
CSE1L in enhancing the effect of defactinib on gastric cancer cells via the inhibition of FAK phosphorylation
Highlight box
Key findings
• Chromosome segregation 1 like (CSE1L) was identified as a prognostic indicator for patients with gastric cancer (GC), capable of influencing the focal adhesion pathway and improving the efficacy of defactinib.
What is known and what is new?
• CSE1L is highly expressed in a variety of tumors, including GC. Defactinib has been shown to improve the prognosis of patients in some tumor types.
• CSE1L was found to affect the level of focal adhesion kinase (FAK) and FAK phosphorylation, suggesting that CSE1L can improve the efficacy of defactinib. This study preliminarily confirmed that inhibition of CSE1L expression could enhance the killing effect of defactinib on GC cells.
What is the implication, and what should change now?
• In order to further confirm the key role of CSE1L in GC, it is necessary to find CSE1L inhibitors or targeted drugs. In addition, comprehensive evaluation of the molecular biological function of CSE1L will help to clarify the mechanism of CSE1L in promoting tumorigenesis and development.
Introduction
Gastric cancer (GC) ranks as the fifth most common malignancy worldwide and is the third leading cause of cancer-related mortality (1). Current therapeutic approaches for GC remain constrained and predominantly include surgical intervention, radiotherapy, and chemotherapy (2). Despite the significant advancements achieved through targeted therapy and immune checkpoint inhibitors in several malignancies, these novel modalities have yet to confer substantial benefits to patients with GC (3,4). Owing to the heterogeneity inherent in GC, monotherapies that target a single molecular pathway frequently demonstrate limited efficacy (5). Consequently, combination therapies that concurrently target multiple pathways should hypothetically yield superior therapeutic outcomes for patients at the expense of increased toxicities (6,7). Overall prognosis is still dismal with survival rarely surpass 1 year in patients with advanced disease.
CSE1L, also known as CAS, CSE1, or XPO2, encodes a protein that is a member of the nuclear pore complex and is responsible for transporting nuclear localization signal (NLS) into the nucleus by importin-α (8). In recent years, CSE1L has been reported to be associated with tumor metastasis and chemotherapeutic drug-induced apoptosis (9). CSE1L promotes cell migration of oral squamous cell carcinoma and increases the proliferation of breast cancer cells (10,11). CSE1L also regulates the expression of the tumor protein P53 (P53) downstream genes by binding to the P53 gene (12). In colon cancer, the high expression of CSE1L indicates a poor prognosis of patients with colon cancer, suggesting that CSE1L may act as an oncogene (13). In addition, it has been reported that CSE1L may serve as a synergistic target with HDAC1/2 inhibitors (mocetinostat) in non-small cell lung cancer (14).
Focal adhesion kinase (FAK), encoded by the protein tyrosine kinase 2 (PTK2) gene, plays a critical role in the regulation of cell-cell interactions, extracellular matrix dynamics, and cellular motility (15). The phosphorylated FAK tyrosine 397 [p-FAK (Y397)] represents a pivotal posttranslational modification that modulates its functional activity (16). P-FAK (Y397) facilitates tumor-associated lymphangiogenesis and lymph node metastasis in ovarian cancer and promotes epithelial-mesenchymal transition in oral squamous cell carcinoma (17,18). In addition, it has been reported that p-FAK positivity is an independent risk factor for GC and can be used as a biomarker to predict the prognosis of GC patients (19). Therefore, p-FAK (Y397) has been proposed as a novel target for cancer therapy (20). Defactinib is an inhibitor targeting p-FAK (Y397) (21), and in clinical studies, defactinib has shown activity in patients with non-small cell lung cancer as a single agent and pancreatic cancer in combination with gemcitabine and immune checkpoint inhibitor but not in merlin-stratified pleural mesothelioma patients (22-24). Most importantly, a study explored the effect of defactinib in diffuse GC and its potential combined targets, confirmed the efficacy of defactinib in GC and the prospect of combined target therapy (25). Based on the effect of defactinib on GC cells, and the role played by CSE1L that this combination worth exploring. In this study, we examine the potential synergistic effects of targeting CSE1L pathway on defactinib. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2049/rc).
