
GCNT3 promotes the proliferation, apoptosis, invasion, and migration of breast cancer through the PI3K/AKT pathway
Highlight box
Key findings
• This research focused on understanding the impact of glucose aminotransferase 3 (GCNT3) on the growth, invasiveness, and spread of breast cancer (BRCA) cells and examined the underlying molecular mechanisms influencing GCNT3 expression.
What is known and what is new?
• GCNT3 has been identified as being elevated and may act as biomarkers for several cancers.
• GCNT3 may be involved in the occurrence and development of BRCA through the PI3K/AKT pathway.
What is the implication, and what should change now?
• The elevated expression of GCNT3 in BRCA tissues found in this study suggests the potential of GCNT3 as a biomarker for predicting BRCA prognosis. The regulation of p-PI3K and p-AKT levels by GCNT3 appears to considerably inhibit BRCA cell development and progression.
Introduction
Breast cancer (BRCA) stands is a common and deadly malignancy among women, and its incidence and mortality rates increasing annually, posing a significant threat to the well-being and life of patients (1). Despite recent advances in treatment strategies for BRCA that have somewhat extended patient survival, the disease remains uncontrollable for many, leading to poor outcomes. Thus, finding new targets for treatment is crucial for enhancing the prognosis of these patients (2).
Located on human chromosome 15q22.2, mucin type core 2,6-N-acetylglucosamine transferase (GCNT3/GNTM/C2GNT/C2GNT2/C2GNTM/C2/4GnT) is a member of the N-acetylglucosamine transferase family. It contributes to mucin biosynthesis by facilitating the formation of core 2 and core 4 O-glycans and I branches, which are essential for protein glycosylation modifications. Previous research has linked GCNT3 to a range of human malignancies, including liver, colon, clear-cell renal cell, lung, melanoma, gastric, and pancreatic cancers (3-9). In high-grade (stages 3 and 4) clear-cell renal cell carcinoma (ccRCC) and tumors with necrosis, GCNT3 has been identified as being overexpressed and may act as a biomarker for these diseases’ invasion and prognosis (5). Furthermore, in highly metastatic melanoma, GCNT3 overexpression has been observed, and inhibiting GCNT3 function has been shown to markedly reduce melanoma cell migration and invasion and to decrease S100A8/A9 reactivity (7). In contrast, GCNT3 shows reduced expression in Epstein-Barr virus (EBV)-related gastric cancers compared non-EBV ones, with its higher expression being closely linked to severe lymph node metastasis in tumors (8). However, the expression pattern of GCNT3 in BRCA remains underexplored.
This study evaluated the expression levels of GCNT3 BRCA through Western blotting and immunohistochemistry (IHC) and further examined the effects of GCNT3 on the proliferation, invasion, and migration of BRCA cells through various cellular function tests. The goal is to determine if GCNT3 gene expression could serve as a prognostic marker in BRCA and to elucidate its potential mechanisms. We present this article in accordance with the ARRIVE and MDAR reporting checklists (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2248/rc).
Methods
Data download
Clinical data concerning the GCNT3 gene were retrieved from the The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov). After integration, standardization, and exclusion of outliers, we were able to identify immune cells and methylation genes linked to GCNT3 across 33 different types of cancer.
