGCNA package (38). To reduce false positives, 50% of genes with the minimum median absolute deviation (MAD) were removed prior to analysis. A weighted adjacency matrix was then constructed using the average linkage method and weighted correlation coefficients calculated from the remaining genes. The weighted adjacency matrix was converted into a topological overlap matrix by calculating the adjacency with the “soft” threshold power (β=8). For the dynamic tree cutting program, the red line was set to 0.9, the module cutting height to 0.35, and the minimum module gene count to 60 (Figure S1A,S1B). Finally, the gene co-expression network was computed after calculating gene significance, module membership, and correlation modules with clinical traits.
Functional enrichment analysis of HNSCC and DLBCL shared genes
Genes shared in the DEGs calculated the from HNSCC dataset and DLBCL related modules were further used for functional enrichment analysis, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Disease Ontology (DO). The enrichment analyses of GO, KEGG, and DO were conducted with ClusterProfiler (39) and DOSE (40) packages, respectively. In all analyses, an adjusted P value <0.05 was defined as the threshold for significance.
Protein-protein interaction (PPI) network construction and identification of prospective genes
To analyze the interactions among proteins of interest, the DEGs and key module genes were uploaded to the STRING database (https://cn.string-db.org) and protein nodes with a composite score exceeding 0.4 were selected to construct a PPI network (41). Additionally, the top 30 closest protein nodes were identified using the CytoHubba plugin in Cytoscape software (version 3.8.2) with six algorithms: BottleNeck, Betweenness, maximum neighborhood component (MNC), density of maximum neighborhood component (DMNC), ClusteringCoefficient, and Stress (42). The intersection genes identified by all the algorithms were considered as prospective genes.
Identification of candidate genes through ML
To investigate the prospective genes, two ML algorithms were employed independently on the separate datasets: support vector machine-recursive feature elimination (SVM-RFE) and random forest (RF). The SVM-RFE algorithm was executed using the e1071, caret, and svmRFE packages, while the RF algorithm was implemented using the RandomForest package.
For the HNSCC dataset, the important nodes identified from the PPI network were used as feature vectors, with tags 1 and 0 indicating tumor and normal samples, respectively. In the SVM-RFE method, the dataset was randomly divided into five equal parts for 5-fold cross-validation. Each part served as a test set in turn, with the remaining four parts combined as the training set. For each feature vector, it was sequentially removed from the dataset, and an SVM model was trained on the remaining features. The error between the predicted results and the true labels was calculated to assess the importance of each feature. Subsequently, the error rate and accuracy of the SVM model with varying numbers of features were calculated and visualized. The genes contained in the model with the highest prediction accuracy and the lowest error rate were selected as the optimal feature set for further analysis. The RF method for the HNSCC dataset involved an input file containing the gene expression profile and grouping information for the feature genes. Seventy percent of the data was used as the training set, with the remaining 30% serving as the test set. The randomForest function created the random forest model based on the training dataset. This model generated several decision trees based on the relationships between the training data’s features and labels, resulting in a set of weak classifiers. The trained random forest model then predicted the samples in the test set. For each new sample, its features were input into each decision tree, and the predictions from all trees were aggregated to obtain the final classification result. The importance of variables was extracted from the final model, ranked by the MeanDecreaseGini index, and the top features were identified.
Using the same criteria and steps, the SVM-RFE and RF models were trained on the DLBCL expression profiles. The feature genes identified by SVM-RFE and RF in both of the two datasets were selected. The intersection genes discovered by both algorithms were recognized as candidate genes due to their excellent capacity to distinguish between tumor and normal data. GO, DO, and KEGG analyses of candidate genes were performed using the ClusterProfiler and DOSE programs, with P<0.05 considered significant (data not shown).
Survival analysis and receiver operating characteristic (ROC) evaluation for identification of hub genes
Utilizing the clinical data from the HNSCC dataset, the survminer and survival packages were used to compute survival analysis results, which were then integrated into a Kaplan-Meier (K-M) survival curve for visualization (43). The survival curves of the prospective geneset were plotted in terms of overall survival, tumor node metastasis (TNM) stage, and grade. Due to limited clinical information available for the DLBCL dataset, an ROC curve was drawn based on the expression profile (44). The area under the ROC curve (AUC) value measured the ability of genes to distinguish between tumor and normal groups, with an AUC value greater than 0.8 indicating effective diagnostic capability. Genes meeting the criteria set by both survival and ROC analyses were classified as hub genes. A co-expression network utilizing these genes was constructed using GeneMANIA (http://genemania.org) (45).
To assess the practical application value of hub genes in clinical diagnosis, the rms package was used to construct a nomogram model incorporating the hub genes (46). Individual prediction outcomes were then computed by mapping the functional relationship between the total score and the probability of the outcome event for 1-, 2-, and 3-year survival.
Drug sensitivity analysis and protein expression
Utilizing the Gene Set Cancer Analysis (GSCA) platform (https://guolab.wchscu.cn/GSCA), which integrates the Gene Set Differential Analysis (GSDA), the potential efficacy of prospective drugs was predicted (47). Additionally, to further evidence the hub genes at the protein level within the HNSCC and DLBCL datasets, the Human Protein Atlas (HPA) database (https://www.proteinatlas.org) was utilized (48).
Pan-cancer analysis
An extensive analysis of the expression levels of the hub genes in tumor and normal tissues across 33 types of cancer in TCGA was conducted using the TIMER 2.0 database (http://timer.cistrome.org). Subsequently, the CancerSEA database (https://biocc.hrbmu.edu.cn/CancerSEA/home.jsp) was utilized to examine the association between their expression levels and 14 cancer functional states, including angiogenesis, apoptosis, cell cycle, cell differentiation, DNA damage, DNA repair, epithelial-mesenchymal transition (EMT), hypoxia, inflammation, invasion, metastasis, proliferation, quiescence, and stem cell-like properties. A correlation heatmap was then created to identify the relationship between the hub genes and tumor immune checkpoints.
Mutation analysis and identification of transcription factors (TFs) with hub genes
To illustrate the mutation landscape, the maftools package was utilized to create waterfall plots by considering the correlation of gene expression between tumor mutational burden (TMB) and microsatellite instability (MSI) (49). Additionally, the TCGA plot package was used to create radar plots demonstrating the correlations between the expression of the hub genes and TMB as well as MSI (50). TF-gene networks were predicted using the ChEA database available on the Network Analyst platform (https://www.networkanalyst.ca). These networks were further refined using Cytoscape software to optimize their structure and function.
Immune infiltration analysis
To examine the association between the hub genes and immune cell infiltration in the HNSCC and DLBCL datasets, the CIBERSORT tool was employed (51). The data were visually represented through a bar chart, detailing the relative proportions of 22 distinct immune cell types in each sample. Additionally, a box plot was used to compare the rankings and differences in the proportions of these immune cells between the tumor and control groups. Further exploration of the interaction between immune infiltration and DLBCL involved analyzing each DLBCL sample’s relationship with immune infiltrating cells using the ImmuneCelAI package. To provide a clear and informative visualization, a lollipop plot of the hub genes in relation to the immune infiltrating cells was generated.