Methods
Patients
Adult patients with a confirmed diagnosis of GC were prospectively enrolled. Consecutive participants referred or admitted to Qinghai University Affiliated Hospital for surgical treatment were screened for eligibility. GC samples were obtained by surgical resection and were fixed using paraformaldehyde. The inclusion criteria included (I) a primary GC tumor that was operable according to presurgical assessments, with no indications of distant metastasis; and (II) adults aged 18 to 75 years. The exclusion criteria were as follows: (I) history of other malignancy or severe concomitant basic disease (cardiopulmonary dysfunction, tuberculosis, Crohn disease, or psychosis); (II) uncontrolled infections except for Helicobacter pylori; (III) emergency surgery due to tumor progression or a history of major abdominal surgery within the last 6 months; (IV) use of corticosteroids, insulin, oral hypoglycemic agent, or other obesity-related medications for an extended period time; and (V) a history of blood transfusions or purification therapy within the previous 3 months. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Medical College of Qinghai University (approval No. 2022-33), and informed consent was taken from all of the patients.
Tissue immunohistochemistry (IHC)
CSE1L protein expression was measured by IHC. Tissue sections were deparaffinize and subsequently underwent antigen retrieval via treatment with a 10-mM sodium citrate buffer solution at a pH of 6.0 that was heated to boiling point for 30 minutes. Afterward, the sections were incubated in a blocking solution containing normal serum to prevent nonspecific binding. Thereafter, the sections were exposed to primary antibodies specific for CSE1L (PA5-21468; Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) at a temperature of 4 ℃ for an extended period, typically overnight. Postincubation, the sections were rinsed and subsequently treated with a secondary antibody (KHC0779; Proteintech, Rosemont, IL, USA) to facilitate the detection of the primary antibody-antigen complex.
The IHC findings were evaluated by two blinded pathologists. The scoring of the IHC results was conducted using a semiquantitative scoring system, where staining in more than 30% of the neoplastic cells was deemed indicative of a positive result. The intensity of staining was characterized as negative or positive.
Cells
The cell lines used in this study were MGC-803, BGC-823, SGC-7901, HGC-27, MKN-45, MKN-74, AGS, and N87. Cell lines were obtained from the Cell Resource Center, Peking Union Medical College [which is part of the National Science and Technology Infrastructure, the National Biomedical Cell-Line Resource (NSTI-BMCR)]. The cells provided by the agency guarantee STR verification and negative detection of mycoplasma. Cells were cultured in a cell incubator containing 5% CO2 at 37 ℃ using RPMI 1640 basic medium (350-005-CL, Wisent Biotechnology, Nanjing, China), 10% fetal bovine serum (FBS) (885-150; Wisent Biotechnology), and 1% penicillin streptomycin (100×) (450-201; Wisent Biotechnology).
Knockout and construction of the overexpression cell lines
Three single-guide RNAs (sgRNAs) were designed (sg1: AATTTGTGAAGCCGATCGAG; sg2: AAGCATCAAGTGCACTATGT; sg3: CGTTTGACTTAAATTCATGA), then cloned using lentiCRISPR (51706, Addgene, Beijing, China). The plasmid was then augmented with Stbl3 Escherichia coli (C737303; Invitrogen) and extracted with a plasmid extraction kit (9760; Takara Bio, Kusatsu, Japan). Lipofectamine 3000 (L3000075; Invitrogen) was used to transfect the vector into MGC-803 cells according to the manufacturer’s instructions. After the transfected cells were screened for resistance, monoclonal cells were selected, and the monoclonal with the best gene knockout efficiency was selected for subsequent experiments. The YOE-LV001-CSE1L plasmid was used for overexpression, and Lipofectamine 3000 was used to transfect the vector into BGC-823 cells according to the manufacturer’s instructions. After the transfected cells passed resistance screening, monoclonal cells were selected, and the monoclonal cell line with the best overexpression efficiency was selected for subsequent experiments.
Cell proliferation and viability assay
The impact of CSE1L on the proliferative capacity of GC cells was assessed using the Cell Counting Kit 8 (CCK8) assay. 1×103 target cells in 100-µL volumes of RPMI 1640 culture medium were placed into 96-well microplates. The cells were incubated for 24, 48, and 72 hours. At each time point, the culture medium was aspirated, and the cells were then subjected to incubation with the CCK8 reagent (C0038; Beyotime Biotech Inc., Haimen, China) for a duration of 1 hour. Subsequently, the absorbance of the plates was measured by spectrophotometer at a wavelength of 450 nm. It was observed that the quantity of viable cells exhibited a positive correlation with the amount of formazan dye produced, which is indicative of cellular metabolic activity.
