
Emerging functions of FMNL1 in myeloid neoplasms: insights from bioinformatics to biological and pharmacological landscapes
Highlight box
Key findings
• Formin-like protein 1 (FMNL1) expression is associated with neutrophil differentiation and function, and its high expression in acute myeloid leukemia (AML) correlates with immune response and inflammatory activity pathways. Additionally, FMNL1 expression influences drug sensitivity in AML, suggesting potential therapeutic implications.
What is known and what is new?
• FMNL1 plays a role in the regulation of the actin cytoskeleton, is predominantly expressed in hematopoietic cells, and its silencing reduces cell proliferation and migration in leukemia cells.
• The potential relationship of FMNL1 with the neutrophil-mediated immune response and the high expression of this gene in AML and chronic myeloid leukemia patients.
What is the implication, and what should change now?
• FMNL1 may play a relevant role in the development of AML. A better understanding of the role of FMNL1 in neutrophil differentiation and function may have implications for disease conditions in which these cells are dysregulated.
Introduction
Myeloid neoplasia describes a collection of disorders marked by the abnormal growth and development of myeloid cells, which are a type of blood cell originating from hematopoietic stem cells in the bone marrow (1-3). These disorders can range from relatively benign conditions to aggressive malignancies and are collectively known as myeloid neoplasms. They include myeloproliferative neoplasms, chronic myeloid leukemia (CML), myelodysplastic syndromes (MDS), and acute myeloid leukemia (AML) (1,3,4).
Formin-like protein 1 (FMNL1) is a member of the formin family of proteins, which are known for their roles in the regulation of the actin cytoskeleton (5). Actin cytoskeleton dynamics are crucial for various cellular processes, including cell shape, migration, division, and signaling (6-9). FMNL1, in particular, is implicated in the regulation of actin assembly and has been studied for its specific functions in various biological contexts (5).
FMNL1 has gained attention in cancer research due to its potential involvement in tumor cell migration and invasion. Altered expression of FMNL1 has been observed in various cancers, suggesting it may contribute to the metastatic potential of cancer cells (10). Understanding the mechanisms by which FMNL1 regulates actin dynamics in cancer cells could provide insights into new therapeutic targets.
Since FMNL1 is predominantly expressed in hematopoietic cells (11,12) and a functional study has revealed an important role of this protein in the proliferation, survival, migration, and invasion of leukemia cells (13), in the present study we investigated the expression of FMNL1 in normal hematopoiesis and samples from patients with myeloid neoplasms. In patients with AML, we investigated potential associations of FMNL1 expression with clinical-laboratory characteristics, mutational status, and survival outcomes. Additionally, we performed a functional genomic analysis to identify molecular and biological processes associated with the expression of FMNL1. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1091/rc).
Methods
Bioinformatics
FMNL1 expression in normal and neoplastic hematopoietic cells was evaluated as previously described (14). Briefly, data from samples from patients with myeloid neoplasia and healthy donors were extracted from the GEO2R, Amazonia! database 2008 (15), cBioPortal (https://www.cbioportal.org) (16,17) and BloodSpot (https://www.fobinf.com/) platforms. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Functional genomics analysis was performed as previously described (14). In brief, the following tools were used for the analysis: Galaxy (https://usegalaxy.org/), ClusterVis (https://biit.cs.ut.ee/clustvis/), ShinyGo 0.77 database (http://bioinformatics.sdstate.edu/go/), GSEA v.4.0.15 [for gene set enrichment analysis (GSEA)]. Drug sensitivity prediction was performed as previously described (14) using the Beat AML study (18,19), Genomics of Drug Sensitivity in Cancer (GDSC) resource (https://www.cancerrxgene.org) (20), and Expression Atlas database (www.ebi.ac.uk/gxa/experiments/E-MTAB-2770). The molecular and biological processes associated with drug response associated with FMNL1 expression were identified using the GeneMANIA software (https://genemania.org/). A P value <0.05 and/or a false discovery rate (FDR) of 25% (q value <0.25) were considered statistically significant.
