
Insight into dipeptidase 1: structure, function, and mechanism in gastrointestinal cancer diseases
Introduction
Cancer continues to be the second most common cause of mortality on a global scale (1). Although significant advancements have been made in medical research regarding prevention, diagnosis, and treatment over the past few decades, more than 50% of cancer patients still ultimately succumb to the disease (2). Identifying potent biomarkers is crucial for facilitating the early detection and diagnosis of cancer, precisely gauging the risks associated with prognosis, and devising more judicious therapeutic strategies (3). In recent years, the dipeptidase (DPEP) family has risen to prominence as novel biomarkers, garnering significant interest due to their intimate association with the etiology and progression of a variety of malignant tumors.
DPEP family members, including DPEP1, DPEP2, and DPEP3, share the same gene locus on the same chromosome and have similar structures (4,5). DPEP1, first identified as “renal dipeptidase”, can hydrolyze a variety of dipeptides; participate in glutathione (GSH), leukotriene, and lipid metabolism; and exert β-lactamase activity and metalloenzyme activity (4). DPEP1 and DPEP2, but not DPEP3, can cleave leukotriene D4 (LTD4) (6), while DPEP3 or DPEP1 can cleave cystinyl-bis-glycine (7). DPEP2 has the highest expression level in the lungs, heart, and testes, while DPEP3 is only present in the testes (8). Three members of the membrane-bound DPEP family differ in substrate specificity and tissue distribution (Table 1), and due to limitations in expression pattern and function, research on DPEP2 and DPEP3 is minimal. In this review, we specifically focus on the biological functions of DPEP1 in cells to clarify their potential regulation mechanisms, with the aim of providing insights into the DPEP1 biology relevant to cancer activity.
Table 1
| Name | DPEP1 | DPEP2 | DPEP3 |
|---|---|---|---|
| Alternate synonyms | MBD1, MDP, RDP | MBD2 | MBD3 |
| Genomic locus | 16q24.3 | 16q22.1 | 16q22.1 |
| Subcellular localization | Apical plasma membrane and serosa | Cell membrane and cytoplasm | Cell membrane and cytoplasm |
| Length (AA) | 411 | 486 | 488 |
| Expression in normal tissues | Kidney, heart, liver, lungs, and testis (9-11) | Lung, spleen, heart, lymph nodes, and testis (12) | Mainly in the testis (13) |
| Hydrolysis activity | Yes | Yes | Yes (14) |
| Zn2+-dependent metalloenzymes | Yes | Yes | Yes |
| Leukotriene metabolism | Yes (15) | Yes (7) | No (14) |
| β-lactamase activity | Penem, carbapenems (16) | ND | ND |
| GPI anchor | Yes | Yes | Yes |
| Special biological function | Physical adhesion (lung, liver) | By acting as a regulator of the NF-κB inflammatory signaling pathway (17) | Cytotoxicity in ovarian tumor (18) |
| Signaling pathway of participation | (I) Regulation of E-cadherin expression in suppressing colon cancer metastasis. (II) TGF-β-induced EMT | Regulation of NF-κB inflammatory signaling | ND |
DPEP, dipeptidase; AA, amino acid; GPI, glycosylphosphatidylinositol; ND, not detectable; TGF-β, transforming growth factor beta; EMT, epithelial-mesenchymal transition; NF-κB, nuclear factor kappa B.
Structure of DPEP1
The DPEP1 gene is located in chromosome 16 (chr16q24.3) and contains 11 exons that encode 411 amino acids (aa). The initial hydrophobic region composed of the first 16 amino acid residues is referred to as the signal peptide (SP). The span from 17aa to 385aa is the main chain of this gene, and DPEP1 contains four N-glycosylation sites linked to asparagine, three disulfide bonding sites, and several Zn2+ binding sites (Figure 1). As a cofactor, Zn2+ stabilizes the secondary structure of DPEP1 and promotes advanced conformation formation (19-21). The glycosylphosphatidylinositol (GPI) site at the carboxyl end of DPEP1 ensures it is covalently anchored on the outer membrane of cells (22). Of note, the carboxyl terminal of DPEP1 contains an extended sequence of hydrophobic amino acids after the GPI anchor, which serves as the propeptide (386aa-411aa), and is usually removed for DPEP1 molecule maturation (20) (Figure 1).