Single-cell RNA sequencing (scRNA-seq) data analysis of HNSCC and lymphoid samples
The scRNA-seq data of GSE172577, comprising five HNSCC samples and five lymph node samples, were downloaded from the GEO database, imported into R, and analyzed with the Seurat package. The PercentageFeatureSet function was used to calculate the percentage of mitochondrial genes in each cell. Next, quality control was performed to filter cells that did not meet specific criteria: a gene count per cell greater than 200 or less than 4,000, the total number of molecules per cell less than 30,000, and the percentage of mitochondrial genes less than 20%. After normalization with the NormalizeData function, the top 2,000 genes with the highest variability were selected for further analysis using the “vst” method within the FindVariableFeatures function. The data were scaled using the ScaleData function before conducting Principal Component Analysis (PCA) analysis. The Harmony algorithm was employed to remove batch effects in the data. The resulting Uniform Manifold Approximation and Projection (UMAP) plot effectively demonstrated the clustering performance of the batch-corrected cells. The singleR package’s integrated HumanPrimaryCellAtlasData database was used to annotate the cells, with the resulting cell clusters visualized using UMAP plots generated by the DimPlot and FeaturePlot functions. Additionally, a sample-cell composition histogram depicted the proportions of cell types in HNSCC tissues and lymphatic tissues. The expression patterns of four hub genes across different cell types in these tissues were investigated and visualized using UMAP plots.
Statistical analysis
Group comparisons were carried out using the t-test to evaluate differences between variables. A P value of less than 0.05 was considered statistically significant. This threshold was rigorously adhered to in order to ensure the reliability of our findings, and to identify meaningful differences and trends within the data. The rigorous application of these methods allows for a robust assessment of the statistical relationships and potential causal connections between the variables studied.
Results
Identification of DEGs and key module-related genes
The PCA result for the HNSCC dataset represented that samples between tumor and normal groups can be separated with the first two principal components (Figure 2A). There were 1,154 and 886 up- and down-regulated DEGs identified using the criteria P value and log2FC (Figure 2B). The 30 DEGs with the largest absolute FCs indicated that there was a significant difference between tumor and normal groups (Figure 2C).

Based on the clinical traits, the genes in the DLBCL dataset were categorized into nine important modules (Figure 2D,2E). The correlation analysis indicated that the turquoise module, including 1,983 genes, was the most prominent module (correlation coefficient =0.76; P=6e−32). The key module-related genes in the turquoise module were then recognized as crucial for further experiments. Additionally, these genes were intersected with the above 2,040 DEGs to identify 85 genes that were common to HNSCC and DLBCL (Figure 2F).
Functional analysis of common genes and PPI network
To explore the potential common biological mechanisms, GO, KEGG, and DO enrichment analyses were conducted on the shared 85 genes. GO enrichment analysis revealed that these genes were mainly enriched in the following categories: response to molecule of bacterial origin, cytoplasmic vesicle lumen, and endopeptidase activity (Figure 3A). KEGG enrichment analysis indicated that shared genes were involved in the interleukin (IL)-17 signaling pathway, transcriptional misregulation in cancer, PI3K-Akt signaling pathway, and JAK-STAT signaling pathway (Figure 3B). DO enrichment analysis showed that common genes were associated with alloimmunization, cerebral childhood acute arthritis, and childhood B acute lymphoblastic leukemia. The top 30 core genes were screened using six algorithms, and by applying the intersection of a Venn diagram (Figure 3C,3D), 21 prospective genes were identified and used to construct the PPI network (Figure 3E).

ML analysis of prospective genes
Two ML algorithms, SVM-REF and RF, were used to examine the new shared geneset including the 21 prospective genes dependently. In the HNSCC dataset, 20 genes were identified by SVM-REF and RF respectively (Figure S1C-S1F). In the DLBCL dataset, SVM-REF identified 19 genes, while RF identified 20 genes (Figure S1G-S1J). The different proposed gene list obtained from above are intersected to obtain a candidate list with 16 genes for further verification (Figure 3F).
Survival analysis and ROC evaluation
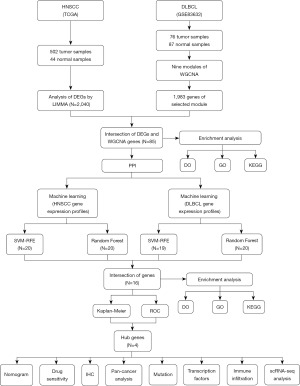
Based on the gene expression profile and clinical information of the HNSCC dataset, K-M survival curves were plotted for the 16 candidate genes, and the hub genes ACACB, MMP8, PAX5 and TNFAIP6 were found to be significantly associated with the overall survival of the patients (Figure 4A-4D). In TNM staging, the presence of M0 status (no distant metastasis) was correlated with the expression of ACACB, MMP8, and TNFAIP6. Additionally, T3 status (tumor greater than 4 cm in greatest dimension) was correlated with both ACACB and MMP8. For early-stage tumors, T1 and T2 statuses (tumor 2 cm or less in greatest dimension and tumor greater than 2 cm but not more than 4 cm, respectively) along with N0 status (no regional lymph node metastasis) were solely correlated with the expression of PAX5 and MMP8. This detailed correlation underscores the potential role these hub genes play in different stages of tumor progression and metastasis in HNSCC (Figure S2).
Subsequently, in the DLBCL dataset, the gene ACACB had an AUC of 0.913, indicating a strong predictive capability. Similarly, MMP8 had an AUC of 0.831, PAX5 had an AUC of 0.820, and TNFAIP6 had an AUC of 0.916 (Figure 4E). These high AUC values suggest that these genes possess significant discriminatory power and are valuable candidates for further downstream analysis. Therefore, the four genes were screened and utilized as a gene panel to serve as potential biomarkers for the diagnosis and prediction of HNSCC and DLBCL diseases. To further investigate the function of these hub genes, a 20-gene interaction network was constructed (Figure 4F).
Diagnosis value evaluation
A nomogram model for disease diagnosis was developed, identifying the gene panel as predictive indicators (Figure 5A). Predictions for 1-, 2-, and 3-year survival probabilities for HNSCC and DLBCL patients were made using a disease diagnosis model. The 1-year calibration curve indicated that the model’s prediction accuracy ranged from 0.76 to 0.86 for the actual situation (Figure 5B). The 2-year calibration curve showed that the model’s prediction accuracy for the actual situation fell between 0.55 and 0.75 (Figure 5C), while the 3-year calibration curve demonstrated that the model’s prediction accuracy could reach 0.65 (Figure 5D). Calibration curves in all three cases suggested that the nomogram model had good predictive ability for HNSCC and DLBCL.

