
Balancing redox stress: anchorage-independent growth requires reductive carboxylation
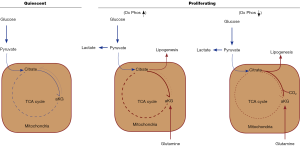
The ability of a cell to exhibit anchorage independent growth is a hallmark of cancer, and has been extensively investigated in hopes of finding new therapeutic strategies. In particular, the processes that give rise to and support this dysregulated growth phenotype, which allows cells to proliferate in the absence of survival signals resulting from their attachment to a basement membrane, has been of interest. The contribution of oncogenes and tumor suppressors on anchorage independent growth has been well studied; however, the alterations in cellular metabolism that support this process are much less defined. Changes in cellular metabolism are among the most notable hallmarks of solid tumors, where glucose is imported at high rates and exported from cancer cells as lactate, instead of being utilized within the citric acid cycle (TCA cycle) (1,2). This process, known as the Warburg effect, represents the primary metabolic phenotype associated with cancer cells. In order to adapt to this altered metabolism, cancer cells have been shown to rely on other primary metabolic pathways to support their increased requirements for biosynthesis and redox control to allow for their sustained proliferation. In particular, glutamine metabolism has been shown to supplement the decrease in glucose-derived carbon entering the TCA cycle in 2D cultured cancer cells (Figure 1, compare quiescent versus proliferating cells). However, it remains unclear how expendable glutamine-fueled anaplerosis is for cells grown in 3D. In their recent Nature article, DeBerardinis and coworkers [2016] set out to describe the metabolic alterations that accompany the anchorage-independent growth of human cancer cells when cultured as a 2D monolayer or as a 3D spheroid, thereby mimicking the detachment from the extracellular matrix that often occurs during tumorigenesis (3). They find that the same cancer cells exhibit drastically different metabolic phenotypes when grown as spheroids than they do in a monolayer culture.
The altered metabolism in cancer has recently gained significant attention as a means to advance therapeutic strategies (4). A great deal of focus has been devoted to the altered primary metabolic pathways, namely glycolysis and glutaminolysis, which have been shown to be significantly altered in cancer cells and tumors. These alterations support the unique biosynthetic requirements of proliferating cells, where glycolytic intermediates feed pathways responsible for nucleotide and NADPH production through the pentose phosphate and one-carbon serine metabolism pathways (5). To compensate for the decreased entry of glucose-derived carbon within the mitochondria, glutamine is imported at high rates to refill TCA cycle intermediates. These TCA cycle intermediates are used for the continued maintenance of oxidative phosphorylation, together with the biosynthesis of amino acid pools and de novo lipid production. Indeed, the increased reliance on glutamine metabolism in cancer cells has presented the gate-keeper enzyme, mitochondrial glutaminase (GLS), as an attractive drug target. Efforts to inhibit GLS have been described, where two distinct classes of allosteric inhibitors have been developed and are currently used to target the increased glutaminolysis in glutamine-addicted cancer cells (6,7). Together, these modifications within primary metabolic pathways represent an attractive druggable chain of events.
The observation of the increased glycolytic rate in neoplasms, first described by the Nobel laureate Otto Warburg in the 1920’s, has been shown to arise from the lesions within tumor driver genes, such as the mutations, deletions, or amplifications of oncogenes and tumor suppressors. For example, expression of the transcription factor MYC, which is commonly up-regulated in a myriad of human cancers, stimulates the expression of glycolytic and glutaminolytic enzymes, giving rise to the Warburg effect and glutamine-fueled anaplerosis (8,9). Similarly, stabilization or over-expression of hypoxia inducible factor (HIF1α), the primary oxygen sensor of the cell, causes a similar effect, by up-regulating genes that act in concert to shift the cellular metabolism away from oxidative phosphorylation, and towards glycolysis (Figure 1, compared increased versus depressed oxidative phosphorylation) (10,11). These two observations suggest that tumors with dysregulated MYC or HIF1α oncoproteins are metabolically distinct from non-transformed cells, making them more viable in the oxygen deprived and metabolically stressed environment of the solid tumor. Indeed, other driver genes, such as mutant Ras, PI3K, or hyperactive Rho-GTPases, have shown similar trends in 2D monolayer and in solid tumors (12,13), thereby demonstrating the metabolic adaptations that accompany oncogenesis represent a conserved mechanism.