According to the structure obtained from X-ray, DPEP1 typically exists as a homodimer in cells. Each dimer consists of 8 α-helix and β-strand structures [(α/β)8] connected by disulfide bonds to form a barrel-like spatial structure (20) (Figure 2). The active site of β-lactamase is located in the barrel-like structure’s middle pocket (Glu141, His168, His214, and His 235), mainly at the β-strand end. A triangular pyramid structure is formed by the combination of binuclear zinc ions bridged at the bottom of the barrel, which provides stability (23). The connection of these two sets of disulfide bonds, Cys71-Cys154 and Cys226-Cys258, further strengthens the pocket structure and enhances the activity of β-lactamase. Renal DPEP is believed to be the only metabolizing enzyme of penem and carbapenem β-lactam antibiotics in mammals. Previous studies have indicated that Glu141, His214, and His235 are involved in the catalysis of dipeptide hydrolase (24,25). Meanwhile, His168 may be essential in the binding of substrates or inhibitors. Finally, cilastatin can change the location of binuclear zinc ions and affect the enzyme’s active site by disrupting their stability, subsequently involving the activity of DPEP1 (20,26).
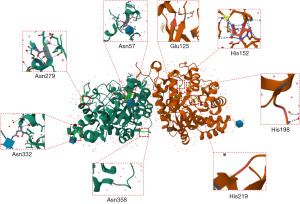
The main chain of DPEP1 contains 369 amino acids (17aa–385aa) with a calculated molecule weight of 42 kDa, but according to Western blot assay, it can shift to 45–70 kDa, potentially due to its glycosylation. DPEP1 contains four N-glycosylation sites (Asn57, Asn279, Asn332, and Asn358), mainly located at the outside junction of the barrel structure of the helix-turn, helix-strand, and helix-helix structural transitions. This allows for carbohydrate compounds to form glycosidic bonds with the amino acid residues of DPEP1 and effectuate the glycosylation process (Figure 2). the N-glycosylation sites Asn57 and Asn332 may also have N-acetylglucosamine structures. Glycosylation is fundamental to the key pathological steps of tumor development and progression, including interference in cell-cell adhesion and promotion of tumor cell invasion (27,28). It is thus reasonable to assume that the carcinogenic effect of DPEP1 in many tumors is closely related to its glycosylation structure, and this should be further verified in future research.
As a renal outer membrane protein, DPEP1 is also distributed in the cytoplasm and nucleus (29). The α/β-barrel structure provides permeability to the outer membrane and maintains the DPEP1 protein-based stability of the outer membrane structure. However, it has not yet been confirmed how many times DPEP1 spans the cell membrane to finally anchor onto the outer membrane.
Biological functions of DPEP1
DPEP1, the first renal DPEP to be discovered, plays an essential role in the hydrolysis of dipeptides and polypeptides, the regulation of GSH degradation, the promotion of the conversion of LTD4 to leukotriene E4 (LTE4), lipid metabolism, and the inactivation of β-lactam antibiotics (30-32). DPEP1 also participates in the cellular myelocytomatosis viral oncogene homolog (c-MYC), LTD4/β-catenin, epithelial-mesenchymal transition (EMT)/E-cadherin, PI3K/Akt/mechanistic target of rapamycin kinase (mTOR), and other signaling pathways, which regulate the transformation of malignant tumors (32-34). In the immune system, DPEP1 can recruit neutrophils to the liver and lungs, which is unrelated to the function of dipeptide hydrolase (11). Moreover, DPEP1 can exert a unique physical adhesion function, with various types of acute inflammation being related to this function, including acute respiratory distress syndrome (ARDS) and multiple organ dysfunction syndrome (MODS). DPEP2 and DPEP3 are similar in physical structure to DPEP1, but their biological functions differ slightly. The structure and functional characteristics of the DPEP family are summarized in Table 1.