Clinical samples
From 2022 to 2023, seven pairs of fresh tumor and corresponding normal tissue samples were gathered from patients undergoing modified radical mastectomy for BRCA at the Affiliated Hospital of Nantong University. Pathological examinations confirmed the identity of both tumor and normal tissues. Additionally, tissue microarrays were conducted at the same center, with samples being collected from patients who had BRCA surgeries between January 2022 and July 2023, with no other primary lesions present. We compiled medical records from 86 patients with BRCA, capturing details such as age, tumor dimensions (cm), TNM (tumor node metastasis) classification, lymph node involvement, and the presence of distant metastasis. This study included only those diagnosed with invasive BRCA via histopathological analysis, and cancerous and adjacent noncancerous tissues were required to be sourced from the same individuals. All participants were newly diagnosed female patients who had not received prior surgical, radiation, chemotherapy, or biological treatments. The use of human tissues in this research was conducted with informed consent, and the ethics committee of the Affiliated Hospital of Nantong University (No. 2024-L111) granted approval for this study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
IHC staining
Four-micron sections of tissue paraffin were baked at 60 ℃ for 2 hours. Following standard dewaxing, endogenous horseradish peroxidase (HRP) was neutralized using hydrogen peroxide. High temperature and pressure were then applied using a sodium citrate solution for tissue antigen restoration. The sections were then treated with 3% H2O2 for 20 minutes, washed, and treated with a dilution of primary GCNT3 antibody. The next day, after the sections were washed three times with phosphate-buffered saline (PBS), a universal secondary antibody was applied dropwise and left to incubate at 37 ℃ for 120 minutes. Subsequent procedures included DAB and hematoxylin staining, followed by gradient dehydration and sealing with neutral gum. Two pathologists independently re-evaluated all specimens. Staining intensity was classified as negative [0], weak [1], moderate [2], or strong [3]. Staining density was scored based on the percentage of positive cells as follows: 0, 0%; 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%. A comprehensive score was calculated by multiplying the intensity and density scores. A score ≤2 points indicated low GCNT3 expression, whereas a score ≥3 points indicated high expression.
Cell transfection
MCF-7 and MDA-MB-231 cell lines were cultured during their growth phase. For transfection, these cells were seeded into a six-well plate and transiently transfected using Lipofectamine 3000. After 48 hours, cells were harvested for various functional assays, and each assay was conducted in triplicate. Proteins were extracted for analysis via Western blotting.
Western blot analysis
For protein analysis, cells were lysed with radioimmunoprecipitation (RIPA) buffer, and 50 µg of protein from each sample was collected for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, the proteins were transfer to a membrane, which was then blocked with 5% nonfat milk for 1 hour. The membrane was incubated with the primary antibody at 4 ℃ overnight, which was followed by incubation with secondary antibody at room temperature. Primary antibodies were GCNT3 (diluted 1:500, cat.no. ab77728, Abcam, Cambridge, UK), p-Akt [diluted 1:1,000, cat.no. 4060, Cell Signaling Technology (CST), Danvers, MA, USA], Akt (diluted 1:1,000, cat.no. 468, CST), p-PI3K (diluted 1:1,000, cat.no. 4228, CST), and PI3K (diluted 1:1,000, cat.no. 4257, CST), with β-actin used as an internal control. Enhanced chemiluminescence was applied to visualize the bands, and the band grayscale values were analyzed to quantify the relative expression levels of the target protein.
Methyl thiazolyl tetrazolium (MTT) assay
Transfected cells with short hairpin RNA (shRNA) sequences and non-transfected cells were gathered, centrifuged, and counted. These were then seeded at an optimal density into a 96-well plate and incubated for 48 hours. MTT reagent was added, and the cells were incubated for an additional 4 hours. Subsequently, the culture medium was removed, dimethyl sulfoxide was added, and the mixture was incubated until the crystals had fully dissolved. The absorbance (optical density) at 490 nm for each well was measured using an enzyme-linked immunosorbent assay.
EdU assay
For the EdU (5-Ethynyl-2'-deoxyuridine) assay using Cell-LightTM EdU Apollo567 kit (Ribo), 100 µL of cell suspension was added to each cell of a 96-well plate and precultured for 24 hours. The cells were incubated at 37 ℃ with EdU for 3 hours and then fixed with 4% paraformaldehyde for 30 minutes. The remaining paraformaldehyde was neutralized with glycine, followed by incubation at room temperature with 0.5% TritonX-100 for 10 minutes. The cells were then rinsed with PBS and then incubated at 37 ℃ with the staining solution for 30 minutes. The excess staining solution was washed off with PBS, and then the nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI) staining solution for 3 minutes. Images were captured using a fluorescence microscope, with red fluorescence indicating EdU-labeled proliferating cells and blue indicating DAPI-stained nuclei. Five random fields of view were selected for analysis, and the average was computed to determine the EdU-positive rate (ratio of red to blue cells) as an indicator of cell proliferation capacity.