Drug sensitivity analysis
ACACB expression was positively associated with the 50% inhibitory concentration (IC50) values of 17-AAG, bleomycin (50 µM), CHIR-99021, dasatinib, docetaxel and Z-LLNle-CHO in Genomics of Drug Sensitivity in Cancer (GDSC) database. Moreover, there was a negative correlation between ACACB expression and IC50 values of 21 types of drugs (Figure 5E). Among them, bleomycin, docetaxel, methotrexate and vorinostat are commonly used for the treatment of DLBCL and HNSCC (52-57). Iop et al. reported that bleomycin could be used as a remedial therapy for patients with recurrent HNSCC after cisplatin/fluorouracil treatment (53). Cottereau et al. reported that bleomycin involved in R-ACVBP could be used for the treatment of DLBCL (58). In the treatment of HNSCC, docetaxel is frequently utilized as part of a combination chemotherapy regimen, particularly in high-grade or recurrent cases. For patients who cannot undergo surgery or radiation therapy, docetaxel, in combination with other drugs such as cisplatin and 5-fluorouracil, may offer a systemic treatment option (54). In the treatment of DLBCL, docetaxel is not the standard treatment of choice. However, in some cases of resistant or relapsed DLBCL, docetaxel may be considered as a second-line or later treatment option (55). Methotrexate can be used in combination with Pembrolizumab for the treatment of recurrent or metastatic HNSCC (54). In DLBCL dataset, for some cases of central nervous system (CNS) invasion or prevention, high doses of methotrexate may be considered for cases involving CNS invasion or prevention due to its ability to effectively penetrate the blood-brain barrier and achieve higher concentrations in the CNS (59). Histone deacetylase (HDAC) inhibitors, such as vorinostat, have been reported to be effective in recurrent or refractory DLBCL (55). In the HNSCC dataset, HDAC inhibitors have been shown to have some anticancer activity in animal models, and there have been clinical trials exploring the use of vorinostat in HNSCC therapy (57).
MMP8 expression was positively associated with IC50 values of CP466722 and TG101348, and a negative correlation between MMP8 expression and IC50 values of 23 types of drugs in GDSC database (Figure 5F). Bortezomib is a proteasome inhibitor that has been approved for the treatment of multiple myeloma but has also shown some activity to DLBCL (60). Selumetinib is an inhibitor that target the BRAF mutation and has been used in the treatment of malignant melanoma, but has also been used as experimental therapies in some DLBCL cases (61).
TNFAIP6 expression showed a positive association with the IC50 values of Phenformin in the GDSC database, suggesting higher drug resistance (Figure 5G). Conversely, TNFAIP6 expression exhibited a negative correlation with the IC50 values of 18 other drugs, indicating enhanced sensitivity to these treatments. The drugs include AG-014699, AMG-706, AZD6482, BEZ235, Bleomycin (50 µM), Camptothecin, CHIR-99021, Cisplatin, JW-7-24-1, LFM-A13, linsitinib, midostaurin, olaparib, pazopanib, SN-38, talazoparib, temsirolimus, and TGX221. This diverse response profile underscores the potential of TNFAIP6 as a biomarker for predicting drug sensitivity and resistance across a range of cancer therapies, thereby aiding in the personalization of treatment plans. Cisplatin is a platinum-based chemotherapy drug that is widely used in the treatment of many cancers, including HNSCC and DLBCL (62-64). Bleomycin is an antibiotic drug that inhibits the growth of tumor cells by binding to DNA and triggering DNA breaks and it is often used to treat a variety of cancers, including HNSCC and DLBCL. Camptothecin is a DNA topoisomerase I inhibitor that causes DNA damage and apoptosis by blocking the DNA replication process and can be used to treat a variety of cancers, including HNSCC and DLBCL (64,65).
PAX5 expression exhibited a positive correlation with the IC50 values of several drugs in the CTRP database, suggesting an increased resistance to these treatments (Figure 5H). Specifically, PAX5 expression was associated with higher IC50 values for drugs such as vincristine, doxorubicin, and etoposide. This indicates that elevated PAX5 levels may contribute to drug resistance in certain therapeutic contexts. Understanding this relationship is crucial for developing more personalized cancer treatments, where PAX5 could serve as a potential biomarker for identifying patients who may benefit from alternative therapies to overcome drug resistance.
Expression validation of hub genes
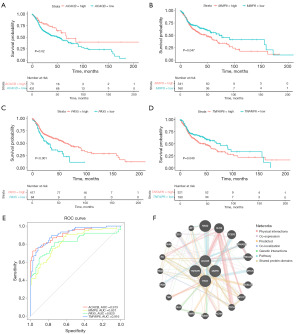
In the TCGA datasets, the Wilcoxon rank-sum test was employed for unpaired comparisons to analyze the expression levels of in the four hub genes. The analysis revealed that in HNSCC tissues, MMP8, TNFAIP6, and PAX5 were significantly upregulated, while ACACB was downregulated (Figure 6A-6D). In DLBCL tissues, MMP8 and TNFAIP6 were upregulated, whereas ACACB and PAX5 showed downregulation (Figure S3A-S3D). The significance of all tests reached a P value of less than 0.001, indicating strong statistical significance in the differential expression of these hub genes.
Using the HPA database to study the protein levels of the hub genes, it was found that there was no significant difference in the expression of MMP8 and TNFAIP6 proteins in both of the datasets (Figure 6E-6G; Figure S3E-S3G). Based on the IHC verification results, it was concluded that PAX5 tended to be highly expressed only in HNSCC tissues (Figure 6H) while in lymphoma tissues, companied by ACACB (Figure S3E,S3H).
Pan-cancer analysis
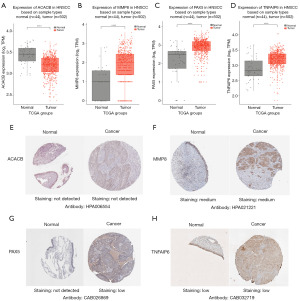
The expression levels of the gene panel in tumor and normal tissues across 33 cancers in TCGA were investigated. ACACB expression differed between the tumor and normal groups in 20 cancers, with significant down-regulation in tumor tissues compared to normal tissues (Figure 7A). MMP8, TNFAIP6 and PAX5 was significantly differentially expressed in 17, 14, 8 cancer types respectively, showing a trend of high expression in tumors (Figure 7B-7D). Besides, in the HNSCC dataset, ACACB, PAX5, and TNFAIP6 showed a high correlation with most immune checkpoints, whereas MMP8 was significantly correlated with many checkpoints (Figure 7E). In the DLBCL dataset, only PAX5 showed significant associations with most immune checkpoints (Figure 7F). These findings suggest that PAX5 may play a role in tumor immunodetection in both of HNSCC and DLBCL tissues.

Mutation and TFs analysis of hub genes
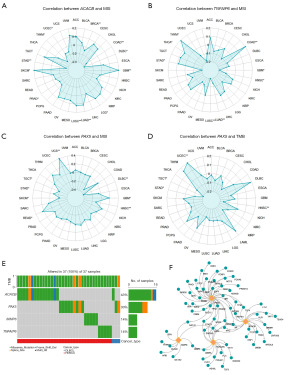
The MSI in DLBCL tissues was significantly negatively correlated with ACACB (P<0.05) and PAX5 (P<0.01), while the MSI in HNSCC tissues was significantly negatively correlated with TNFAIP6 (P<0.05) and PAX5 (P<0.01). In terms of TMB, only PAX5 (P<0.01) exhibited a significant negative correlation in HNSCC tissues (Figure 8A-8D). The waterfall plot revealed that the majority of mutation types in DLBCL and HNSCC tissues were missense mutations (Figure 8E). As depicted in Figure 8F, the TF-gene interaction network consists of 67 nodes and 81 edges. Key TFs such as EGR1, MYC, BACH1, and AR were found to regulate multiple hub genes within the network, highlighting their potential roles in gene regulation and the underlying molecular mechanisms.