The cytotoxic effect of defactinib on GC cells was similarly evaluated with the CCK8 assay. In this instance, 6×103 target cells were plated in 100-µL volumes of RPMI 1640 medium into 96-well microplates. After a 6-hour incubation period to allow for cell attachment, the cells were treated with of 10 µM of defactinib for a period of 18 hours. Following treatment, the culture medium was removed, and the cells were incubated with the CCK8 reagent for 1 hour. The absorbance was then measured by spectrophotometer at 450 nm wavelength, with the absorbance values being directly proportional to the number of viable cells and the formazan product formed. Defactinib (HY-12289; MedChemExpress, Monmouth Junction, NJ, USA) was used in accordance with the guidelines provided by the manufacturer.
Transwell migration and invasion assays
Cell migration was assessed using a Transwell chamber with 8-µm pore membranes (3422; Corning, Corning, NY, USA). GC cells (2×105) in serum-free medium were added to the upper chamber, while the lower chamber contained medium with 20% FBS. After 24 hours, the upper chamber was fixed with methanol and stained with crystal violet, and the nonmigratory cells were removed. Migratory cells on the underside were photographed.
For the invasion assay, the Transwell membrane was precoated with Matrigel (356234; Corning). The procedure was similar to that of the migration assay, with the addition of Matrigel to assess cell invasion through a matrix.
Clone formation assay
For clone formation assays, 5,000 cells were seeded per well in 6-well cell culture plates. Dulbecco’s modified Eagle medium, supplemented with 10% FBS, was refreshed every 4 days. Following a 12-day incubation period, the cells were fixed using 100% methanol and subsequently stained with crystal violet (G1063; Solarbio, Beijing, China) for a duration of 20 minutes.
Cell adhesion assay
Cells (6×104) were seeded into 96-well plates on coverslips that were precoated with 1% sterile gelatin (G-2500; Sigma-Aldrich, St. Louis, MO, USA) and exposed to different treatments. After 3 hours of culturing, the plates were gently washed with phosphate-buffered saline (PBS) to remove the nonadherent cells. The attached cells were then detected via CCK8 assay. Plates were scanned spectrophotometrically at 450 nm, and the number of viable cells was positively correlated with the formazan product.
RNA sequencing (RNA-seq) and analysis
Total RNA was extracted from cells using TRIzol reagent and was used for RNA-seq with the Agilent 2100 bioanalyzer system (Agilent Technologies, Inc., Santa Clara, CA, USA). The RNA was converted to complementary DNA (cDNA) using the GeneChip WT Plus Kit (Thermo Fisher Scientific). The cDNA was then purified, fragmented, and hybridized with the microarray probes. Posthybridization, a microarray chip (GeneChem, Shanghai, China) was processed by washing and staining. Genes of the MGC-803 CSE1L-knockout (CSE1LKO) and MGC-803 CSE1L-negative control (CSE1LNC) cell line with corrected P values of <0.05 and a fold change of >2.0 were considered significantly differentially expressed genes (DEGs). All DEGs were included in The Cancer Genome Atlas (TCGA) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses.
Western blotting
Cells were rinsed with cold PBS and lysed using RIPA buffer (C500005; Sangon Biotech, Shanghai, China) with a protease inhibitor cocktail being added (C600381; Sangon Biotech). The lysate was briefly sonicated and then underwent sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to polyvinylidene fluoride (PVDF) membranes, blocked, and incubated with primary antibodies overnight at 4 ℃ [all dilutions were 1:1,000 dilution, except for that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was 1:3,000]. After incubation with secondary antibodies and washing, the blots were developed using an enhanced chemiluminescence (ECL) assay. Antibodies against FAK (66258-1-Ig; Proteintech), p-FAK (Y397) (T55587; Abmart, Shanghai, China), CSE1L (PA5-21468; Invitrogen), β-actin (66009-1-Ig; Proteintech), GAPDH (60004-1-Ig; Proteintech), integrin beta-4 (ITGB4) (21738-1-AP; Proteintech), and myosin regulatory light polypeptide 9 (MYL9) (15354-1-AP; Proteintech) were used according to respective the manufacturer’s instructions.