Experimental validation
The cell lines NB4, MOLM-13, MV4-11, OCI-AML3, Kasumi-1, THP-1, and HL-60 were provided by Prof. Eduardo Magalhães Rego from the University of São Paulo in Ribeirão Preto, Brazil. The SET-2 cells were received from Prof. Fabíola Attié de Castro, also at the University of São Paulo in Ribeirão Preto, Brazil. Sara Teresinha Olalla Saad from the University of Campinas in Campinas, Brazil, supplied the HEL, K-562, U-937, and KU812 cells. These cells were cultured following the guidelines provided by the American Type Culture Collection (ATCC) or the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). All-trans retinoic acid (ATRA) was sourced from Sigma-Aldrich (St. Louis, MO, USA). Induction of cellular differentiation in NB4 cells induced by ATRA, validation by flow cytometry, and morphological analysis were performed as previously described (14). Gene expression analysis by quantitative polymerase chain reaction (qPCR) was performed as previously described (14), but using the primers for FMNL1 [forward (F): GAAGCTGAAGAGCTATGTGG, reverse (R): CCTGCGTGGACTCCTGAACT] and HPRT1 (F: GAACGTCTTGCTCGAGATGTGA, R: TCCAGCAGGTCAGCAAAGAAT). The relative expression levels were calculated using the 2−ΔΔCT method (21). Analysis of protein expression by Western blot was performed as previously detailed (14) and using antibodies against FMNL1 (sc-81274, Santa Cruz Biotechnology, Santa Cruz, CA, USA), tubulin (#2144, Cell Signaling Technology, Danvers, MA, USA) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (#5174, Cell Signaling Technology).
Statistical analysis
The parameters of overall survival (OS) and disease-free survival (DFS) for the AML cohort (n=173) (22) were previously defined (14). Statistical analyzes were carried out using SPSS Statistics for Windows, version 21.0 (SPSS, Chicago, IL, USA), Stata Statistic/Data Analysis 14.1 (Stata Corp., College Station, TX, USA), GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA) and R package Cutpointr (23,24). Kaplan-Meier curves were used to estimate survival, with comparisons made using the Cox regression and/or log-rank test. For continuous variables, statistical tests such as Student’s t-test, Mann-Whitney, Kruskal-Wallis, or analysis of variance (ANOVA) were employed. For categorical variables, the Fisher’s exact test or Chi-squared test was used. A P value of <0.05 was considered statistically significant.
Results
FMNL1 is highly expressed upon granulocytic cell differentiation
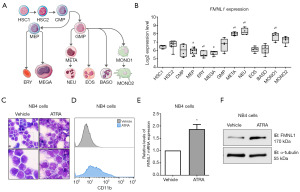
Firstly, we investigated FMNL1 expression in different cell populations of normal hematopoietic origin. FMNL1 was highly expressed in metamyelocytes, neutrophils, and monocytes when evaluated against hematopoietic stem cells (all P<0.05). On the other hand, FMNL1 expression was reduced in megakaryocyte-erythroid progenitors, erythrocytes, and megakaryocytes in relation to hematopoietic stem cells (all P<0.05) (Figure 1A,1B). In NB4 cells, FMNL1 expression increased after ATRA-induced granulocytic differentiation (Figure 1C-1F).
FMNL1 expression in myeloid neoplasms
Next, we investigated the expression of FMNL1 in myeloid neoplasms. FMNL1 expression is elevated in AML and CML patients with relative to expression in samples from healthy donors (all P<0.01, Figure 2A). No significant differences were observed in FMNL1 expression according to the molecular risk of AML patients (Figure 2B). The stratification of AML patients into different cytogenetic risk groups indicated a trend towards higher FMNL1 expression in patients with complex karyotype AML (Figure 2C). In AML patients, FMNL1 expression did not significantly impact survival outcomes (Figure 2D) and was not associated with relevant clinical and laboratory characteristics (Table 1). Mutational analysis revealed that reduced FMNL1 expression is linked to an increased frequency of WT1 mutations (P=0.02, Table 2). In cellular models of myeloid neoplasia, the expression of FMNL1 is varied and its high expression stands out in Kasumi-1 cells (a t(8;21) AML cell line), MOLM-13 (a FLT3-ITD AML cell line), KU812 (a CML-derived cell line), and HEL (a JAK2V617F AML cell line) (Figure 2E,2F). In U-937 cells, a smaller band was observed for the expected intensity for FMNL1, suggesting a truncated form or a smaller variant transcript as predominant in this cell line (Figure S1).