Recently, Mullen and colleagues [2012] reported that cancer cells with defective mitochondria, through the depletion of the respiratory complex III component cytochrome b (CytB), exhibit not only an increased rate of glutamine uptake and metabolism, but utilize glutamine for de novo lipid biosynthesis, as distinct from non-transformed cells (14). In these cancer cells, glutamine was first converted to glutamate, and subsequently deamidated to give α-ketoglutarate (αKG), which was then incorporated into the TCA cycle. However, instead of entering the TCA cycle and being metabolized through the normal series of oxidative reactions to form succinate, this glutamine-derived αKG was metabolized in the reverse direction (i.e., through a reductive pathway) to form citrate. This process, termed reductive carboxylation, converts glutamine-derived αKG to citrate through the incorporation of CO2 by the cytosolic or mitochondrial NADP+/NADPH-dependent enzyme isocitrate dehydrogenase 1 or 2 (IDH1/IDH2). Mitochondrial citrate is then transported to the cytosol through the citrate-specific transport protein (CTP) to be converted to the lipogenic precursor, acetyl-CoA, by the ATP-dependent citrate lyase (ACL). The reductive carboxylation of glutamine was shown to be essential for maintaining the proliferation of cancer cells with defective mitochondria. Similarly, this reductive carboxylation pathway was found to be upregulated and critical for sustained proliferation of cancer cells either grown in a hypoxic environment or driven by the deletion of the von-Hippel Lindau factor (VHL) (i.e., stabilization of HIF1α), a tumor suppressor commonly repressed in renal cell carcinomas (RCC) (10,11). Fendt et al. [2013] described the driving force of this reductive glutamine metabolism that supports lipogenesis to be dependent on the relative concentrations of the metabolites αKG and citrate, rather than a specific signaling pathway. Therefore, this metabolic phenotype is uncoupled from specific cell signaling events, demonstrating more generally that this pathway is a response to the overall metabolic status of a cell which can be manipulated by the outcome of cancer-driving genes (15). Together, as demonstrated in Figure 1 above, these studies show that reductive glutamine metabolism is primarily upregulated upon decreases in oxidative phosphorylation and increases in glycolysis, characteristic of many tumor cells.
Recent effort has been focused on translating the observations of the altered metabolism in cultured cancer cells to the targeting of human tumors. The metabolic alterations that occur within human tumors have been shown to be dependent on both the genetic lesions driving the neoplasm, the tissue of origin, and the perfusion of the physical tumor. For example, Yuneva and colleagues [2012] described the head-to-head comparison of the metabolism of both MYC- and MET-driven tumors arising from liver or lung tissues, and found that the utilization of both glucose and glutamine was not only unique between the two oncogenes, but also when the neoplasms driven by the same oncogene arise from different tissues (16). These results suggest that targeting the metabolic alterations within tumors, driven by the same oncogene, or arising within the same tissue but driven by different driver gene mutations, can be quite unique.
More recent studies investigating the observed metabolic consequences of Kras-driven non-small cell lung cancer (NSCLC) cells grown in vitro, or as tumors formed in vivo, find significant differences when the cells are cultured in 2D versus their 3D tumor counterparts (17). Interestingly, the changes in the metabolic phenotypes of these cells were shown to be specific for their being grown in vitro or in vivo, where transition of the same cells from one condition to another stimulated the changes in the cells metabolic response (i.e., glutamine-fueled anaplerosis in vitro switches to glucose-fueled anaplerosis in vivo, and vice versa). To further complicate the translation of studies highlighting the metabolic adaptations resulting from oncogene-driven cells to clinical relevance, Hensley et al. [2016] reported the metabolic heterogeneity among tumors within patients with NSCLC, using stable-isotope tracing methods, and related this heterogeneity to the perfusion of the tumor tissue among patients (18). This study proposes that the microenvironment shapes the balance of the availability of nutrients and the signaling networks that are initiated to sustain proliferation in different perfusion environments to promote malignancy. Together, these studies suggest that the targeting of the altered glucose and glutamine metabolism within tumors is dependent on a variety of factors, including the tumor-driving gene, the tissue origin of the neoplasm, and the microenvironment of the growth within the body.
Given the challenges of broadly targeting the metabolic alterations that arise from a specific oncogene, opportunities have developed for targeting a cell’s ability to overcome a significant growth barrier, namely anchorage-independence. Studies have demonstrated that mammary epithelial cells require metabolic adaptation upon detachment from their basement membrane and subsequent malignant transformation, presenting a potential strategy for targeting this adaptation. It was found that normal mammary epithelial cells follow programmed cell death upon their detachment, whereas aggressive breast cancer cells thrive by avoiding this process. The avoidance of this cell-growth control was dependent on the maintenance of reactive oxygen species (ROS) that accumulate as a result from depressed oxidative metabolism (i.e., decrease in ATP levels), acquired through either the upregulation of oncogenes and anti-oxidant enzymes or by exogenously supplied anti-oxidants (19,20). Furthermore, it was found that the upregulation of the ErbB2 oncoprotein, commonly found in breast cancers, rescued ROS-dependent apoptosis through the stimulation of the pentose phosphate pathway (PPP), resulting in the production of the important anti-oxidant NADPH (19). However, given that the PPP functions in the cytoplasm, it remained unclear how cytosolic NADPH prevented the build-up of ROS within the mitochondria.