Immune infiltration analysis
In the DLBCL dataset, the associations between 22 types of immune cells and the four key hub genes were examined. As illustrated in Figure 9A the predominant immune cells included neutrophils, monocytes, CD4 T cells, NK cells, CD8 T cells, B cells, mast cells, and M0 macrophages. Detailed immune infiltration for each sample is presented in Figure S4A. Notably, the first four cell types demonstrated significant differences between the normal and tumor groups (Figure 9B). Furthermore, significant correlations were observed between the expression of the four hub genes and several immune cells, specifically CD4 T cells, B cells, NK cells, CD8 T cells, neutrophils, M0 macrophages, M1 macrophages, and M2 macrophages, as depicted in Figures 9C-9F.
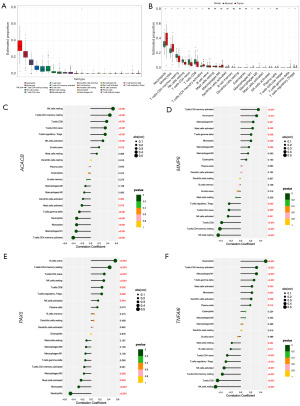
In the HNSCC dataset, the top ten predominant immune cells as follows: CD4 T cells, M0 macrophages, M2 macrophages, M1 macrophages, NK cells, dendritic cells, plasma cells, CD8 T cells, monocytes, and mast cells (Figure S4B). A comprehensive overview of immune infiltration for each sample is depicted in Figure S4C. Significant differences in immune cell populations between the tumor and normal groups were also observed. The immune cells that showed marked variations include CD4 T cells, M0 macrophages, M2 macrophages, M1 macrophages, NK cells, monocytes, mast cells, B cells, and dendritic cells (Figure S4D).
Single-cell subpopulations and transcriptome landscape in HNSCC and lymphoid tissues
After batch correction, the cells were clustered into 13 cell clusters (Figure 10A). Using the HumanPrimaryCellAtlasData database for automatic annotation, the cells were further classified into six cell clusters: mononuclear phagocyte system (MPs), epithelial cells, lymphocytes, fibroblasts, endothelial cells, and mast cells (Figure 10B). Differences between these six types of cells in HNSCC and lymphoid tissue were explored, showing that HNSCC tissues had more fibroblasts, epithelial cells, and MPs compared with lymphoid tissues (Figure 10C). The difference in cell proportion between the two tissues was more evidently demonstrated by the sample-cell composition histogram (Figure 10D).
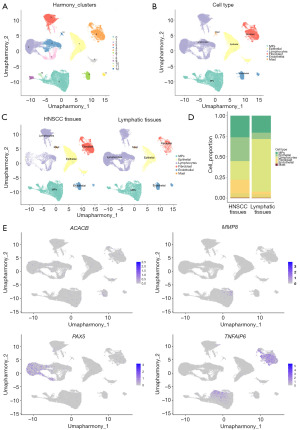
The expression of the hub genes in six cell clusters was comprehensively explored and compared. In terms of expression levels across all samples, the results indicated that PAX5 was highly expressed in lymphocytes, while TNFAIP6 was predominantly expressed in fibroblasts and MPs (Figure 10E). When comparing the expression between HNSCC and lymph samples, no significant differences in the expression of ACACB were detected (Figure S5A). However, it was notable that MMP8 showed higher expression in most cell types of tumor samples (Figure S5B). Furthermore, as shown in Figure S5C,S5D, PAX5 expression in the lymphocytes of lymphoid tissue was higher than in HNSCC tissue, whereas TNFAIP6 expression was higher in fibroblasts and MPs compared to lymphoid tissue.
Discussion
Head and neck tumors represent a significant proportion of global cancer incidences, with HNSCC comprising about 90% of these cases. A major challenge in treating HNSCC is the late detection due to vague early symptoms, which diminishes the effectiveness of early intervention strategies (66,67). Similarly, DLBCL, accounting for a substantial share of non-Hodgkin lymphomas, is characterized by its aggressive nature and rapid progression. Both diseases share critical challenges in treatment, notably the inefficacy of standard therapies such as immunotherapy for certain patients (68). The urgency for developing personalized and more effective treatment options is clear, as both diseases exhibit complexities in their pathophysiology, often requiring the identification of reliable biomarkers to aid in targeted therapy (69-72). Previous research has predominantly focused on each disease separately without exploring the shared pathological features and potential common therapeutic targets. This gap highlights the necessity for integrated studies that consider both HNSCC and DLBCL, aiming to uncover mutual pathways and biomarkers that could lead to better diagnostic and therapeutic approaches.
In this work, we intersected the key genes linked to DLBCL which were identified using WGCNA with the DEGs from the HNSCC dataset. Consequently, enrichment analyses with GO, KEGG, and DO were performed on the shared genes. The most enriched GO terms in biological processes suggested an abnormal enhancement of cell activation and positive regulation of cell activation in tumor cells, which may be involved in the proliferation, metastasis, and invasion of cancer cells (73). Meanwhile, the regulation of leukocyte activation and differentiation was implicated in modulating the immune response and tumor immunity (74). Plus, lymphocyte activation played a role in recognizing and eliminating tumor cells (75,76). In the KEGG enrichment analysis, Hematopoietic cell lineage refers to the development process of different cell types in the blood cell lineage. Hematopoietic stem cells differentiate and mature to form various types of blood cells, including red blood cells, white blood cells, and platelets. B cells are an important part of the hematopoietic cell lineage and play a crucial role in the immune response (77). In DLBCL, B cells proliferate abnormally and form malignant tumors consisting of large B cell clones. These malignant cells lose their normal cell differentiation and regulatory mechanisms, leading to uncontrolled proliferation and survival. The JAK-STAT signaling pathway is closely associated with the occurrence and development of various tumors, including hematologic tumors and solid tumors (78). Dysregulation of this pathway can contribute to abnormal cell growth and survival in DLBCL and HNSCC (79,80). The IL-17 signaling pathway involves the pro-inflammatory cytokine IL-17, which is involved in inflammatory responses and immune regulation. Abnormal activation of the IL-17 pathway has been observed in various tumors, such as breast cancer and prostate cancer, and may contribute to tumor formation and progression (81). Th1 and Th2 cells are two different subtypes of helper T cells in the immune system. Dysregulated differentiation of Th1 and Th2 cells can lead to an abnormal immune response, which has been implicated in the development and progression of various tumors (82).