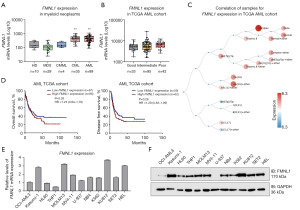
Table 1
| Clinicopathological factors† | Total | FMNL1 | ||
|---|---|---|---|---|
| Low‡ | High‡ | P value¶ | ||
| Total, n [%] | 173 [100] | 87 [50.3] | 86 [49.7] | |
| Age (years) | 0.54 | |||
| <60 | 91 [53] | 48 [55.2] | 43 [50.0] | |
| ≥60 | 82 [47] | 39 [44.8] | 43 [50.0] | |
| Age (years), median [range] | 58 [18–88] | 57 [21–82] | 60 [18–88] | 0.25 |
| Bone marrow blasts (%), median [range] | 72 [30–100] | 72 [30–100] | 72 [30–99] | 0.88 |
| White blood cell count (×109/L), median [range] | 17 [0.4–297.4] | 17.5 [0.5–297] | 19.5 [0.4–224] | 0.12 |
| Gender, n [%] | 0.76 | |||
| Male | 92 [53] | 45 [51.7] | 47 [54.7] | |
| Female | 81 [47] | 42 [48.3] | 39 [45.3] | |
| Molecular risk§, n [%] | 0.18 | |||
| Good | 33 [20] | 19 [22.4] | 14 [16.5] | |
| Intermediate | 95 [55] | 50 [58.8] | 45 [52.9] | |
| Poor | 42 [25] | 16 [18.8] | 26 [30.6] | |
†, the clinical and laboratorial data of TCGA AML cohort were obtained from cBioPortal for Cancer Genomics (http://www.cbioportal.org); ‡, gene expression values were dichotomized by ROC curve and C statistic; §, molecular risk was stratified according TCGA study; 3 AML patients were not classified; ¶, for statistical analyzes, Mann-Whitney test was used for measured factors, and Fisher’s exact test or Chi-squared test was used for categorical factors. FMNL1, formin-like protein 1; TCGA, The Cancer Genome Atlas; AML, acute myeloid leukemia; ROC, receiver operating characteristic.
Table 2
| Gene† | TCGA AML cohort (n=173), n (%) | High FMNL1‡ (n=86), n (%) | Low FMNL1‡ (n=87), n (%) | P value§ |
|---|---|---|---|---|
| FLT3 | 49 (28.3) | 20 (23.3) | 29 (33.3) | 0.17 |
| NPM1 | 48 (27.7) | 21 (24.4) | 27 (31.0) | 0.39 |
| DNMT3A | 42 (24.3) | 21 (24.4) | 21 (24.1) | >0.99 |
| IDH2 | 17 (9.8) | 11 (12.8) | 6 (6.9) | 0.21 |
| IDH1 | 16 (9.2) | 9 (10.5) | 7 (8.0) | 0.61 |
| RUNX1 | 15 (8.7) | 7 (8.1) | 8 (9.2) | >0.99 |
| TET2 | 15 (8.7) | 5 (5.8) | 10 (11.5) | 0.28 |
| TP53 | 14 (8.1) | 5 (5.8) | 9 (10.3) | 0.41 |
| NRAS | 12 (6.9) | 7 (8.1) | 5 (5.7) | 0.57 |
| CEBPA | 13 (7.5) | 6 (7.0) | 7 (8.0) | >0.99 |
| WT1 | 10 (5.8) | 1 (1.2) | 9 (10.3) | 0.02 |
| PTPN11 | 8 (4.6) | 3 (3.5) | 5 (5.7) | 0.72 |
| KIT | 7 (4.0) | 6 (7.0) | 1 (1.1) | 0.06 |
| KRAS | 7 (4.0) | 3 (3.5) | 4 (4.6) | >0.99 |
| U2AF1 | 7 (4.0) | 4 (4.7) | 3 (3.4) | 0.72 |
†, mutations with frequency >4% in the total cohort (TCGA AML study) were included in the analysis. Genes are described according to the HUGO Gene Nomenclature Committee; ‡, the frequency of mutations was estimated in each group of patients (i.e., high and low FMNL1 expressors); §, Fisher’s exact test. FMNL1, formin-like protein 1; TCGA, The Cancer Genome Atlas; AML, acute myeloid leukemia; HUGO, Human Genome Organisation.