Cell immunofluorescence
Cells were seeded at a density of 2×105 per well in 24-well plates on 12-mm gelatin-coated coverslips. After 24 hours, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS. Nonspecific binding was blocked with 5% bovine serum albumin (BSA) in PBS for 2 hours. The cells were then incubated with anti-p-FAK (Y397) antibody diluted to 1:200 overnight at 4 ℃. This was followed by a 2-hour incubation with fluorescent secondary antibody (1:100) and phalloidin (1:100; CD4683; Coolaber Science & Technology Co., Ltd., Beijing, China) to stain the actin cytoskeleton. Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI) (SL7100; Coolaber). Finally, the coverslips were mounted onto microscope slides using an antifading medium, and images were captured using a confocal microscope at 400× magnification.
Bioinformatics analysis
RNA-seq CSE1L gene expression in GC and another 32 cancers was retrieved from the UCSC Cancer Genomics Browser via TCGA data portal (https://www.cancer.gov/ccg/research/genome-sequencing/tcga). An additional 27 samples with paired tissues were extracted from the database to assess CSE1L gene expression for GC. The messenger RNA (mRNA) gene chip data of GC in the Kaplan-Meier plotter database (https://kmplot.com/analysis/) were used to evaluate the association of CSE1L expression with the 5-year (60 months) overall survival (OS) of patients with GC (26).
Statistical analysis
GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, USA) was used to generate graphs for the experiments. Data were shown as the mean ± standard deviation (SD). Statistical comparisons were made using one-way analysis of variance for multiple groups or the Student’s t-test for two groups. Survival rates of patients with GC were plotted with the Kaplan-Meier method, and differences were assessed using the log-rank test. A P value less than 0.05 was considered to indicate statistical significance.
Results
CSE1L was highly expressed in GC tissues and suggests a poor prognosis
Transcriptome data from TCGA database were utilized to examine the expression levels of CSE1L mRNA in various tumor and normal tissues. The analysis revealed that CSE1L mRNA expression levels were higher in tumor tissues in comparaison to normal tissues in 16 out of 33 tumors (Figure 1A). Further comparison of CSE1L mRNA expression in unpaired and paired adjacent GC samples confirmed a marked increase in CSE1L mRNA levels within the database GC tissues (Figure 1B). IHC staining for CSE1L was conducted on 83 GC tissues and adjacent noncancerous tissues from patients diagnosed with GC. The results indicated significant upregulation of CSE1L protein expression in the GC samples (Figure 1C, Table 1). The Kaplan-Meier curve suggested that high CSE1L expression is associated with a worse prognosis in patients with GC (Figure 1D).
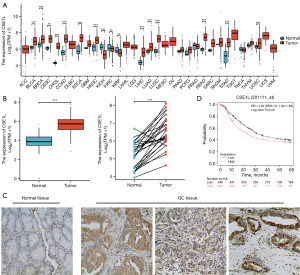
Table 1
| Histology | Expression, n [%] | Total cases | P value | |
|---|---|---|---|---|
| Positive | Negative | |||
| Normal | 22 [27] | 61 [73] | 83 | |
| Tumor | 51 [61] | 32 [39] | 83 | |
| Total | 73 | 93 | 166 | <0.001 |
CSE1L, chromosome segregation 1 like; GC, gastric cancer.
In conclusion, our data confirmed the high expression of CSE1L in GC tissues compared to normal tissue and high expression is associated with poor prognosis.