ROC analysis and K-M studies have demonstrated that ACACB, MMP8, PAX5, and TNFAIP6 were associated with favorable prognoses in HNSCC and possessed high diagnostic value in DLBCL, highlighting their potential as biomarkers. ACACB is traditionally associated with fatty acid metabolism. However, within the tumor microenvironment, it is posited to modulate the metabolic status and functionality of immune cells, thus exerting an indirect influence on the immunological response to tumors. It has been identified as a biomarker for oral squamous cell carcinoma, focusing on the role in fatty acid metabolism and the regulation of energetic homeostasis (83). MMP8 participates in the remodeling of the extracellular matrix. During tumorigenesis, alterations in the extracellular matrix mediated by MMP8 may condition the tumor milieu, potentially hindering or facilitating immune cell infiltration and subsequent antitumor immunity. PAX5 serves as a transcriptional overseer crucial for the ontogeny and maturation of B-cell lymphocytes. Its regulatory clout extends to modulating the transcriptional equipoise of other genes implicated in oncogenesis, hence contributing to the transcriptional aberrancies encountered in cancer. TNFAIP6 operates as an effector downstream of tumor necrosis factor (TNF) signaling, implicated in cellular adhesion, motility, and the inflammatory cascade. Notably, MMP8 and PAX5 have also previously been reported as biomarkers for DLBCL, whereas TNFAIP6 has not yet been explored (84-87). Using the four hub genes as a gene panel, we developed a nomogram to enhance disease diagnosis and prognostic predictions in patients with HNSCC and DLBCL. Additionally, we conducted a drug sensitivity analysis, utilizing the predictive power of these hub genes to tailor treatment options for patients, thereby enhancing the potential for personalized medicine in these cancers.
Recently, a substantial body of evidence has demonstrated that the complex interaction among TMB and MSI plays a crucial role in determining the effectiveness of immunotherapy (88,89). Tumor mutation burden refers to the number of mutations accumulated in a fixed length of the coding region of the tumor genome. High TMB implies that more neoantigens may be expressed on the surface of tumor cells (90), potentially enhancing the immune system’s capacity to identify and attack tumor cells. MSI refers to variations in the length of repetitive sequences in tumor DNA. High MSI (MSI-H) is often associated with defects in DNA repair mechanisms, particularly in the mismatch repair (MMR) system. MSI-H tumors also tend to produce more neoantigens, which activate the immune system (91). Both high TMB and MSI-H status can be used as biomarkers to predict tumor response to specific immunotherapies. For example, immune checkpoint inhibitors, such as PD-1/PD-L1 and CTLA-4 inhibitors, have been shown to have a positive therapeutic impact on tumors with high TMB or MSI-H status (92,93). Our study found that the expression of four hub genes was significantly correlated with CTLA-4 immunosuppressive pathway-related checkpoints (CTLA4, CD80, CD86) in the HNSCC dataset, and PD-1/PD-L1 immunosuppressive pathway-related checkpoints (CD274, PDCD1) in the DLBCL dataset. Moreover, ACACB exhibited high MSI in DLBCL, while TNFAIP6 and PAX5 showed high MSI in HNSCC. Notably, PAX5 also demonstrated high TMB in HNSCC. Therefore, based on these findings, clinical trials can be designed and conducted for patients with DLBCL and HNSCC who exhibit these genetic traits. These trials aim to test new treatments or improved protocols for existing treatments.
In addition to TMB and MSI, immune cell infiltration is also an important factor. These cells can significantly influence tumor growth, spread, and response to therapy. In HNSCC, analyzing and comparing the infiltration of various immune cell types between tumor tissue and normal tissue can provide insights into tumor immune evasion, tumor antigen presentation, and the impact of inflammatory responses on disease progression and treatment outcomes. Our study found significant differences in the proportions of CD4 T cells, macrophages, monocytes, mast cells, B cells, dendritic cells, and NK cells in tumor tissues compared to normal tissues, suggesting avenues for future research. For example, high levels of CD4+ T cell infiltration are often associated with a better prognosis, while the presence of macrophages, especially M2-type macrophages, may promote tumor growth and lead to poor treatment outcomes (94). To enhance the tumor-specific T cell responses, strategic combination therapies can be applied. This entails the concurrent application of immune checkpoint inhibitors alongside adjunctive treatments such as chemotherapy, targeted therapy, or radiotherapy, which collectively serve to amplify the immunogenicity of tumors, thereby bolstering T cell-mediated anti-tumor activity. Furthermore, adoptive T cell transfer represents a cutting-edge approach, wherein T cells are ex vivo engineered to possess cancer-specific receptors, for instance, chimeric antigen receptors (CARs), followed by their subsequent reinfusion into patients. Such methodologies have shown considerable promise, particularly within the realm of hematological malignancies, including DLBCL. In the tumor microenvironment, the function of immune infiltrating cells is also affected by local factors such as inhibitory cytokines, metabolites, and signaling molecules released by tumor cells. Understanding how these factors interact and influence the efficacy of immune cells is crucial for developing novel therapeutic approaches.
Regarding the scRNA data analysis, the increased abundance of fibroblasts in HNSCC might be attributed to significant changes in the tumor microenvironment, such as heightened inflammation, hypoxia, and altered extracellular matrix composition (95,96). These changes could promote the recruitment and activation of fibroblasts, crucial for tissue remodeling and tumor progression. Cancer-associated fibroblasts have been reported to lead to poor prognosis in HPV-positive HNSCC patients (97). EMT is a process where epithelial cells acquire mesenchymal characteristics, allowing cells to migrate and invade surrounding tissue, promoting tumor growth and metastasis (98). MPs are immunosuppressive cells originating from bone marrow, having immunosuppressive and immune tolerance effects. In the tumor microenvironment, MPs can promote tumor growth and progression and inhibit the immune system’s tumor-killing effect (99).
Moreover, MMP8 shows higher expression in five cell types in HNSCC tissues, indicating its involvement in disease processes such as tumor metastasis (85). TNFAIP6 exhibits higher expression specifically in fibroblasts and MPs. This gene, initially cloned from TNF-stimulated diploid human fibroblasts, encodes a secreted protein with a hyaluronic acid-binding domain (100). This protein is crucial for stabilizing the extracellular matrix and regulating cell migration, making it a key modulator of extracellular matrix organization during tissue restructuring (101). Conversely, PAX5 shows higher expression in lymphoid tissues compared to HNSCC tissues. The gene is predominantly expressed in B lymphocytes and B cell lymphoma, encoding members of the paired-box family of TFs. These factors play a crucial role in early development and are implicated in tumor transformation through altered gene expression (102).
One major limitation in this study is that the analysis was confined to data from only three datasets (TCGA, GSE83632 and GSE172577). The small DLBCL sample size used in this study, as well as a lack of clinical information, may have an impact on the findings’ generalizability and statistical power. Targets identified through our analysis of gene expression levels may not accurately reflect their function and importance in actual biological processes, necessitating further functional trials and clinical validation. Moving forward, our immediate goal is to champion or contribute to clinical trials aimed at evaluating novel therapeutic interventions or drug combinations emergent from preliminary investigations. This entails fostering collaborative efforts with multidisciplinary teams to spearhead innovations in treatment modalities, diagnostic methodologies, and patient care protocols. The prevailing challenge within current research paradigms stems from the extensive genetic and phenotypic heterogeneity manifested by both HNSCC and DLBCL, which invariably complicates the delineation of effective treatment frameworks.