High expression of gene FMNL1 in patients with AML correlates with immune response and inflammatory activity
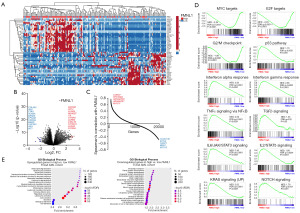
When stratified by FMNL1 expression levels, AML patients exhibit distinct genetic signatures (Figure 3A-3C) that are linked to crucial biological and molecular processes underlying AML development and progression (Figure 3D,3E). Specifically, high FMNL1 expression is associated with a negative enrichment of genes involved in MYC proto-oncogene (MYC) and transcription factor E2F (E2F) targets, as well as the G2/M checkpoint. Conversely, it shows positive enrichment for pathways including the p53 pathway, interferon alpha and gamma responses, TNF signaling via nuclear factor kappa B (NFκB), transforming growth factor beta (TGFβ) signaling, interleukin 6 (IL6)/janus kinase (JAK)/signal transducer and activator of transcription 3 (STAT3) signaling, IL2/STAT5 signaling, KRAS proto-oncogene GTPase (KRAS) signaling, and notch receptor (NOTCH) signaling (all FDR <0.25 and P<0.05, Figure 3D). Gene ontology analysis further identifies several relevant biological processes, notably highlighting upregulated pathways associated with immune response and inflammation such as neutrophil degranulation and granulocyte/neutrophil activation. Collectively, these findings underscore a potential role for FMNL1, particularly in granulocytes, particularly neutrophils, during both homeostasis and disease contexts.
Investigating gene FMNL1 mRNA expression and its impact on drug sensitivity in models of AML
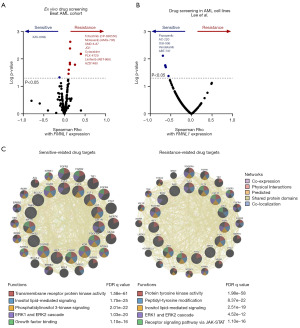
Finally, we investigated whether FMNL1 mRNA levels may impact the effectiveness of antineoplastic agents in AML models. The mRNA levels of FMNL1 showed positive correlations with eight drugs (tofacitinib, motesanib, XMD 8-87, JQ1, cytarabine, PLX-4720, linifanib, and AZD1480; all P<0.05) and negative correlation with one drug (XAV-939b, P<0.05) in assays using AML primary cells from the Beat AML study (Figure 4A). In AML cell lines, FMNL1 expression exhibited negative correlations with five drugs (pazopanib, AC-220, OSI-906, vandetanib, and ABT-737; all P<0.05, Figure 4B). To elucidate the molecular and biological processes associated with drug response based on FMNL1 expression, we analyzed the molecular targets of these drugs in a network. Targets of drugs sensitive to FMNL1 expression were linked with transmembrane receptor protein kinase activity, inositol lipid-mediated signaling, phosphatidylinositol 3-kinase signaling, ERK1 and ERK2 cascade, and growth factor binding (all FDR <0.01, Figure 4C). On the other hand, targets of drugs resistant to FMNL1 expression were associated with protein tyrosine kinase activity, peptidyl-tyrosine modification, inositol lipid-mediated signaling, ERK1 and ERK2 cascade, and receptor signaling pathway via JAK-STAT (all FDR <0.01, Figure 4C).