CSE1L promotes multiple malignant phenotypes of GC cells
To elucidate the role of CSE1L in GC cells, this study evaluated the expression levels of CSE1L across eight GC cell lines (MGC-803, BGC-823, SGC-7901, HGC-27, MKN-45, MKN-74, AGS, and N87). The results indicated that MGC-803 cells exhibited the highest expression of CSE1L, while BGC-823 cells demonstrated the lowest expression (Figure 2A). To further investigate, a stable MGC-803 CSE1LKO cell line was generated using CRISPR/Cas9 technology, and a stable BGC-823 CSE1L-overexpression (CSE1LOE) cell line was established using an overexpression plasmid. The results of gene knockout and overexpression are illustrated in Figure 2B,2C. Various cell phenotype assays were employed to assess the biological function of CSE1L. Specifically, the CCK8 assay was adopted to determine the impact of CSE1L on the proliferative capacity of GC cells (Figure 2D,2E). While transwell assay was used to evaluate the influence of CSE1L on the migratory and invasive capabilities of these cells (Figure 2F,2G). Additionally, adhesion assay was performed to determine the effect of CSE1L on the adhesive properties of GC cells (Figure 2H,2I). Finally, plate clone formation assay was used to assess the effect of CSE1L on the clonogenic potential of GC cells (Figure 2J,2K). These experiments demonstrated that MGC-803 CSE1LKO cells exhibited significantly reduced proliferation, migration, invasion, clone formation, and adhesion capabilities compared to MGC-803 CSE1LNC cells. Additionally, BGC-823 CSE1LOE cells displayed enhanced migration, invasion, clone formation, and adhesion abilities relative to BGC-823 CSE1LNC cells.
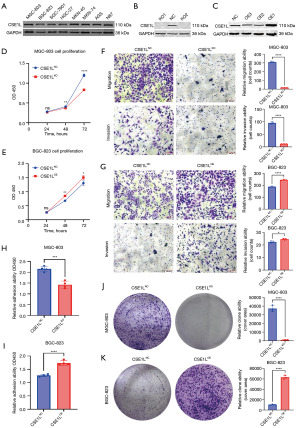
The adhesion ability of tumor cells is often neglected; however, adhesion ability is a key behavior during solid tumor metastasis. Research suggests (27) that the focal adhesion pathway is the key to regulating cell adhesion ability. P-FAK can promote cell adhesion. Fortunately, drugs targeting p-FAK, including defactinib and 1,2,4,5-benzenetetramine tetrahydrochloride (Y15), can inhibit the proliferation, migration, and adhesion of tumor cells. However, whether the ability of CSE1L to promote the adhesion of GC cells through the focal adhesion pathway is unknown.
CSE1L upregulates the focal adhesion pathway
To elucidate the molecular mechanisms underlying CSE1L’s promotion malignant phenotypes, a comparative analysis of the transcriptome data from MGC-803 CSE1LKO cells and MGC-803 CSE1LNC cells was conducted. Subsequent volcano plots illustrated the differential genes between CSE1LKO and CSE1LNC (Figure 3A), suggesting that 447 genes are downregulated and 194 genes are upregulated in CESIL knockout cells. KEGG enrichment analysis showed that CSE1L is involved in multiple tumor-related signaling pathways, including PI3K-Akt, Wnt, and focal adhesion signaling pathways (Figure 3B). The focal adhesion pathway being one of the enriched pathways may be able to explain why CSE1L promotes proliferation, invasion, and adhesion of GC cells. A heatmap showed the mRNA levels of the genes that were regulated by CSE1L: 12 genes in the focal adhesion pathway were all upregulated by CSE1L (Figure 3C). The expression of genes in the focal adhesion pathway, including ITGB4, PTK2, MYL9, and β-actin, was verified. Real-time quantitative polymerase chain reaction confirmed the regulation of the focal adhesion pathway by CSE1L at the mRNA level (Figure 3D,3E). Western blotting confirmed that CSE1L could upregulate the protein expression of ITGB4, FAK, MYL9, β-actin, and CSE1L and promote the phosphorylation of FAK (Y397) (Figure 3F,3G).
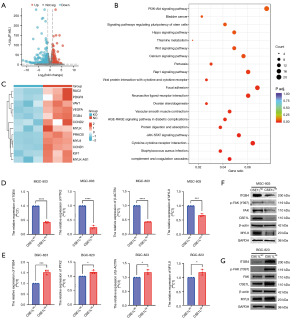
By analyzing the RNA-seq data, we found that CSE1L upregulates the focal adhesion pathway. In addition, CSE1L could upregulate p-FAK (Y397), which is a novel antitumor target. Whether the downregulation of CSE1L could increase the efficacy of drugs targeting p-FAK (Y397) was subsequently investigated.