DPEP1 in ferroptosis
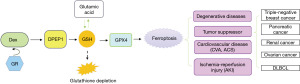
Ferroptosis is an iron-dependent form of programmed cell death, which, in essence, is GSH depletion (35). DPEP1 can participate in the hydrolysis of GSH. When GSH depletion results in the loss of GSH peroxidase (GPX4) activity, lipid oxides cannot be metabolized by GPX4-catalyzed GSH reductase, prompting mitochondria to produce an excess of lipid peroxides, thereby inducing the occurrence of mitochondria-dependent ferroptosis (36,37) (Figure 3). The reduction of free ferric ions directly affects the synthesis of heme and hemoglobin, resulting in a range of ischemia-related diseases such as stroke (38), cardiovascular disease (39), intracerebral hemorrhage (37), ischemia-reperfusion injury (40), and renal damage (41). Ferroptosis is mainly associated with the suppression of certain cancers, such as triple-negative breast cancer (42), diffuse large B-cell lymphoma (43), ovarian cancer (44), and pancreatic ductal adenocarcinoma (10) and even with cisplatin resistance in cancer (45). This seems to explain DPEP1’s tumor-inhibiting function in these cancer diseases. In addition, dexamethasone, as a positive regulator of ferroptosis, can upregulate the expression of DPEP1 in a glucocorticoid receptor (GR)-dependent manner, aggravate GSH deficiency, and then increase ferroptosis susceptibility and promote the occurrence of related diseases (46) (Figure 3).
DPEP1 in inflammation
Associations with osteoarthritis (OA)
In genetics, DPEP1 is associated with the risk of OA (47). Zhang et al. identified and genotyped 14 single-nucleotide polymorphisms (SNPs) associated with the DPEP1 gene and reported that SNP rs1126464 is a missense mutation of the DPEP1 gene, significantly increasing the risk of OA (47). The DPEP1 protein has the effect of hydrolyzing β-lactam antibiotics, which also increases the difficulty of OA treatment (48). Cilastatin, an inhibitor of DPEP1, has been approved for use in combination with imipenem in the treatment of OA, which not only can target DPEP1 but also prolong the antibacterial effect (48).
Inhibiting β-lactam antibiotics
DPEP1 participates in the hydrolysis metabolism of carbapenem β-lactam antibiotics (20). In the DPEP1 structure, the active site of β-lactamase is located in the middle of the barrel structure formed by the DPEP1 dimer, which maintains the activity and stability of the enzyme. As an inhibitor of DPEP1, cilastatin can change the position of DPEP1 binuclear Zn2+ and destroy the barrel structure, which destroys the stability of the β-lactamase active site, thus inhibiting the function of DPEP1 to hydrolyze β-lactam antibiotics. Therefore, cilastatin is usually combined with imipenem to prolong the effective time of imipenem and to reduce renal toxicity (49), which is the most meaningful application of DPEP1 for β-lactam antibiotics.
Neutrophil recruitment
A significant feature of inflammation is the accumulation of neutrophils from the bloodstream to the inflammatory tissue. Neutrophils are the first line of defense against foreign pathogens, and it is essential to effectively control inflammation to recruit them to the site of infection (50). Lack of classic recruitment leads to recurrent and persistent infection, while inappropriate recruitment of neutrophils leads to MODS. Indeed, ARDS and MODS accompanied by severe infection are associated with abnormal accumulation of neutrophils (51). As a physical adhesion receptor, DPEP1 promotes the recruitment of neutrophils from the blood to the inflamed liver and lung. The effect of this physical adhesion receptor is not dependent on its enzyme activity. DPEP1 merely exerts a physical adhesive effect, driving an excessive accumulation of neutrophils in the lungs and liver, thereby inducing an inflammatory response (11). DPEP1 is present in the endothelium of the capillaries in human lung tissue, which is the site of neutrophil sequestration during sepsis. Stimulated by bacterial endotoxin lipopolysaccharide (LPS), neutrophils can cross the very thin lung capillary wall and gather at the inflamed site of the lung. It exerts a similar effect in the kidneys, which is one of the reasons why DPEP1 causes a series of renal-related diseases, such as colitis-associated colon (52) and acute kidney injury (53). We believe that DPEP1 can serve as an anti-inflammatory therapeutic target for lung and liver diseases and renal diseases. Exploring targeted drugs for DPEP1 may profoundly improve clinical treatment.