Conclusions
In this study, we have successfully utilized bioinformatics and ML algorithms to identify four key hub genes (ACACB, MMP8, PAX5, and TNFAIP6). Our survival analysis indicates that these genes hold promise as diagnostic markers for DLBCL in patients with HNSCC. We have developed a diagnostic nomogram that integrates these biomarkers, providing a valuable tool for clinical decision-making and potentially improving patient outcomes. Moreover, our findings shed new light on the complex pathogenesis of DLBCL in the context of HNSCC, offering fresh perspectives that could guide future research and therapeutic strategies. While our results are promising, further validation studies are essential to firmly establish the clinical utility of these biomarkers in enhancing the diagnosis and treatment of DLBCL with HNSCC. Future research should focus on longitudinal studies and clinical trials to confirm these genes’ roles and efficacy in clinical settings, aiming to translate these insights into actionable, patient-specific therapeutic interventions.
Acknowledgments
Funding: This research received financial support from
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1064/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1064/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1064/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin 2024;74:12-49. [Crossref] [PubMed]
- Solomon B, Young RJ, Rischin D. Head and neck squamous cell carcinoma: Genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin Cancer Biol 2018;52:228-40. [Crossref] [PubMed]
- Li JSZ, Abbasi A, Kim DH, et al. Chromosomal fragile site breakage by EBV-encoded EBNA1 at clustered repeats. Nature 2023;616:504-9. [Crossref] [PubMed]
- Johnson DE, Burtness B, Leemans CR, et al. Head and neck squamous cell carcinoma. Nat Rev Dis Primers 2020;6:92. [Crossref] [PubMed]
- Ludwig R, Schubert A, Barbatei D, et al. A multi-centric dataset on patient-individual pathological lymph node involvement in head and neck squamous cell carcinoma. Data Brief 2023;52:110020. [Crossref] [PubMed]
- Hamoir M, Schmitz S, Gregoire V. The role of neck dissection in squamous cell carcinoma of the head and neck. Curr Treat Options Oncol 2014;15:611-24. [Crossref] [PubMed]
- Wang X, Guo J, Yu P, et al. The roles of extracellular vesicles in the development, microenvironment, anticancer drug resistance, and therapy of head and neck squamous cell carcinoma. J Exp Clin Cancer Res 2021;40:35. [Crossref] [PubMed]
- Iosselevitch I, Tabibian-Keissar H, Barshack I, et al. Gastric DLBCL clonal evolution as function of patient age. Front Immunol 2022;13:957170. [Crossref] [PubMed]
- Decruyenaere P, Offner F, Vandesompele J. Circulating RNA biomarkers in diffuse large B-cell lymphoma: a systematic review. Exp Hematol Oncol 2021;10:13. [Crossref] [PubMed]
- Lamy T, Damaj G, Soubeyran P, et al. R-CHOP 14 with or without radiotherapy in nonbulky limited-stage diffuse large B-cell lymphoma. Blood 2018;131:174-81. [Crossref] [PubMed]
- Beltran BE, Castro D, Paredes S, et al. EBV-positive diffuse large B-cell lymphoma, not otherwise specified: 2020 update on diagnosis, risk-stratification and management. Am J Hematol 2020;95:435-45. [Crossref] [PubMed]
- Karbalaie Niya MH, Safarnezhad Tameshkel F, Keyvani H, et al. Epstein-Barr virus molecular epidemiology and variants identification in head and neck squamous cell carcinoma. Eur J Cancer Prev 2020;29:523-30. [Crossref] [PubMed]
- Núñez-Acurio D, Bravo D, Aguayo F. Epstein-Barr Virus-Oral Bacterial Link in the Development of Oral Squamous Cell Carcinoma. Pathogens 2020;9:1059. [Crossref] [PubMed]
- Galizia D, Minei S, Maldi E, et al. How Risk Factors Affect Head and Neck Squamous Cell Carcinoma (HNSCC) Tumor Immune Microenvironment (TIME): Their Influence on Immune Escape Mechanisms and Immunotherapy Strategy. Biomedicines 2022;10:2498. [Crossref] [PubMed]
- Rahman R, Shaikh MH, Gopinath D, et al. Human papillomavirus and Epstein-Barr virus co-infection in oral and oropharyngeal squamous cell carcinomas: A systematic review and meta-analysis. Mol Oral Microbiol 2023;38:259-74. [Crossref] [PubMed]
- Du Q, Guo X, Wang M, et al. The application and prospect of CDK4/6 inhibitors in malignant solid tumors. J Hematol Oncol 2020;13:41. [Crossref] [PubMed]
- Nan Z, Dou Y, Chen A, et al. Identification and validation of a prognostic signature of autophagy, apoptosis and pyroptosis-related genes for head and neck squamous cell carcinoma: to imply therapeutic choices of HPV negative patients. Front Immunol 2023;13:1100417. [Crossref] [PubMed]
- Han H, Fan G, Song S, et al. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood 2021;137:1603-14. [Crossref] [PubMed]
- William WN Jr, Zhao X, Bianchi JJ, et al. Immune evasion in HPV(-) head and neck precancer-cancer transition is driven by an aneuploid switch involving chromosome 9p loss. Proc Natl Acad Sci U S A 2021;118:e2022655118. [Crossref] [PubMed]
- Jhunjhunwala S, Hammer C, Delamarre L. Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer 2021;21:298-312. [Crossref] [PubMed]
- Quah HS, Cao EY, Suteja L, et al. Single cell analysis in head and neck cancer reveals potential immune evasion mechanisms during early metastasis. Nat Commun 2023;14:1680. [Crossref] [PubMed]
- Nie M, Ren W, Ye X, et al. The dual role of CD70 in B-cell lymphomagenesis. Clin Transl Med 2022;12:e1118. [Crossref] [PubMed]
- Ghorani E, Swanton C, Quezada SA. Cancer cell-intrinsic mechanisms driving acquired immune tolerance. Immunity 2023;56:2270-95. [Crossref] [PubMed]
- Kobayashi Y, Lim SO, Yamaguchi H. Oncogenic signaling pathways associated with immune evasion and resistance to immune checkpoint inhibitors in cancer. Semin Cancer Biol 2020;65:51-64. [Crossref] [PubMed]
- Wu SY, Fu T, Jiang YZ, et al. Natural killer cells in cancer biology and therapy. Mol Cancer 2020;19:120. [Crossref] [PubMed]
- Song JY, Nwangwu M, He TF, et al. Low T-cell proportion in the tumor microenvironment is associated with immune escape and poor survival in diffuse large B-cell lymphoma. Haematologica 2023;108:2167-77. [Crossref] [PubMed]
- Elmusrati A, Wang J, Wang CY. Tumor microenvironment and immune evasion in head and neck squamous cell carcinoma. Int J Oral Sci 2021;13:24. [Crossref] [PubMed]