Knockdown of CSE1L enhances the efficacy of defactinib
CCK8 assay was used to evaluate the effect of CSE1L on defactinib’s antitumor ability. After CSE1L was knocked out, the killing effect of defactinib was enhanced but was inhibited after overexpression of CSE1L (Figure 4A). Following this, Western blotting was used for evaluating the effect of CSE1L on p-FAK (Y397) and indicated that knockout of CSE1L could downregulate the expression of p-FAK (Y397); meanwhile, the overexpression of CSE1L could upregulate the expression of p-FAK (Y397) (Figure 4B). Subsequently, immunofluorescence was used to clarify the effect of CSE1L on p-FAK (Y397) and actin proteins. The localization of p-FAK (Y397) protein in MGC-803 CSE1LNC cells was mainly at the edge of the cell, and F-actin protein showed typical characteristics of microfilaments. The expression of p-FAK (Y397) was significantly decreased in MGC-803 CSE1LKO cells, and the filamentous structure of F-actin was disordered. p-FAK (Y397) was mainly expressed in the nucleus of BGC-823 CSE1LNC cells, and F-actin had no typical filamentous structure. p-FAK (Y397) was located at the edge of BGC-823 CSE1LOE cells, and F-actin showed a typical microfilament structure (Figure 4C). Subsequently, Western blotting was used to evaluate the protein expression levels of FAK and CSE1L in GC cells, which showed a positive correlation between CSE1L and FAK (Figure 4D,4E). Western blotting further indicated that CSE1L could upregulate the expression of FAK and p-FAK (Y397) in MGC-803 and BGC-823 cells, but whether this also applied to other GC cells and tissues was not examined. Bioinformatics analysis of RNA-seq data from GC samples in TCGA showed that CSE1L mRNA and FAK mRNA (PTK2) were positively correlated (Figure 4F). These results suggest that the expression level of CSE1L can affect the efficacy of defactinib. Therefore, defactinib was added GC cells, and defactinib was found to have a weak killing effect on MGC-803 cells but a strong killing effect on BGC-823 cells (Figure 4G).
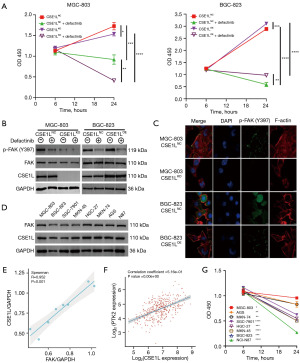
Taken together, these results confirm that CSE1LKO enhances the cytotoxicity of defactinib in GC cells. A positive correlation was found between CSE1L expression and FAK expression, suggesting that defactinib may be more effective in patients with GC who have a low CSE1L expression.
Discussion
Targeted therapy and immune checkpoint inhibitors have sparked optimism in cancer therapy but unfortunately, in GC, the benefits have been limited and the overall prognosis has not improved significantly (28,29). The heterogeneity of GC is one of the main reasons for the poor outcome even with the addition of immunotherapy and targeted therapy. Identification of different molecular subtypes of GC and customize novel therapeutic targets based on that might improve the efficacy of targeted therapy (5). In this study, we found that GC cells have a high expression of CSE1L and it correlates with poor progress. CSE1L promotes the proliferation, migration, invasion, adhesion, and clone formation of GC cells. These results suggest that CSE1L can promote the malignant progression of GC. A study found that silencing CSE1L can downregulate the transcription factor MITF and inhibit the growth and metastasis of GC (30). It has been reported that CSE1L has a potential synergistic effect with other agents (i.e., HDAC inhibitors) (14).
The results of transcriptome sequencing suggested that CSE1L could upregulate the focal adhesion pathway in GC cells. This pathway is not only related to tumor cell adhesion, proliferation, and migration ability but also tumor drug resistance (31,32). Several agents can antagonize or inhibit the key targets in the focal adhesion pathway: FAK and p-FAK like defactinib, however its efficacy has been limited as a single agent. In this study, we found that CSE1L could upregulate FAK and p-FAK (Y397) and that silencing CSE1L could enhance the efficacy of defactinib. However, the regulatory mechanism of CSE1L on FAK and p-FAK (Y397) remains unclear. One possibility would be via P53 as it has been reported that mutant P53 can promote FAK expression (33), and interestingly, CSE1L is able to promote the expression of its downstream genes by binding to P53 (12). We thus speculate that CSE1L upregulates FAK and p-FAK (Y397) via P53.