DPEP1 in gastrointestinal cancer
DPEP1 functions in regulating tumor transformation and malignancy in two ways. DPEP1 can act as an oncogene; promote tumor proliferation, survival, invasion, adhesion, and migration; and even induce drug resistance (54,55). Alternatively, DPEP1 can maintain genomic stability, inhibit tumorigenesis, and enhance chemotherapy sensitivity (10,56). Nevertheless, the biological mechanism of DPEP1 in cancer is related to its unique structure and enzyme activity.
DPEP1 enhance the stemness of tumor cells
DPEP1 may promote the proliferation and invasion of colorectal cancer cells through interaction with signaling pathways such as c-MYC and PI3K/AKT/mTOR, and form a positive feedback loop with ASCL2 to enhance the drug resistance of colorectal cancer cells. Although the specific mechanisms of drug resistance are not yet clear, it may be related to DPEP1’s involvement in regulating the behavior of immune cells in the tumor microenvironment, including their infiltration, activation status, and cytotoxic capabilities towards tumor cells, thereby indirectly participating in the regulation of tumor cell drug resistance. These research findings provide important information for a deeper understanding of the role of DPEP1 in tumor biology and offer potential directions for the development of new therapeutic strategies.
Transcriptional regulation of DPEP1 in gastrointestinal cancer
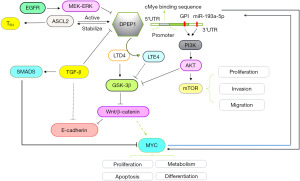
c-Myc, as a transcription factor, regulates cell differentiation and proliferation through various mechanisms, including transcriptional amplification of target genes, and can even alter the susceptibility of cancer cells to immunotherapy (33,57). Within the promoter region of DPEP1, c-Myc binds to the specific DNA motif and transcriptionally active DPEP1 expression in colorectal cancer cells (33). Moreover, DPEP1 can inhibit ubiquitin-mediated degradation of c-Myc, extending the half-life of the c-MYC protein in colorectal cancer cells and thus enhancing protein stability. Therefore, DPEP1 and c-MYC protein may form a positive feedback loop, maintaining their high expression levels in colorectal cancer cells, which promotes the invasion and proliferation of cancer cells and even induces their drug resistance (33). Similarly, miR-193a-5p can directly target DPEP1 by binding to its messenger RNA 3 prime untranslated region in hepatoblastoma (HB); miR-193a-5p suppression promotes cell proliferation and metastasis though the activation of PI3K/AKT/mTOR signaling (58). The miR-193a-5p-DPEP1 axis appears to be an attractive therapeutic target in HB (Figure 4).
