
BIRC3 as a yet underestimated prognostic marker of malignancies?
Working in the field of human genetics since >20 years the announcement and publication of a discovery of a new candidate gene for a specific genetic disorder is not that unusual to me. In contrary, even though the whole exome of human was announced to be sequenced in 2001 already (1), candidate genes for inborn as well as acquired diseases were not becoming less since that time (Figure 1). Knowing the field, this is not surprising. Even though human genome project (HUGO) told us where genes may be located within the 46 human chromosomes, HUGO per se was never able to tell us what the function of all these genes is. To find out about this, studies in patients are necessary to identify which genes are impaired in connection with which disorder or disease; and such studies need to be followed or accompanied by functional studies.
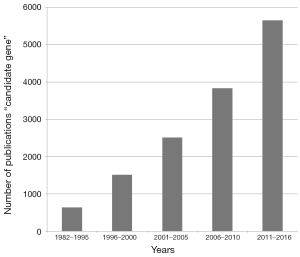
The study of Gressot and coworkers published in the present issue of Oncotarget (2) is a good example for this kind of so-called ‘post-genomic’ research. Using a combination of clinical studies, database analyses, meta-analyses and functional studies they could nicely provide evidence that Baculoviral IAP Repeat Containing 3 gene (BIRC3, also earlier denominated as apoptosis inhibitory protein IAP2) seems to play a crucial role in malignant transformation of low grade gliomas to glioblastoma, as also recently found by others (3). Besides it seems to be new prognostic marker of glioblastoma.
As Gressot and coworkers (2) also mentioned, BIRC3 being a negative regulator of the non-canonical NF-κB protein, has yet been shown to play a role also in other malignancies, too.
It was known already since 2012 that BIRC3 disruption can be observed in fludarabine refractory chronic lymphocytic leukemia (4), and just recently we could show that it is also involved either as deletion or duplication event in a subset of acute lymphocytic leukemia patients (5); BIRC3 is a translocation partner in MALT lymphoma (6); altered expression of BIRC3 was recently seen in breast (7,8) as well as pancreatic cancer (9); BIRC3 amplification and or upregulation of its expression were observed in gastrointestinal stromal tumors (10), and bladder cancer (cell lines) (11); involvement of BIRC3 is suggested in melanoma (12), colorectal cancer (13,14) and nasopharyngeal carcinoma (15); interestingly, a predictive value of BIRC3 has been postulated in oesophageal adenocarcinoma patients, as well (16).
Besides, BIRC3 also is suspected to play a role in childhood asthma (17), in human herpesvirus 6 (HHV-6) infection and associated neurologic diseases (18), as well as in age-related macular degeneration (19).
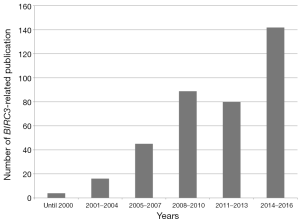
In conclusion the study of Gressot and coworkers (2) is one more puzzle stone which highlights the importance of BIRC3 in tumor development and progression. Still, it remains surprising that BIRC3 alterations were recognized already in 2004 e.g., in neuroblastoma (20), but most studies involving this gene came out just lately (Figure 2). It seems BIRC3 somehow escaped from the focus of research attention until recently. Overall, it remains true what Yamato et al. stated in 2015 (21) for BIRC3: “BIRC3 mutations are present in a wide range of epithelial tumors and most nonsense or frameshift mutations confer direct transforming potential.” Besides, amplification and deletions were observed recently. “This oncogenic function of BIRC3 mutants is largely independent of their ability to activate NF-κB signaling. In addition to the BIRC3-NIK-NF-κB signaling pathway, BIRC3-NIK signaling targets effectors other than NF-κB and thereby contributes directly to carcinogenesis. Identification (and further characterization) of these effectors may provide a basis for the development of targeted agents for the treatment of lymphoid malignancies and other cancers with BIRC3 alterations.” Finally, for practical patient care, the potential of BIRC3 as a prognostic marker should be delineated for more detail in near future.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Ning Huang (Department of Neurosurgery, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.09.29). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Marshall E. NIH to produce a 'working draft' of the genome by 2001. Science 1998;281:1774-5. [Crossref] [PubMed]