Our study also indicates that CSE1L could affect the microfilaments and cytoskeleton of GC cells. In the immunofluorescence experiments, the expression level of CSE1L influenced the expression of p-FAK and cellular localization, as well as the cell microfilaments composed of F-actin. FAK phosphorylation can promote actin polymerization, which in turn promotes cell motility and adhesion (30,34). The above results reasonably explain why CSE1L can promote the migration, invasion, and adhesion ability of GC cells. Specifically, the high expression of CSE1L can promote the formation of F-actin microfilaments, which promotes cell adhesion via the formation of stress fibers and facilitates cell movement through the formation of pseudopodia (35-37).
Conclusions
This study confirmed the oncogenic role of CSE1L in GC. Silencing CSE1L was found to increase the effect of the FAK phosphorylation inhibitor defactinib. CSE1L has the potential to serve as a synergistic target of defactinib.
Acknowledgments
The authors thank the Department of Pathology of Qinghai University Affiliated Hospital for immunohistochemical analysis.
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2049/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2049/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2049/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2049/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Medical College of Qinghai University (approval No. 2022-33), and informed consent was taken from all of the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet 2020;396:635-48. [Crossref] [PubMed]
- Sexton RE, Al Hallak MN, Diab M, et al. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev 2020;39:1179-203. [Crossref] [PubMed]
- Riley RS, June CH, Langer R, et al. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discov 2019;18:175-96. [Crossref] [PubMed]
- Zeng Y, Jin RU. Molecular pathogenesis, targeted therapies, and future perspectives for gastric cancer. Semin Cancer Biol 2022;86:566-82. [Crossref] [PubMed]
- Chia NY, Tan P. Molecular classification of gastric cancer. Ann Oncol 2016;27:763-9. [Crossref] [PubMed]
- Guan WL, He Y, Xu RH. Gastric cancer treatment: recent progress and future perspectives. J Hematol Oncol 2023;16:57. [Crossref] [PubMed]
- Passaro A, Al Bakir M, Hamilton EG, et al. Cancer biomarkers: Emerging trends and clinical implications for personalized treatment. Cell 2024;187:1617-35. [Crossref] [PubMed]
- Brinkmann U, Brinkmann E, Bera TK, et al. Tissue-specific alternative splicing of the CSE1L/CAS (cellular apoptosis susceptibility) gene. Genomics 1999;58:41-9. [Crossref] [PubMed]
- Tai CJ, Hsu CH, Shen SC, et al. Cellular apoptosis susceptibility (CSE1L/CAS) protein in cancer metastasis and chemotherapeutic drug-induced apoptosis. J Exp Clin Cancer Res 2010;29:110. [Crossref] [PubMed]
- Li G, Li R, Wang W, et al. DDX27 regulates oral squamous cell carcinoma development through targeting CSE1L. Life Sci 2024;340:122479. [Crossref] [PubMed]
- Ye M, Chen Y, Liu J, et al. Interfering with CSE1L/CAS inhibits tumour growth via C3 in triple-negative breast cancer. Cell Prolif 2022;55:e13226. [Crossref] [PubMed]
- Tanaka T, Ohkubo S, Tatsuno I, et al. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell 2007;130:638-50. [Crossref] [PubMed]
- Wang D, Liufu J, Yang Q, et al. Identification and validation of a novel signature as a diagnostic and prognostic biomarker in colorectal cancer. Biol Direct 2022;17:29. [Crossref] [PubMed]
- Duan L, Tadi MJ, Maki CG. CSE1L is a negative regulator of the RB-DREAM pathway in p53 wild-type NSCLC and can be targeted using an HDAC1/2 inhibitor. Sci Rep 2023;13:16271. [Crossref] [PubMed]
- Alcaraz J, Otero J, Jorba I, et al. Bidirectional mechanobiology between cells and their local extracellular matrix probed by atomic force microscopy. Semin Cell Dev Biol 2018;73:71-81. [Crossref] [PubMed]
- Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol 2005;6:56-68. [Crossref] [PubMed]