Signaling transduction of DPEP1 in gastrointestinal cancer
DPEP1 primarily transforms LTD4 into LTE4 (30). LTD4, as a proinflammatory mediator in the tumor microenvironment, can induce the inhibition of GSK-3β activity, which can target β-catenin to reduce the activity of β-catenin and the formation of the β-catenin-E-cadherin complex, thereby destroying cell-cell adhesion and promoting tumor cell metastasis (59,60). GSK-3β and β-catenin can mediate the Wnt/β-catenin signal transduction pathway (61), which is crucial for the malignant transformation of cells. When the activity of DPEP1 changes, the function of the Wnt/β-catenin signal pathway is also affected (60). DPEP can act both as an upstream factor and downstream target of the Wnt/β-catenin signaling pathway. For instance, achaete scute-like 2 (ASCL2), a helix-loop-helix transcription factor, can drive tumor cell processes, inhibit apoptosis, and even increase the resistance of cancer cells to chemotherapeutic drugs through Wnt/β-catenin signaling (62-64). In colon cancer, DPEP1 can enhance the stability of ASCL2 protein by inhibiting its ubiquitination; in turn, ASCL2 acts as a transcription factor to activate the transcription activity of the DPEP1 gene and promote its expression (55). Additionally, DPEP1 can enhance the expression of colon cancer cell stemness markers, such as LGR5, CD133, and CD44 (55). Overall, DPEP1 is able to form a positive feedback loop with ASCL2 to enhance the stemness of tumor cells, thereby improving the resistance to chemotherapeutic drugs, at least in colon cancer (Figure 4).
In the MEK/ERK pathway, the expression level of DPEP1 can be increased upon stimulation with EGF, and the sensitivity to gemcitabine is enhanced in pancreatic cancer (10). TGF-β is a transcriptional repressor of DPEP1 and can suppress its expression. A study has shown that in colorectal cancer cell lines, TGF-β can regulate the expression of E-cadherin through affecting DPEP1, promoting cell invasion, but such effects are not observed in cells lacking DPEP1 (30). However, its specific mechanism needs to be further illustrated.
DPEP1 is also involved in the metabolism of GSH (65), an antioxidant that maintains the redox state in the cellular microenvironment and protects cells from pathological stress (66). Decreased DPEP1 expression leads to GSH homeostasis disorder, which may promote the tumorigenesis related to the mechanism of GSH-induced ferroptosis.
DPEP1 and tumor-infiltrating immune
A recent spatial multi-omics profiling study has shed light on the co-evolution of sporadic colorectal tumors and their microenvironment, where DPEP1, as an extracellular matrix regulator, is implicated in immune exclusion reactions alongside other regulators such as DDR1, TGFBI, and PAK4. This immune exclusion signature (IEX) is associated with tumor progression in CIN+ (chromosomal instability positive) tumors, correlates with a reduction in cytotoxic cell infiltration, and exhibits prognostic value in an independent cohort (67). The study indicates that DPEP1 may influence the behavior of immune cells within the tumor microenvironment, including their infiltration, activation status, and cytotoxic capabilities towards tumor cells, thereby promoting tumor immune evasion. This further emphasizes the potential role of DPEP1 in the tumor immune microenvironment. The relationship between DPEP1 and tumor-infiltrating immune cells is primarily reflected in its potential involvement in modulating the function and state of immune cells within the tumor microenvironment, affecting the cytotoxic capabilities of immune cells against tumors. These findings provide potential targets and a theoretical foundation for future tumor immunotherapy strategies targeting DPEP1.
DPEP1 and prognosis of different malignant tumors
High expression of DPEP1 in colorectal cancer is associated with parameters such as pathological invasiveness and poor prognosis (68). As a membrane-bound metalloproteinase, DPEP1 may participate in the degradation of the extracellular matrix, cell invasion, and metastasis in colorectal cancer. Similarly, DPEP1 is significantly upregulated in HB and is associated with poor prognosis in HB patients (69). In contrast, in breast cancer, Wilms tumors, and pancreatic ductal adenocarcinoma, the expression of DPEP1 is reduced, and it is even considered a potential tumor suppressor, possibly being negatively correlated with the invasiveness of tumor cells. The above studies indicate that the expression patterns and functions of DPEP1 vary in multiple types of tumors, and it may play different roles in different types of cancer.