Discussion
Herein, we explored the role of FMNL1 in samples from patients with myeloid neoplasia, especially AML, under clinical characteristics, biological functions, and potential impacts on pharmacological therapies. In the present study, we observed a higher expression of FMNL1 in patients with CML and AML. In previous studies, high expression of FMNL1 has been associated with neoplastic phenotype, being reported in chronic lymphocytic leukemia (11), T non-Hodgkin’s lymphomas (25), nasopharyngeal carcinoma (26), non-small cell lung cancer (27), clear cell renal cell carcinoma (28,29). In glioma, high FMNL1 expression was an independent predictor of poor prognosis (30). In relation to drugs whose effectiveness was associated with the expression of FMNL1, cytarabine stands out. Cytarabine is part of the “3+7 regimen” used for decades in the treatment of AML (31,32). In our analysis, FMNL1 expression was not associated with relevant clinical and laboratory characteristics or with survival outcomes in patients with AML. Despite these findings, a previous study highlights its participation in the proliferation, survival, migration, and invasion of leukemia cells, highlighting its role in the biology of the disease (13). Thus, the observation that FMNL1 is expressed at higher levels in patients with AML, without affecting prognosis, suggests that FMNL1 may be involved in the disease’s pathophysiology but not in the therapeutic response to the chemotherapy regimen used.
A result that deserves to be highlighted is the elevated expression of FMNL1 in granulocytic and neutrophil precursors as well as the association of high expression of this gene with molecular signatures of immune response and inflammation in patients with AML. FMNL1 is predominantly expressed in hematopoietic cells and plays a significant role in the immune system (11,12,33). It regulates leukocyte shape and motility, which are critical for immune surveillance and response. There are pieces of evidence that this protein helps in the formation of lamellipodia and filopodia, structures that enable leukocytes to move and navigate through tissues (12,34). The role of FMNL1 in macrophages has been the target of several studies, in which this protein participates in adhesion, migration, podosome formation, and phagocytosis (35-37), but future studies investigating the contributions of FMNL1 to the functioning of neutrophils are of interest.
Among the AML-related mutations, low FMNL1 expression was associated with WT1 gene mutations. There is no direct and well-established relationship between FMLN1 and WT1 in terms of their pathways or interactions. They are involved in different cellular processes and are typically studied in different contexts within cancer research. However, both genes can contribute to cancer development through their respective roles in cell structure and regulation (13,38-40), suggesting that any overlap in the pathways they influence could be an area of interest for further research.
Our study provides new insights into the role of FMNL1 in myeloid neoplasms, though it has certain limitations. The available data for CML, MDS, and chronic myelomonocytic leukemia (CMML) are limited for the cohorts analyzed, preventing a thorough clinical-laboratory correlation with FMNL1 expression. This is particularly important for MDS, given its highly heterogeneous nature of the disease. Additionally, our study focused on gene expression; future research should include validation through protein expression analyses.
Conclusions
FMNL1 plays a potential role in granulocyte differentiation and function. FMNL1’s differential expression is associated with key signaling pathways implicated in leukemogenesis and inflammation. This study sheds light on potential therapeutic implications and warrants further investigation.