Colony formation assay
For colony the formation assay, BRCA cells were treated with trypsin, thoroughly mixed, and transferred to a culture medium. These cells were seeded at a suitable density in a culture dish and allowed to grow for 2 weeks to enable colony formation. After this period, the colonies were stained with crystal violet for 30 minutes, the dish was turned upside down, and the number of cell colonies was counted using a low-power microscope.
Transwell cell invasion assay
Migration or invasion assays were conducted by Transwell chambers (Corning, 8 µm pore size) uncoated or coated with Matrigel, respectively. Firstly, 1×105 cells per well suspended in 100 µL FBS-free culture medium was seeded to the upper chamber, with a lower chamber filled with 600 µL 30% FBS culture medium. After 24 hours, the filters were fixed with methanol, and stained with crystal violet, and then the number of cells that had migrated to the lower chamber was examined with microscope photography. The number of cells per field was calculated.
Cell apoptosis experiment
An annexin V-FITC cell apoptosis detection kit (Invitrogen) was used to assess apoptosis in cells. MCF-7 and MDA-MB-231 cells were inoculated into a six-well plate at a density of 3×105 cells per well and allowed to adhere for 24 hours. Subsequently, the supernatant was removed, and the cells were harvested and resuspend in 0.1 mL of annexin V binding buffer. The cells were incubated with FITC-conjugated annexin V at room temperature in the dark for 15 minutes to determine the percentage of apoptotic cells based on the annexin V:propidium iodide ratio of membrane-associated proteins. This procedure was repeated three times.
Animal experiments
A protocol was prepared before the study without registration. Animal experiments were performed under a project license (No. P20230306-002) approved by Animal Ethics Committee of Nantong University, in compliance with Nantong University guidelines for the care and use of animals. The negative control (sh-NC) and sh-GCNT3 transfected 4T1 cells were harvested and suspended in a mixture of culture medium and Matrigel. Female BALB/c nude mice (weighing 18–20 g) were purchased from the Animal Laboratory Center of Nantong University. This cell suspension was subcutaneously injected into five pairs of nude mice. Four weeks postinoculation, the mice were killed, and the tumors were excised and weighed to assess the growth.
Statistical analysis
All statistical analyses were conducted using SPSS 25 (IBM Corp., Armonk, NY, USA). Data were presented as the mean ± standard deviation derived from a minimum of three independent experiments. Both the Student t-test (two-tailed) and one-way analysis of variance (ANOVA) were used to evaluate the significance of differences between groups, with statistical significance defined as a P value <0.05.
Results
GCNT3 in TCGA database
Through analysis of TCGA database, we identified the significant overexpression of the GCNT3 gene in a variety of tumors such as BRCA, lung adenocarcinoma, colon cancer, head-and-neck squamous cell carcinoma, gastric adenocarcinoma, lung squamous cell carcinoma, and hepatocellular carcinoma, among others (Figure 1A). Specifically, GCNT3 levels were elevated in BRCA tissues compared to normal breast tissue (Figure 1B). A paired difference analysis further substantiated the elevated expression of GCNT3 in BRCA specimens (Figure 1C). We downloaded clinical information related to the GCNT3 gene from TCGA database and performed integration, standardization, and removal of extreme values. This allowed us to obtain information on the immune cells (Figure 1D) and methylation genes (Figure 1E) associated with GCNT3 across 33 types of cancer.
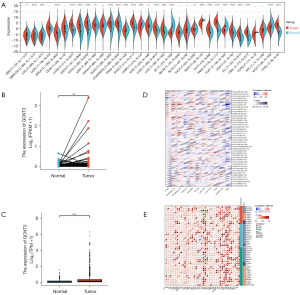
High expression of GCNT3 in BRCA
To more deeply examine the expression patterns of GCNT3 in BRCA, we employed Western blot analysis to measure GCNT3 protein levels in both non-tumor and tumor tissues from seven BRCA patient pairs. The findings indicated a pronounced overexpression of GCNT3 in the tumor tissues (Figure 2A). Additionally, we assessed GCNT3 expression in 86 BRCA cases through IHC, identifying a high expression in 52 cases and a low expression in 34 cases (Figure 2B). We also examined the relationship between GCNT3 expression and various clinicopathological parameters, discovering a significant association of GCNT3 expression with tumor size (P=0.001) and TNM stage (P=0.25) but no significant correlations with age, lymph node metastasis, or distant metastasis were noted (Table 1).