- Lainé A, Labiad O, Hernandez-Vargas H, et al. Regulatory T cells promote cancer immune-escape through integrin αvβ8-mediated TGF-β activation. Nat Commun 2021;12:6228. [Crossref] [PubMed]
- Kumari N, Choi SH. Tumor-associated macrophages in cancer: recent advancements in cancer nanoimmunotherapies. J Exp Clin Cancer Res 2022;41:68. [Crossref] [PubMed]
- Wu Y, Yi M, Niu M, et al. Myeloid-derived suppressor cells: an emerging target for anticancer immunotherapy. Mol Cancer 2022;21:184. [Crossref] [PubMed]
- Topper MJ, Vaz M, Marrone KA, et al. The emerging role of epigenetic therapeutics in immuno-oncology. Nat Rev Clin Oncol 2020;17:75-90. [Crossref] [PubMed]
- Griffin GK, Wu J, Iracheta-Vellve A, et al. Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature 2021;595:309-14. [Crossref] [PubMed]
- Maier B, Leader AM, Chen ST, et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 2020;580:257-62. [Crossref] [PubMed]
- Chen W. TGF-β Regulation of T Cells. Annu Rev Immunol 2023;41:483-512. [Crossref] [PubMed]
- Budimir N, Thomas GD, Dolina JS, et al. Reversing T-cell Exhaustion in Cancer: Lessons Learned from PD-1/PD-L1 Immune Checkpoint Blockade. Cancer Immunol Res 2022;10:146-53. [Crossref] [PubMed]
- Azzaoui I, Uhel F, Rossille D, et al. T-cell defect in diffuse large B-cell lymphomas involves expansion of myeloid-derived suppressor cells. Blood 2016;128:1081-92. [Crossref] [PubMed]
- Clough E, Barrett T, Wilhite SE, et al. NCBI GEO: archive for gene expression and epigenomics data sets: 23-year update. Nucleic Acids Res 2024;52:D138-44. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Yu G, Wang LG, Yan GR, et al. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 2015;31:608-9. [Crossref] [PubMed]
- Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023;51:D638-46. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Ranstam J, Cook JA. Kaplan-Meier curve. Br J Surg 2017;104:442. [Crossref] [PubMed]
- Park SH, Goo JM, Jo CH. Receiver operating characteristic (ROC) curve: practical review for radiologists. Korean J Radiol 2004;5:11-8. [Crossref] [PubMed]
- Franz M, Rodriguez H, Lopes C, et al. GeneMANIA update 2018. Nucleic Acids Res 2018;46:W60-4. [Crossref] [PubMed]
- Park SY. Nomogram: An analogue tool to deliver digital knowledge. J Thorac Cardiovasc Surg 2018;155:1793. [Crossref] [PubMed]
- Basu A, Bodycombe NE, Cheah JH, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 2013;154:1151-61. [Crossref] [PubMed]
- Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 2010;28:1248-50. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Liao C, Wang X. TCGAplot: an R package for integrative pan-cancer analysis and visualization of TCGA multi-omics data. BMC Bioinformatics 2023;24:483. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Rossi D, Spina V, Gaidano G. Biology and treatment of Richter syndrome. Blood 2018;131:2761-72. [Crossref] [PubMed]
- Iop A, Cartei G, Isaia A. Vinorelbine, bleomycin and methotrexate as a salvage therapy for patients with head and neck squamous carcinoma in relapse after cisplatin/fluorouracil. Ann Oncol 1998;9:225-7. [Crossref] [PubMed]
- Cohen EEW, Soulières D, Le Tourneau C, et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet 2019;393:156-67. [Crossref] [PubMed]
- Lancman G, Steinberg A, Gabrilove J. Serpentine Supravenous Hyperpigmentation in an HIV Patient Receiving R-CHOP for DLBCL. Int J Hematol Oncol Stem Cell Res 2018;12:1-3. [PubMed]
- Miyazaki K, Asano N, Yamada T, et al. DA-EPOCH-R combined with high-dose methotrexate in patients with newly diagnosed stage II-IV CD5-positive diffuse large B-cell lymphoma: a single-arm, open-label, phase II study. Haematologica 2020;105:2308-15. [Crossref] [PubMed]
- Tanaka N, Patel AA, Tang L, et al. Replication Stress Leading to Apoptosis within the S-phase Contributes to Synergism between Vorinostat and AZD1775 in HNSCC Harboring High-Risk TP53 Mutation. Clin Cancer Res 2017;23:6541-54. [Crossref] [PubMed]
- Cottereau AS, Nioche C, Dirand AS, et al. (18)F-FDG PET Dissemination Features in Diffuse Large B-Cell Lymphoma Are Predictive of Outcome. J Nucl Med 2020;61:40-5. [Crossref] [PubMed]
- Wilson MR, Eyre TA, Martinez-Calle N, et al. Timing of high-dose methotrexate CNS prophylaxis in DLBCL: an analysis of toxicity and impact on R-CHOP delivery. Blood Adv 2020;4:3586-93. [Crossref] [PubMed]
- Godfrey J, Mei M, Chen L, et al. Results from a phase I trial of pembrolizumab plus vorinostat in relapsed/refractory B-cell non-Hodgkin lymphoma. Haematologica 2024;109:533-42. [Crossref] [PubMed]
- Galanina N, Smith SM, Liao C, et al. University of Chicago phase II consortium trial of selumetinib (MEKi) demonstrates low tolerability and efficacy in relapsed DLBCL. Br J Haematol 2018;181:264-7. [Crossref] [PubMed]
- Cui L, Lu Y, Zheng J, et al. ACTN1 promotes HNSCC tumorigenesis and cisplatin resistance by enhancing MYH9-dependent degradation of GSK-3β and integrin β1-mediated phosphorylation of FAK. J Exp Clin Cancer Res 2023;42:335. [Crossref] [PubMed]
- van Imhoff GW, McMillan A, Matasar MJ, et al. Ofatumumab Versus Rituximab Salvage Chemoimmunotherapy in Relapsed or Refractory Diffuse Large B-Cell Lymphoma: The ORCHARRD Study. J Clin Oncol 2017;35:544-51. [Crossref] [PubMed]
- Niitsu N, Kohuri M, Higashihara M, et al. Phase II study of the CPT-11, mitoxantrone and dexamethasone regimen in combination with rituximab in elderly patients with relapsed diffuse large B-cell lymphoma. Cancer Sci 2006;97:933-7. [Crossref] [PubMed]
- Bhattacharya A, Tóth K, Mazurchuk R, et al. Lack of microvessels in well-differentiated regions of human head and neck squamous cell carcinoma A253 associated with functional magnetic resonance imaging detectable hypoxia, limited drug delivery, and resistance to irinotecan therapy. Clin Cancer Res 2004;10:8005-17. [Crossref] [PubMed]
- Wilson WH, Wright GW, Huang DW, et al. Effect of ibrutinib with R-CHOP chemotherapy in genetic subtypes of DLBCL. Cancer Cell 2021;39:1643-1653.e3. [Crossref] [PubMed]
- Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 2018;24:679-90. [Crossref] [PubMed]
- Cheson BD, Nowakowski G, Salles G. Diffuse large B-cell lymphoma: new targets and novel therapies. Blood Cancer J 2021;11:68. [Crossref] [PubMed]