Acknowledgments
The authors would like to thank Prof. Patricia Favaro (Federal University of São Paulo, Diadema, SP, Brazil) and Prof. Matheus Rodrigues Lopes (Federal University of Vale do São Francisco, Paulo Afonso, BA, Brazil) for past studies that inspire these analyses. The authors would also like to acknowledge all the research participants contributing to The Cancer Genome Atlas (TCGA) resource for providing high quality data for analyses. Figure 1A was adapted with permission from Carvalho et al. “Comprehensive analysis of the HCK gene in myeloid neoplasms: Insights into biological functions, prognosis, and response to antineoplastic agents”. This article was published in Hematol Transfus Cell Ther, Vol number 46, pages 273-82, Copyright Elsevier (2024).
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1091/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1901/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1091/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1091/coif). J.A.M.N. serves as an unpaid editorial board member of Translational Cancer Research from September 2023 to August 2025. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391-405. [Crossref] [PubMed]
- Korn C, Méndez-Ferrer S. Myeloid malignancies and the microenvironment. Blood 2017;129:811-22. [Crossref] [PubMed]
- Arber DA, Orazi A, Hasserjian RP, et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood 2022;140:1200-28. [Crossref] [PubMed]
- Parmar K, Kundu R, Maiti A, et al. Updates in biology, classification, and management of acute myeloid leukemia with antecedent hematologic disorder and therapy related acute myeloid leukemia. Leuk Res 2024;144:107546. [Crossref] [PubMed]
- Colón-Franco JM, Gomez TS, Billadeau DD. Dynamic remodeling of the actin cytoskeleton by FMNL1γ is required for structural maintenance of the Golgi complex. J Cell Sci 2011;124:3118-26. [Crossref] [PubMed]
- Sun J, Zhong X, Fu X, et al. The Actin Regulators Involved in the Function and Related Diseases of Lymphocytes. Front Immunol 2022;13:799309. [Crossref] [PubMed]
- Gibieža P, Petrikaitė V. The regulation of actin dynamics during cell division and malignancy. Am J Cancer Res 2021;11:4050-69. [PubMed]
- Heng YW, Koh CG. Actin cytoskeleton dynamics and the cell division cycle. Int J Biochem Cell Biol 2010;42:1622-33. [Crossref] [PubMed]
- Pradeau-Phélut L, Etienne-Manneville S. Cytoskeletal crosstalk: A focus on intermediate filaments. Curr Opin Cell Biol 2024;87:102325. [Crossref] [PubMed]
- Labat-de-Hoz L, Alonso MA. Formins in Human Disease. Cells 2021;10:2554. [Crossref] [PubMed]
- Favaro PM, de Souza Medina S, Traina F, et al. Human leukocyte formin: a novel protein expressed in lymphoid malignancies and associated with Akt. Biochem Biophys Res Commun 2003;311:365-71. [Crossref] [PubMed]
- Gardberg M, Heuser VD, Iljin K, et al. Characterization of Leukocyte Formin FMNL1 Expression in Human Tissues. J Histochem Cytochem 2014;62:460-70. [Crossref] [PubMed]
- Favaro P, Traina F, Machado-Neto JA, et al. FMNL1 promotes proliferation and migration of leukemia cells. J Leukoc Biol 2013;94:503-12. [Crossref] [PubMed]
- Carvalho MFL, de Almeida BO, Bueno MLP, et al. Comprehensive analysis of the HCK gene in myeloid neoplasms: Insights into biological functions, prognosis, and response to antineoplastic agents. Hematol Transfus Cell Ther 2024;46:273-82. [Crossref] [PubMed]
- Carrour TL, Assou S, Tondeur S, et al. Amazonia!: An Online Resource to Google and Visualize Public Human whole Genome Expression Data. The Open Bioinformatics Journal 2010;4:5-10. [Crossref]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Bottomly D, Long N, Schultz AR, et al. Integrative analysis of drug response and clinical outcome in acute myeloid leukemia. Cancer Cell 2022;40:850-864.e9. [Crossref] [PubMed]
- Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526-31. [Crossref] [PubMed]
- Lee SI, Celik S, Logsdon BA, et al. A machine learning approach to integrate big data for precision medicine in acute myeloid leukemia. Nat Commun 2018;9:42. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059-74. [Crossref] [PubMed]
Thiele C Hirschfeld G. cutpointr: Improved Estimation and Validation of Optimal Cutpoints in R. arXiv:2002.09209.- Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics 2005;61:92-105. [Crossref] [PubMed]
- Favaro PM, Traina F, Vassallo J, et al. High expression of FMNL1 protein in T non-Hodgkin's lymphomas. Leuk Res 2006;30:735-8. [Crossref] [PubMed]
- Chen WH, Cai MY, Zhang JX, et al. FMNL1 mediates nasopharyngeal carcinoma cell aggressiveness by epigenetically upregulating MTA1. Oncogene 2018;37:6243-58. [Crossref] [PubMed]
- Yang XY, Liao JJ, Xue WR. FMNL1 down-regulation suppresses bone metastasis through reducing TGF-β1 expression in non-small cell lung cancer (NSCLC). Biomed Pharmacother 2019;117:109126. [Crossref] [PubMed]
- Zhang MF, Li QL, Yang YF, et al. FMNL1 Exhibits Pro-Metastatic Activity via CXCR2 in Clear Cell Renal Cell Carcinoma. Front Oncol 2020;10:564614. [Crossref] [PubMed]
- Ma G, Wang Z, Liu J, et al. Quantitative proteomic analysis reveals sophisticated metabolic alteration and identifies FMNL1 as a prognostic marker in clear cell renal cell carcinoma. J Cancer 2021;12:6563-75. [Crossref] [PubMed]
- Higa N, Shinsato Y, Kamil M, et al. Formin-like 1 (FMNL1) Is Associated with Glioblastoma Multiforme Mesenchymal Subtype and Independently Predicts Poor Prognosis. Int J Mol Sci 2019;20:6355. [Crossref] [PubMed]
- Rowe JM. The "7+3" regimen in acute myeloid leukemia. Haematologica 2022;107:3. [Crossref] [PubMed]
- Rahmé R, Braun T. Venetoclax Combined with Intensive Chemotherapy: A New Hope for Refractory and/or Relapsed Acute Myeloid Leukemia? J Clin Med 2024;13:549. [Crossref] [PubMed]
- Lu G, Wang H, Xu R, et al. Formin protein FMNL1 is a biomarker for tumor-infiltrating immune cells and associated with well immunotherapeutic response. J Cancer 2023;14:2978-89. [Crossref] [PubMed]
- Thompson SB, Sandor AM, Lui V, et al. Formin-like 1 mediates effector T cell trafficking to inflammatory sites to enable T cell-mediated autoimmunity. Elife 2020;9:e58046. [Crossref] [PubMed]
- Naj X, Hoffmann AK, Himmel M, et al. The formins FMNL1 and mDia1 regulate coiling phagocytosis of Borrelia burgdorferi by primary human macrophages. Infect Immun 2013;81:1683-95. [Crossref] [PubMed]
- Miller MR, Blystone SD. Human Macrophages Utilize the Podosome Formin FMNL1 for Adhesion and Migration. Cellbio (Irvine, Calif) 2015;4:1-11. [Crossref] [PubMed]
- Mersich AT, Miller MR, Chkourko H, et al. The formin FRL1 (FMNL1) is an essential component of macrophage podosomes. Cytoskeleton (Hoboken) 2010;67:573-85. [Crossref] [PubMed]
- Ma G, Zhang B, Fu S, et al. Formin-related protein 1 facilitates proliferation and aggressive phenotype of clear cell renal cell carcinoma through MAPK/MMP2 pathway. Mol Cell Probes 2023;71:101921. [Crossref] [PubMed]
- Wu J, Yan H, Xiang C. Wilms' tumor gene 1 in hematological malignancies: friend or foe? Hematology 2023;28:2254557. [Crossref] [PubMed]
- Goel H, Rahul E, Gupta AK, et al. Molecular update on biology of Wilms Tumor 1 gene and its applications in acute myeloid leukemia. Am J Blood Res 2020;10:151-60. [PubMed]