DPEP1 glycosylation in gastrointestinal cancer and nonneoplastic diseases
Protein glycosylation is one of the most common posttranslational modifications involved in various cellular activities, including migration, proliferation, adhesion, apoptosis, protein stability, and drug resistance (70,71). Protein glycosylation is closely related to inflammation, immune responses, and tumorigenesis (27,72,73). The protein structure of DPEP1 consists of four glycosylation sites, which are located on the outside of the spatial structure of DPEP1, which is conducive to the binding of other carbohydrates. It has been posited that two sites of DPEP1 may have carbohydrate chain connections, Ser57 and Ser332, but this has not been confirmed. In DPEP1, GPI is the only anchor point between DPEP1 and the cell membrane. Once a genetic mutation causes a defect in GPI, DPEP1 will not be able to attach to the cell membrane. This may increase the risk of paroxysmal nocturnal hemoglobinuria leukemia (29). Additionally, it has been reported that GPI anchoring is the principal effect of PI3K/AKT signaling pathway activation, which promotes gastrointestinal tumor proliferation, invasion, and metastasis (69) (Figure 4).
The relationship between DPEP1 and drug resistance in gastrointestinal cancer
Research has indicated that DPEP1 may be associated with drug resistance in colorectal cancer cells. DPEP1 can form a positive feedback loop with ASCL2, enhancing the chemoresistance of these cells, which is a significant factor affecting the efficacy of chemotherapy in colorectal cancer patients. Additionally, DPEP1 can promote the proliferation of colorectal cancer cells by forming a positive feedback loop with c-MYC, a mechanism that may amplify DPEP1 expression in colorectal cancer cells, thereby increasing their resistance to chemotherapeutic agents. Furthermore, the expression of DPEP1 in colorectal cancer is linked to various signaling pathways, including the PI3K/AKT/mTOR, MEK/ERK, and Wnt/β-catenin pathways, all of which are implicated in the development of tumor cell resistance. DPEP1 may also indirectly modulate the chemoresistance of tumor cells by participating in the behavior of immune cells within the tumor microenvironment. In summary, DPEP1 may be involved in the formation and regulation of drug resistance in colorectal cancer through diverse mechanisms, including the formation of positive feedback loops with transcription factors and the modulation of immune cell behavior. These findings provide a potential target and theoretical foundation for future therapeutic strategies targeting DPEP1 in cancer treatment.
Conclusions
DPEP1 is strongly associated with tumor metastasis, drug resistance, and poor prognosis, especially in gastrointestinal cancer (55). Targeting DPEP1 may reduce the adverse effects of anticancer drugs by inducing ferroptosis. As a DPEP1 inhibitor, cilastatin can inhibit DPEP1’s β-lactamase function and has been widely used in clinical practice (47). However, whether cilastatin’s inhibition of DPEP1 in malignant tumors is of value and whether it can inhibit the carcinogenic or treatment resistance caused by DPEP1 when combined with current therapeutic drugs remains to be explored. As a glycosylated protein, DPEP1 glycosylation may confer itself special functions, but the mechanism related to this effect has not been extensively examined, particularly in colorectal cancer. Therefore, further research into DPEP1 may contribute to the diagnosis, prognostic assessment, and therapy of gastrointestinal cancer.
Acknowledgments
Funding: None.