- He J, Chen Z, Xue Q, et al. Identification of molecular subtypes and a novel prognostic model of diffuse large B-cell lymphoma based on a metabolism-associated gene signature. J Transl Med 2022;20:186. [Crossref] [PubMed]
- Cai H, Liang J, Jiang Y, et al. KLF7 regulates super-enhancer-driven IGF2BP2 overexpression to promote the progression of head and neck squamous cell carcinoma. J Exp Clin Cancer Res 2024;43:69. [Crossref] [PubMed]
- Yin J, Zheng S, He X, et al. Identification of molecular classification and gene signature for predicting prognosis and immunotherapy response in HNSCC using cell differentiation trajectories. Sci Rep 2022;12:20404. [Crossref] [PubMed]
- Liu X, Liu Y, Liu J, et al. Correlation between the gut microbiome and neurodegenerative diseases: a review of metagenomics evidence. Neural Regen Res 2024;19:833-45. [Crossref] [PubMed]
- Gadwa J, Amann M, Bickett TE, et al. Selective targeting of IL2Rβγ combined with radiotherapy triggers CD8- and NK-mediated immunity, abrogating metastasis in HNSCC. Cell Rep Med 2023;4:101150. [Crossref] [PubMed]
- Yang S, Jia J, Wang F, et al. Targeting neutrophils: Mechanism and advances in cancer therapy. Clin Transl Med 2024;14:e1599. [Crossref] [PubMed]
- Hodgins JJ, Khan ST, Park MM, et al. Killers 2.0: NK cell therapies at the forefront of cancer control. J Clin Invest 2019;129:3499-510. [Crossref] [PubMed]
- Vignali PDA, DePeaux K, Watson MJ, et al. Hypoxia drives CD39-dependent suppressor function in exhausted T cells to limit antitumor immunity. Nat Immunol 2023;24:267-79. [Crossref] [PubMed]
- Ng AP, Coughlan HD, Hediyeh-Zadeh S, et al. An Erg-driven transcriptional program controls B cell lymphopoiesis. Nat Commun 2020;11:3013. [Crossref] [PubMed]
- Quintás-Cardama A, Verstovsek S. Molecular pathways: Jak/STAT pathway: mutations, inhibitors, and resistance. Clin Cancer Res 2013;19:1933-40. [Crossref] [PubMed]
- Lai SY, Childs EE, Xi S, et al. Erythropoietin-mediated activation of JAK-STAT signaling contributes to cellular invasion in head and neck squamous cell carcinoma. Oncogene 2005;24:4442-9. [Crossref] [PubMed]
- Radke J, Ishaque N, Koll R, et al. The genomic and transcriptional landscape of primary central nervous system lymphoma. Nat Commun 2022;13:2558. [Crossref] [PubMed]
- Wu N, Wang Y, Wang K, et al. Cathepsin K regulates the tumor growth and metastasis by IL-17/CTSK/EMT axis and mediates M2 macrophage polarization in castration-resistant prostate cancer. Cell Death Dis 2022;13:813. [Crossref] [PubMed]
- Ruterbusch M, Pruner KB, Shehata L, et al. In Vivo CD4(+) T Cell Differentiation and Function: Revisiting the Th1/Th2 Paradigm. Annu Rev Immunol 2020;38:705-25. [Crossref] [PubMed]
- Fan Y, Wang J, Wang Y, et al. Development and Clinical Validation of a Novel 5 Gene Signature Based on Fatty Acid Metabolism-Related Genes in Oral Squamous Cell Carcinoma. Oxid Med Cell Longev 2022;2022:3285393. [Crossref] [PubMed]
- Yushi Q, Li Z, Von Roemeling CA, et al. Osteopontin is a multi-faceted pro-tumorigenic driver for central nervous system lymphoma. Oncotarget 2016;7:32156-71. [Crossref] [PubMed]
- Liu M, Huang L, Liu Y, et al. Identification of the MMP family as therapeutic targets and prognostic biomarkers in the microenvironment of head and neck squamous cell carcinoma. J Transl Med 2023;21:208. [Crossref] [PubMed]
- Kurimoto K, Hayashi M, Guerrero-Preston R, et al. PAX5 gene as a novel methylation marker that predicts both clinical outcome and cisplatin sensitivity in esophageal squamous cell carcinoma. Epigenetics 2017;12:865-74. [Crossref] [PubMed]
- Balasenthil S, Gururaj AE, Talukder AH, et al. Identification of Pax5 as a target of MTA1 in B-cell lymphomas. Cancer Res 2007;67:7132-8. [Crossref] [PubMed]
- Hu C, Zhao L, Liu W, et al. Genomic profiles and their associations with TMB, PD-L1 expression, and immune cell infiltration landscapes in synchronous multiple primary lung cancers. J Immunother Cancer 2021;9:e003773. [Crossref] [PubMed]
- Fong W, Li Q, Ji F, et al. Lactobacillus gallinarum-derived metabolites boost anti-PD1 efficacy in colorectal cancer by inhibiting regulatory T cells through modulating IDO1/Kyn/AHR axis. Gut 2023;72:2272-85. [Crossref] [PubMed]
- McGrail DJ, Pilié PG, Rashid NU, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol 2021;32:661-72. [Crossref] [PubMed]
- Marabelle A, Le DT, Ascierto PA, et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J Clin Oncol 2020;38:1-10. [Crossref] [PubMed]
- Weng J, Li S, Zhu Z, et al. Exploring immunotherapy in colorectal cancer. J Hematol Oncol 2022;15:95. [Crossref] [PubMed]
- Kasi PM, Budde G, Krainock M, et al. Circulating tumor DNA (ctDNA) serial analysis during progression on PD-1 blockade and later CTLA-4 rescue in patients with mismatch repair deficient metastatic colorectal cancer. J Immunother Cancer 2022;10:e003312. [Crossref] [PubMed]
- Schroeder BA, LaFranzo NA, LaFleur BJ, et al. CD4+ T cell and M2 macrophage infiltration predict dedifferentiated liposarcoma patient outcomes. J Immunother Cancer 2021;9:e002812. [Crossref] [PubMed]
- Schwörer S, Cimino FV, Ros M, et al. Hypoxia Potentiates the Inflammatory Fibroblast Phenotype Promoted by Pancreatic Cancer Cell-Derived Cytokines. Cancer Res 2023;83:1596-610. [Crossref] [PubMed]
- Affo S, Yu LX, Schwabe RF. The Role of Cancer-Associated Fibroblasts and Fibrosis in Liver Cancer. Annu Rev Pathol 2017;12:153-86. [Crossref] [PubMed]
- Kürten CHL, Kulkarni A, Cillo AR, et al. Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing. Nat Commun 2021;12:7338. [Crossref] [PubMed]
- Lambert AW, Weinberg RA. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat Rev Cancer 2021;21:325-38. [Crossref] [PubMed]
- Olingy CE, Dinh HQ, Hedrick CC. Monocyte heterogeneity and functions in cancer. J Leukoc Biol 2019;106:309-22. [Crossref] [PubMed]
- Wisniewski HG, Vilcek J. Cytokine-induced gene expression at the crossroads of innate immunity, inflammation and fertility: TSG-6 and PTX3/TSG-14. Cytokine Growth Factor Rev 2004;15:129-46. [Crossref] [PubMed]
- Kuznetsova SA, Mahoney DJ, Martin-Manso G, et al. TSG-6 binds via its CUB_C domain to the cell-binding domain of fibronectin and increases fibronectin matrix assembly. Matrix Biol 2008;27:201-10. [Crossref] [PubMed]
- Feldman AL, Dogan A. Diagnostic uses of Pax5 immunohistochemistry. Adv Anat Pathol 2007;14:323-34. [Crossref] [PubMed]