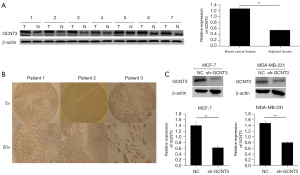
Table 1
| Clinical parameter | All (N=86) | GCNT3 expression | P value | |
|---|---|---|---|---|
| Low expression (n=34) | High expression (n=52) | |||
| Age (years) | 0.24 | |||
| <45 | 34 | 16 | 18 | |
| ≥45 | 52 | 18 | 34 | |
| Tumor size (cm) | 0.001* | |||
| ≤2 | 28 | 18 | 10 | |
| >2 and ≤5 | 41 | 14 | 27 | |
| >5 | 17 | 2 | 15 | |
| TNM | 0.025 | |||
| I | 22 | 14 | 8 | |
| II | 39 | 15 | 24 | |
| III | 20 | 4 | 16 | |
| IV | 5 | 1 | 4 | |
| Lymph node metastasis | 0.11 | |||
| No | 60 | 27 | 33 | |
| Yes | 26 | 7 | 19 | |
| Distant metastasis | 0.35 | |||
| No | 81 | 33 | 48 | |
| Yes | 5 | 1 | 4 | |
*, statistically significance. TNM, tumor node metastasis.
To evaluate the effects of GCNT3 on the behavior of BRCA cells, we analyzed the effect of a GCNT3-specific interference plasmid (sh-GCNT3) in MCF-7 and MDA-MB-231 cell lines using Western blotting. Transient transfection was performed with Lipofectamine 3000, confirming effective knockdown by the shRNA vector. Cell proliferation was monitored 48 hours after transfection, which indicated significant suppression of GCNT3 expression in the BRCA cells transfected with sh-GCNT3 as compared to controls. Additionally, colony formation assays revealed that colonies in the sh-GCNT3 group were smaller than those in the NC group (Figure 2C).
Effects of GCNT3 on the proliferation of BRCA cells
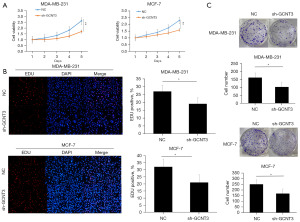
Further examination of the effect of GCNT3 knockdown on BRCA cell proliferation and viability was carried out through MTT, EdU, and colony formation assays. The MTT assays showed that sh-GCNT3 notably reduced the proliferation of MCF-7 and MDA-MB-231 cells relative to the NC group (Figure 3A). Consistent findings were observed in the EdU proliferation assays (Figure 3B). Additionally, the colony formation experiments indicated that colonies were smaller in size in the sh-GCNT3 group compared to the NC group (Figure 3C). Taken together, the MTT, EdU, and colony formation assays confirmed that the knockdown of GCNT3 significantly inhibited the growth of BRCA cells.
Effects of GCNT3 on the invasion and migration of BRCA cells
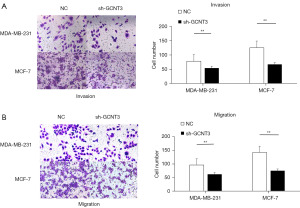
To determine the effect of GCNT3 influence on the migration and invasion of BRCA cells, we transfected MCF-7 and MDA-MB-231 cells with a GCNT3 interference plasmid. We then assessed the impact of GCNT3 on these processes using Transwell assays. The results from the Transwell assays indicated that the knockdown of GCNT3 led to a reduction in both the migration and invasion of MCF-7 and MDA-MB-231 cells (Figure 4A,4B).