Footnote
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2436/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2436/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sahin TK, Ayasun R, Rizzo A, et al. Prognostic Value of Neutrophil-to-Eosinophil Ratio (NER) in Cancer: A Systematic Review and Meta-Analysis. Cancers (Basel) 2024;16:3689. [Crossref] [PubMed]
- Sahin TK, Rizzo A, Aksoy S, et al. Prognostic Significance of the Royal Marsden Hospital (RMH) Score in Patients with Cancer: A Systematic Review and Meta-Analysis. Cancers (Basel) 2024;16:1835. [Crossref] [PubMed]
- Guven DC, Erul E, Kaygusuz Y, et al. Immune checkpoint inhibitor-related hearing loss: a systematic review and analysis of individual patient data. Support Care Cancer 2023;31:624. [Crossref] [PubMed]
- Austruy E, Jeanpierre C, Antignac C, et al. Physical and genetic mapping of the dipeptidase gene DPEP1 to 16q24.3. Genomics 1993;15:684-7. [Crossref] [PubMed]
- Huber M, Keppler D. Inhibition of leukotriene D4 catabolism by D-penicillamine. Eur J Biochem 1987;167:73-9. [Crossref] [PubMed]
- Zhang X, Zhang Y, Gao X, et al. Integrated Single-Cell and Transcriptome Sequencing Analyses Identify Dipeptidase 2 as an Immune-Associated Prognostic Biomarker for Lung Adenocarcinoma. Pharmaceuticals (Basel) 2023;16:871. [Crossref] [PubMed]
- Habib GM, Shi ZZ, Cuevas AA, et al. Identification of two additional members of the membrane-bound dipeptidase family. FASEB J 2003;17:1313-5. [Crossref] [PubMed]
- Schiza C, Korbakis D, Panteleli E, et al. Discovery of a Human Testis-specific Protein Complex TEX101-DPEP3 and Selection of Its Disrupting Antibodies. Mol Cell Proteomics 2018;17:2480-95. [Crossref] [PubMed]
- Allison SJ. DPEP1 and CHMP1A in kidney ferroptosis. Nat Rev Nephrol 2021;17:707. [Crossref] [PubMed]
- Zhang G, Schetter A, He P, et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One 2012;7:e31507. [Crossref] [PubMed]
- Choudhury SR, Babes L, Rahn JJ, et al. Dipeptidase-1 Is an Adhesion Receptor for Neutrophil Recruitment in Lungs and Liver. Cell 2019;178:1205-1221.e17. [Crossref] [PubMed]
- Wang Y, Zhang T, Du H, et al. Dipeptidase 2 is a prognostic marker in lung adenocarcinoma that is correlated with its sensitivity to cisplatin. Oncol Rep 2023;50:161. [Crossref] [PubMed]
- Xie Y, Khan R, Wahab F, et al. The testis-specifically expressed Dpep3 is not essential for male fertility in mice. Gene 2019;711:143925. [Crossref] [PubMed]
- Hayashi K, Longenecker KL, Koenig P, et al. Structure of human DPEP3 in complex with the SC-003 antibody Fab fragment reveals basis for lack of dipeptidase activity. J Struct Biol 2020;211:107512. [Crossref] [PubMed]
- Yamaya M, Sekizawa K, Terajima M, et al. Dipeptidase inhibitor and epithelial removal potentiate leukotriene D4-induced human tracheal smooth muscle contraction. Respir Physiol 1998;111:101-9. [Crossref] [PubMed]
- Nakagawa H, Inazawa J, Inoue K, et al. Assignment of the human renal dipeptidase gene (DPEP1) to band q24 of chromosome 16. Cytogenet Cell Genet 1992;59:258-60. [Crossref] [PubMed]
- Yang X, Yue Y, Xiong S. Dpep2 Emerging as a Modulator of Macrophage Inflammation Confers Protection Against CVB3-Induced Viral Myocarditis. Front Cell Infect Microbiol 2019;9:57. [Crossref] [PubMed]
- Hamilton E, O'Malley DM, O'Cearbhaill R, et al. Tamrintamab pamozirine (SC-003) in patients with platinum-resistant/refractory ovarian cancer: Findings of a phase 1 study. Gynecol Oncol 2020;158:640-5. [Crossref] [PubMed]
- Daniel AG, Farrell NP. The dynamics of zinc sites in proteins: electronic basis for coordination sphere expansion at structural sites. Metallomics 2014;6:2230-41. [Crossref] [PubMed]