To address the dependence on redox balance during anchorage-independent growth more broadly, Jiang and coworkers [2016] set out to describe the changes in metabolism that are observed when cancer cells are grown in a 2D monolayer, or during their growth as 3D spheroids. They found an interesting connection between the upregulation of reductive glutamine metabolism, and the shuttling of NADPH from the cytoplasm to the mitochondrion to abrogate ROS stress (Figure 2). In particular, they describe the specific roles of the cytosolic and mitochondrial isocitrate dehydrogenase enzymes, IDH1 and IDH2, respectively, in reducing glutamine-derived αKG to citrate at the expense of cytosolic NADPH, which is then transported into the mitochondrion through the CTP and subsequently oxidized back to αKG, producing NADPH. This process effectively shuttles NADPH from the cytoplasm to the mitochondrial matrix using reductive glutamine metabolism (Figure 2) (3). These findings serve to connect the cooperation of reductive glutamine metabolism with redox control within cells that require the maintenance of oxidative stress to withstand their detachment from the extracellular matrix, thereby adding important detail on the portrait of metabolic changes that tumor cells develop to maintain their malignant state.
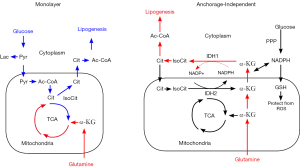
In conclusion, the work of Jiang, DeBerardinis and colleagues present essential details that describe the changes in metabolism accompanying anchorage-independent growth, a transformed phenotype that leads to highly aggressive malignancies. Drug targeting strategies are underway to inhibit metabolic enzymes, specifically, which represent new and attractive therapeutics. It remains unclear under what circumstances these therapies will prove most effective, thereby highlighting the importance for defining these metabolic aberrations in great detail.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Decai Yu (Hepatobiliary Institute of Nanjing University, Department of Hepatobiliary Surgery, Drum Tower Hospital, the Affiliated Medical School of Nanjing University, Nanjing, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.09.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011;11:325-37. [Crossref] [PubMed]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012;21:297-308. [Crossref] [PubMed]
- Jiang L, Shestov AA, Swain P, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 2016;532:255-8. [Crossref] [PubMed]
- Tennant DA, Durán RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer 2010;10:267-77. [Crossref] [PubMed]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 2011;27:441-64. [Crossref] [PubMed]
- Stalnecker CA, Ulrich SM, Li Y, et al. Mechanism by which a recently discovered allosteric inhibitor blocks glutamine metabolism in transformed cells. Proc Natl Acad Sci U S A 2015;112:394-9. [Crossref] [PubMed]
- DeLaBarre B, Gross S, Fang C, et al. Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry 2011;50:10764-70. [Crossref] [PubMed]
- Li B, Simon MC. Molecular Pathways: Targeting MYC-induced metabolic reprogramming and oncogenic stress in cancer. Clin Cancer Res 2013;19:5835-41. [Crossref] [PubMed]
- Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A 2008;105:18782-7. [Crossref] [PubMed]
- Gameiro PA, Yang J, Metelo AM, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab 2013;17:372-85. [Crossref] [PubMed]
- Wise DR, Ward PS, Shay JE, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A 2011;108:19611-6. [Crossref] [PubMed]
- Weinberg F, Hamanaka R, Wheaton WW, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A 2010;107:8788-93. [Crossref] [PubMed]
- Lukey MJ, Greene KS, Erickson JW, et al. The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy. Nat Commun 2016;7:11321. [Crossref] [PubMed]
- Mullen AR, Wheaton WW, Jin ES, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 2011;481:385-8. [PubMed]
- Fendt SM, Bell EL, Keibler MA, et al. Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nat Commun 2013;4:2236. [Crossref] [PubMed]
- Yuneva MO, Fan TW, Allen TD, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab 2012;15:157-70. [Crossref] [PubMed]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab 2016;23:517-28. [Crossref] [PubMed]
- Hensley CT, Faubert B, Yuan Q, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell 2016;164:681-94. [Crossref] [PubMed]
- Schafer ZT, Grassian AR, Song L, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 2009;461:109-13. [Crossref] [PubMed]
- Davison CA, Durbin SM, Thau MR, et al. Antioxidant enzymes mediate survival of breast cancer cells deprived of extracellular matrix. Cancer Res 2013;73:3704-15. [Crossref] [PubMed]