Effects of GCNT3 on the apoptosis of BRCA cells
To examine the effect of GCNT3 on the apoptosis of BRCA cells, MCF-7 and MDA-MB-231 cells were transfected with a GCNT3 interference plasmid. Subsequently, apoptosis was assessed using flow cytometry. The findings demonstrated that the knockdown of GCNT3 considerably enhanced apoptosis in both the MCF-7 and MDA-MB-231 cells (Figure 5A,5B).
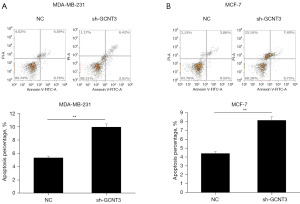
GCNT3 regulated the occurrence and development of BRCA through the PI3K/AKT pathway
To determine the role of GCNT3 in modulating the PI3K/AKT signaling pathway in BRCA cells, GCNT3 interference plasmids were transfected into MCF-7 and MDA-MB-231 cells. Western blot analysis was employed to examine the variations in PI3K/p-PI3K and AKT/p-AKT protein levels between the experimental and control groups. The results indicated that in both the MCF-7 and MDA-MB-231 cells, the reduction of GCNT3 led to decreased levels of p-PI3K and p-AKT, with no notable changes in the levels of PI3K and AKT (Figure 6A,6B). These findings imply that GCNT3 influences BRCA cell progression through its regulatory effects on p-PI3K and p-AKT expression.
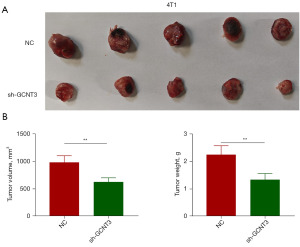
Effects of GCNT3 knockdown on the growth of subcutaneous transplanted tumors in nude mice
To assess the effect of GCNT3 knockdown on tumor growth, a nude mouse xenograft model was constructed. We transfected 4T1 cells with both sh-NC and sh-GCNT3 and implanted them into nude mice. On day 30 postinoculation, the mice were euthanized, and the tumors were excised, measured, and weighed. The findings indicated a substantial decrease in both the weight and size of tumors derived from 4T1 cells transfected with sh-GCNT3 (P<0.01, Figure 7). This demonstrates that reducing GCNT3 expression significantly curtails the tumorigenic capacity of BRCA cells.
Discussion
There is growing body of evidence indicating that molecular targeted therapy is a promising field in cancer treatment (10,11). It is therefore vital to examine the complex molecular mechanisms underlying BRCA’s onset and progression. All mucins are characterized by having one or more core glycans; C1GNT is crucial for core 1 formation. While C3GNT is crucial for core 3, and C2GNT facilitates the transition of core 1 to core 2 and from core 3 to core 4 (12). Core 2 β1,6 N-acetylglucosamine transferase (GCNT3/C2GNT) is pivotal to the biosynthesis of mucoglycans (13,14), and its aberrant expression leads to excessive mucin production (14). GCNT3’s role extends to vital physiological functions such as inflammation and immune responses (15), and it is recognized as an independent indicator of adverse clinical outcomes, exhibiting numerous oncogenic traits (16).
A recent study has shown that GCNT3 is markedly expressed in both lung cancer tissues and cells (6), and high GCNT3 levels correlate with poorer outcomes in patients undergoing radiotherapy. The suppression of GCNT3 in the A549 lung cancer cell line reduces proliferation, migration, and invasion and notably curtails epithelial-mesenchymal transition (17). Similarly, Li et al. reported elevated GCNT3 expression in lung cancer, noting that miR-302b-3p can suppress the proliferation, migration, and invasion of non-small cell lung cancer (NSCLC) cells by targeting GCNT3 (6). Other study has linked increased GCNT3 expression with heightened sensitivity to radiation therapy in hepatocellular carcinoma cells and xenograft tumors (3). Moreover, Rao et al. provided new insights into GCNT3’s role in pancreatic cancer, indicating a clinical association between GCNT3 overexpression and enhanced malignancy (9). Their in vitro research showed that GCNT3 contributes to mucin synthesis in pancreatic cancer, with aberrant expression linked to increased mucin production, invasive tumor growth, and decreased patient survival. These observations align with our findings in BRCA and suggest broader implications for other cancers such as lung, skin, melanoma, and mesothelioma, which also display elevated GCNT3 levels. However, the link between GCNT3 overexpression and increased tumor malignancy remains a subject of debate.