- Gressot LV, Doucette T, Yang Y, et al. Analysis of the inhibitors of apoptosis identifies BIRC3 as a facilitator of malignant progression in glioma. Oncotarget 2016; [Epub ahead of print]. [PubMed]
- Wang D, Berglund A, Kenchappa RS, et al. BIRC3 is a novel driver of therapeutic resistance in Glioblastoma. Sci Rep 2016;6:21710. [Crossref] [PubMed]
- Rossi D, Fangazio M, Rasi S, et al. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia. Blood 2012;119:2854-62. [Crossref] [PubMed]
- Alhourani E, Othman MA, Melo JB, et al. BIRC3 alterations in chronic and B-cell acute lymphocytic leukemia patients. Oncol Lett 2016;11:3240-3246. [PubMed]
- Du MQ. MALT lymphoma: A paradigm of NF-κB dysregulation. Semin Cancer Biol 2016;39:49-60. [Crossref] [PubMed]
- Wang H, Xu B, Zhang X, et al. PADI2 gene confers susceptibility to breast cancer and plays tumorigenic role via ACSL4, BINC3 and CA9 signaling. Cancer Cell Int 2016;16:61. [Crossref] [PubMed]
- Lu W, Ning H, Gu L, et al. MCPIP1 Selectively Destabilizes Transcripts Associated with an Antiapoptotic Gene Expression Program in Breast Cancer Cells That Can Elicit Complete Tumor Regression. Cancer Res 2016;76:1429-40. [Crossref] [PubMed]
- Gan H, Liu H, Zhang H, et al. SHh-Gli1 signaling pathway promotes cell survival by mediating baculoviral IAP repeat-containing 3 (BIRC3) gene in pancreatic cancer cells. Tumour Biol 2016;37:9943-50. [Crossref] [PubMed]
- Falkenhorst J, Grunewald S, Mühlenberg T, et al. Inhibitor of Apoptosis Proteins (IAPs) are commonly dysregulated in GIST and can be pharmacologically targeted to enhance the pro-apoptotic activity of imatinib. Oncotarget 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Kim SH, Ho JN, Jin H, et al. Upregulated expression of BCL2, MCM7, and CCNE1 indicate cisplatin-resistance in the set of two human bladder cancer cell lines: T24 cisplatin sensitive and T24R2 cisplatin resistant bladder cancer cell lines. Investig Clin Urol 2016;57:63-72. [Crossref] [PubMed]
- Jin L, Chen J, Liu XY, et al. The double life of RIPK1. Mol Cell Oncol 2015;3:e1035690 [Crossref] [PubMed]
- Kobayashi T, Masaki T, Nozaki E, et al. Microarray Analysis of Gene Expression at the Tumor Front of Colon Cancer. Anticancer Res 2015;35:6577-81. [PubMed]
- Ji QX, Liu LL, Li L, et al. Roles of glucose-regulated protein 78 in proliferation and migration of human colorectal carcinoma cell line RKO. Zhonghua Bing Li Xue Za Zhi 2016;45:401-6. [PubMed]
- Song Y, Peng X, Wang M, et al. Gene expression profiling of taxol-resistant nasopharyngeal carcinoma cells with siRNA-mediated FOLR1 downregulation. Int J Clin Exp Pathol 2015;8:11314-22. [PubMed]
- Piro G, Giacopuzzi S, Bencivenga M, et al. TAK1-regulated expression of BIRC3 predicts resistance to preoperative chemoradiotherapy in oesophageal adenocarcinoma patients. Br J Cancer 2015;113:878-85. [Crossref] [PubMed]
- Gao XM. A network approach predicts NFKBIA and BIRC3 as pathogenic genes in childhood asthma. Genet Mol Res 2016;15. [PubMed]
- Shao Q, Lin Z, Wu X, et al. Transcriptome sequencing of neurologic diseases associated genes in HHV-6A infected human astrocyte. Oncotarget 2016; [Epub ahead of print]. [PubMed]
- Telegina DV, Korbolina EE, Ershov NI, et al. Identification of functional networks associated with cell death in the retina of OXYS rats during the development of retinopathy. Cell Cycle 2015;14:3544-56. [Crossref] [PubMed]
- Takita J, Ishii M, Tsutsumi S, et al. Gene expression profiling and identification of novel prognostic marker genes in neuroblastoma. Genes Chromosomes Cancer 2004;40:120-32. [Crossref] [PubMed]
- Yamato A, Soda M, Ueno T, et al. Oncogenic activity of BIRC2 and BIRC3 mutants independent of nuclear factor-κB-activating potential. Cancer Sci 2015;106:1137-42. [Crossref] [PubMed]