- Liu X, Li J, Yang X, et al. Carcinoma-associated fibroblast-derived lysyl oxidase-rich extracellular vesicles mediate collagen crosslinking and promote epithelial-mesenchymal transition via p-FAK/p-paxillin/YAP signaling. Int J Oral Sci 2023;15:32. [Crossref] [PubMed]
- Sun R, Yuan L, Jiang Y, et al. ALKBH5 activates FAK signaling through m6A demethylation in ITGB1 mRNA and enhances tumor-associated lymphangiogenesis and lymph node metastasis in ovarian cancer. Theranostics 2023;13:833-48. [Crossref] [PubMed]
- Peng K, Li S, Li Q, et al. Positive Phospho-Focal Adhesion Kinase in Gastric Cancer Associates With Poor Prognosis After Curative Resection. Front Oncol 2022;12:953938. [Crossref] [PubMed]
- Chuang HH, Zhen YY, Tsai YC, et al. FAK in Cancer: From Mechanisms to Therapeutic Strategies. Int J Mol Sci 2022;23:1726. [Crossref] [PubMed]
- Hu HH, Wang SQ, Shang HL, et al. Roles and inhibitors of FAK in cancer: current advances and future directions. Front Pharmacol 2024;15:1274209. [Crossref] [PubMed]
- Gerber DE, Camidge DR, Morgensztern D, et al. Phase 2 study of the focal adhesion kinase inhibitor defactinib (VS-6063) in previously treated advanced KRAS mutant non-small cell lung cancer. Lung Cancer 2020;139:60-7. [Crossref] [PubMed]
- Wang-Gillam A, Lim KH, McWilliams R, et al. Defactinib, Pembrolizumab, and Gemcitabine in Patients with Advanced Treatment Refractory Pancreatic Cancer: A Phase I Dose Escalation and Expansion Study. Clin Cancer Res 2022;28:5254-62. [Crossref] [PubMed]
- Fennell DA, Baas P, Taylor P, et al. Maintenance Defactinib Versus Placebo After First-Line Chemotherapy in Patients With Merlin-Stratified Pleural Mesothelioma: COMMAND-A Double-Blind, Randomized, Phase II Study. J Clin Oncol 2019;37:790-8. [Crossref] [PubMed]
- Peng K, Zhang F, Wang Y, et al. Development of Combination Strategies for Focal Adhesion Kinase Inhibition in Diffuse Gastric Cancer. Clin Cancer Res 2023;29:197-208. [Crossref] [PubMed]
- Győrffy B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation (Camb) 2024;5:100625. [Crossref] [PubMed]
- Li X, Combs JD 3rd, Salaita K, et al. Polarized focal adhesion kinase activity within a focal adhesion during cell migration. Nat Chem Biol 2023;19:1458-68. [Crossref] [PubMed]
- Li K, Zhang A, Li X, et al. Advances in clinical immunotherapy for gastric cancer. Biochim Biophys Acta Rev Cancer 2021;1876:188615. [Crossref] [PubMed]
- Patel TH, Cecchini M. Targeted Therapies in Advanced Gastric Cancer. Curr Treat Options Oncol 2020;21:70. [Crossref] [PubMed]
- Li Y, Yuan S, Liu J, et al. CSE1L silence inhibits the growth and metastasis in gastric cancer by repressing GPNMB via positively regulating transcription factor MITF. J Cell Physiol 2020;235:2071-9. [Crossref] [PubMed]
- Tapial Martínez P, López Navajas P, Lietha D. FAK Structure and Regulation by Membrane Interactions and Force in Focal Adhesions. Biomolecules 2020;10:179. [Crossref] [PubMed]
- Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol 2015;31:65-75. [Crossref] [PubMed]
- Pifer PM, Yang L, Kumar M, et al. FAK Drives Resistance to Therapy in HPV-Negative Head and Neck Cancer in a p53-Dependent Manner. Clin Cancer Res 2024;30:187-97. [Crossref] [PubMed]
- Shinde AV, Humeres C, Frangogiannis NG. The role of α-smooth muscle actin in fibroblast-mediated matrix contraction and remodeling. Biochim Biophys Acta Mol Basis Dis 2017;1863:298-309. [Crossref] [PubMed]
- Salgado-Lucio ML, Ramírez-Ramírez D, Jorge-Cruz CY, et al. FAK regulates actin polymerization during sperm capacitation via the ERK2/GEF-H1/RhoA signaling pathway. J Cell Sci 2020;133:jcs239186. [Crossref] [PubMed]
- Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys 2011;40:169-86. [Crossref] [PubMed]
- Bisaria A, Hayer A, Garbett D, et al. Membrane-proximal F-actin restricts local membrane protrusions and directs cell migration. Science 2020;368:1205-10. [Crossref] [PubMed]
(English Language Editor: J. Gray)