In 2015, González-Vallinas et al. found that patients with colon cancer and lower GCNT3 expression levels had a significantly increased risk of recurrence than did those with higher levels (4). This discrepancy might stem from the type-specific intracellular mechanisms of the GCNT3 pathway influencing cancer progression or regression. Therefore, GCNT3 expression in metabolic genes could serve as a valuable prognostic molecular biomarker for early-stage colon cancer and may also help monitor the response to chemotherapy. Similarly, a previous study discovered that elevated GCNT3 levels in epithelial ovarian cancer correlate with improved clinical outcomes and responses to initial treatments (18).
In BRCA, particularly within metastatic tissues, GCNT3 expression is notably higher compared to that in normal tissues, indicating a potential link between GCNT3 and the development and spread of BRCA. In our study, we used protein imprinting techniques to measure GCNT3 protein levels, revealing substantially higher levels in BRCA cancer tissues than in normal tissues. IHC confirmed a relationship between GCNT3 expression, tumor size, and TNM staging in patients with BRCA. Moreover, reducing GCNT3 expression influenced cell proliferation, underscoring its critical role in tumor growth, invasion, migration, and apoptosis. The PI3K/AKT pathway, crucial for cancer metastasis, involves GCNT3; activation of this pathway can disrupt cellular growth and survival controls, leading to competitive growth advantages, metastatic potential, angiogenesis, and resistance to treatment (19-21). These observations suggest that GCNT3 could regulate BRCA development through the PI3K/AKT pathway, directly affecting the cancer’s ability to spread.
Conclusions
This report showed that knockdown of GCNT3 also notably hindered tumorigenic capabilities in vivo, indicating that GCNT3 is markedly upregulated in BRCA tissues and thus potentially a viable biomarker for predicting the prognosis of patients with BRCA. By regulating the levels of p-PI3K and p-AKT in breast cells, GCNT3 effectively inhibits the initiation and progression of BRCA cells. Thus, GCNT3 may be a novel therapeutic target for curtailing BRCA metastasis and may lead to significant advancements in future cancer treatment.
Acknowledgments
Funding: This research was supported by
Footnote
Reporting Checklist: The authors have completed the ARRIVE and MDAR reporting checklists. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2248/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2248/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2248/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2248/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics committee of the Affiliated Hospital of Nantong University (No. 2024-L111) and informed consent was taken from all the patients. Animal experiments were performed under a project license (No. P20230306-002) approved by Animal Ethics Committee of Nantong University, in compliance with Nantong University guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Britt KL, Cuzick J, Phillips KA. Key steps for effective breast cancer prevention. Nat Rev Cancer 2020;20:417-36. [Crossref] [PubMed]
- Pashayan N, Antoniou AC, Ivanus U, et al. Personalized early detection and prevention of breast cancer: ENVISION consensus statement. Nat Rev Clin Oncol 2020;17:687-705. [Crossref] [PubMed]
- Liu T, Zhang S, Chen J, et al. The transcriptional profiling of glycogenes associated with hepatocellular carcinoma metastasis. PLoS One 2014;9:e107941. [Crossref] [PubMed]
- González-Vallinas M, Vargas T, Moreno-Rubio J, et al. Clinical relevance of the differential expression of the glycosyltransferase gene GCNT3 in colon cancer. Eur J Cancer 2015;51:1-8. [Crossref] [PubMed]
- Batai K, Imler E, Pangilinan J, et al. Whole-transcriptome sequencing identified gene expression signatures associated with aggressive clear cell renal cell carcinoma. Genes Cancer 2018;9:247-56. [Crossref] [PubMed]
- Li Q, Ran P, Zhang X, et al. Downregulation of N-Acetylglucosaminyltransferase GCNT3 by miR-302b-3p Decreases Non-Small Cell Lung Cancer (NSCLC) Cell Proliferation, Migration and Invasion. Cell Physiol Biochem 2018;50:987-1004. [Crossref] [PubMed]
- Sumardika IW, Youyi C, Kondo E, et al. β-1,3-Galactosyl-O-Glycosyl-Glycoprotein β-1,6-N-Acetylglucosaminyltransferase 3 Increases MCAM Stability, Which Enhances S100A8/A9-Mediated Cancer Motility. Oncol Res 2018;26:431-44. [Crossref] [PubMed]
- Liu J, Zhang Y, Liu W, et al. MiR-BART1-5p targets core 2β-1,6-acetylglucosaminyltransferase GCNT3 to inhibit cell proliferation and migration in EBV-associated gastric cancer. Virology 2020;541:63-74. [Crossref] [PubMed]
- Rao CV, Janakiram NB, Madka V, et al. Small-Molecule Inhibition of GCNT3 Disrupts Mucin Biosynthesis and Malignant Cellular Behaviors in Pancreatic Cancer. Cancer Res 2016;76:1965-74. [Crossref] [PubMed]
- Barzaman K, Karami J, Zarei Z, et al. Breast cancer: Biology, biomarkers, and treatments. Int Immunopharmacol 2020;84:106535. [Crossref] [PubMed]
- Zuo Z, Zhou Z, Chang Y, et al. Ribonucleotide reductase M2 (RRM2): Regulation, function and targeting strategy in human cancer. Genes Dis 2022;11:218-33. [Crossref] [PubMed]
- Tan S, Cheng PW. Mucin biosynthesis: identification of the cis-regulatory elements of human C2GnT-M gene. Am J Respir Cell Mol Biol 2007;36:737-45. [Crossref] [PubMed]
- Stone EL, Ismail MN, Lee SH, et al. Glycosyltransferase function in core 2-type protein O glycosylation. Mol Cell Biol 2009;29:3770-82. [Crossref] [PubMed]
- Yeh JC, Ong E, Fukuda M. Molecular cloning and expression of a novel beta-1, 6-N-acetylglucosaminyltransferase that forms core 2, core 4, and I branches. J Biol Chem 1999;274:3215-21. [Crossref] [PubMed]
- Wang M, She W, Yang J, et al. Gene Expression Analysis by Real-Time PCR in Nasal Brushings of Adult Patients with Allergic Rhinitis, Suspected Allergic Rhinitis, and Nonallergic Rhinitis. Int Arch Allergy Immunol 2021;182:301-10. [Crossref] [PubMed]
- Wang Y, Fang X, Xie H, et al. GCNT3 Promotes Hepatocellular Carcinoma Progression and EMT by Activating the PI3K/AKT Pathway. Biochem Genet 2024; Epub ahead of print. [Crossref] [PubMed]
- Zhang Y, Xiao P, Hu X. LINC00511 enhances LUAD malignancy by upregulating GCNT3 via miR-195-5p. BMC Cancer 2022;22:389. [Crossref] [PubMed]
- Kang Q, Tingting W, Bingzi D, et al. GCNT3 regulated MUC13 to promote the development of hepatocellular carcinoma through the GSK3β/β-catenin pathway. Dig Liver Dis 2024;56:1572-81. [Crossref] [PubMed]
- Fernández LP, Sánchez-Martínez R, Vargas T, et al. The role of glycosyltransferase enzyme GCNT3 in colon and ovarian cancer prognosis and chemoresistance. Sci Rep 2018;8:8485. [Crossref] [PubMed]
- Miricescu D, Totan A, Stanescu-Spinu II, et al. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int J Mol Sci 2020;22:173. [Crossref] [PubMed]
- Glaviano A, Foo ASC, Lam HY, et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer 2023;22:138. [Crossref] [PubMed]
(English Language Editor: J. Gray)