- Nitanai Y, Satow Y, Adachi H, et al. Crystal structure of human renal dipeptidase involved in beta-lactam hydrolysis. J Mol Biol 2002;321:177-84. [Crossref] [PubMed]
- Satoh S, Ohtsuka K, Keida Y, et al. Gene structural analysis and expression of human renal dipeptidase. Biotechnol Prog 1994;10:134-40. [Crossref] [PubMed]
- Mitselou A, Batistatou A, Nakanishi Y, et al. Comparison of the dysadherin and E-cadherin expression in primary lung cancer and metastatic sites. Histol Histopathol 2010;25:1257-67. [PubMed]
- Armstrong DJ, Mukhopadhyay SK, Campbell BJ. Physicochemical characterization of renal dipeptidase. Biochemistry 1974;13:1745-50. [Crossref] [PubMed]
- Adachi H, Katayama T, Nakazato H, et al. Importance of Glu-125 in the catalytic activity of human renal dipeptidase. Biochim Biophys Acta 1993;1163:42-8. [Crossref] [PubMed]
- Keynan S, Hooper NM, Turner AJ. Identification by site-directed mutagenesis of three essential histidine residues in membrane dipeptidase, a novel mammalian zinc peptidase. Biochem J 1997;326:47-51. [Crossref] [PubMed]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 1991;11:281-96. [Crossref] [PubMed]
- Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 2015;15:540-55. [Crossref] [PubMed]
- Lumibao JC, Tremblay JR, Hsu J, et al. Altered glycosylation in pancreatic cancer and beyond. J Exp Med 2022;219:e20211505. [Crossref] [PubMed]
- Kinoshita T. Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol 2020;10:190290. [Crossref] [PubMed]
- Park SY, Lee SJ, Cho HJ, et al. Dehydropeptidase 1 promotes metastasis through regulation of E-cadherin expression in colon cancer. Oncotarget 2016;7:9501-12. [Crossref] [PubMed]
- Peng D, Fu M, Wang M, et al. Targeting TGF-β signal transduction for fibrosis and cancer therapy. Mol Cancer 2022;21:104. [Crossref] [PubMed]
- Li K, Zhu Y, Cheng J, et al. A novel lipid metabolism gene signature for clear cell renal cell carcinoma using integrated bioinformatics analysis. Front Cell Dev Biol 2023;11:1078759. [Crossref] [PubMed]
- Liu Q, Deng J, Yang C, et al. DPEP1 promotes the proliferation of colon cancer cells via the DPEP1/MYC feedback loop regulation. Biochem Biophys Res Commun 2020;532:520-7. [Crossref] [PubMed]
- Li Y, Wu M, Xu S, et al. Colorectal cancer stem cell-derived exosomal long intergenic noncoding RNA 01315 (LINC01315) promotes proliferation, migration, and stemness of colorectal cancer cells. Bioengineered 2022;13:10827-42. [Crossref] [PubMed]
- Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol 2021;22:266-82. [Crossref] [PubMed]
- Guan Y, Liang X, Ma Z, et al. A single genetic locus controls both expression of DPEP1/CHMP1A and kidney disease development via ferroptosis. Nat Commun 2021;12:5078. [Crossref] [PubMed]
- von Mässenhausen A, Zamora Gonzalez N, Maremonti F, et al. Dexamethasone sensitizes to ferroptosis by glucocorticoid receptor-induced dipeptidase-1 expression and glutathione depletion. Sci Adv 2022;8:eabl8920. [Crossref] [PubMed]
- Zheng D, Liu J, Piao H, et al. ROS-triggered endothelial cell death mechanisms: Focus on pyroptosis, parthanatos, and ferroptosis. Front Immunol 2022;13:1039241. [Crossref] [PubMed]
- Zhang Y, Xin L, Xiang M, et al. The molecular mechanisms of ferroptosis and its role in cardiovascular disease. Biomed Pharmacother 2022;145:112423. [Crossref] [PubMed]
- Wang K, Chen XZ, Wang YH, et al. Emerging roles of ferroptosis in cardiovascular diseases. Cell Death Discov 2022;8:394. [Crossref] [PubMed]
- Friedmann Angeli JP, Schneider M, Proneth B, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 2014;16:1180-91. [Crossref] [PubMed]
- Yang F, Xiao Y, Ding JH, et al. Ferroptosis heterogeneity in triple-negative breast cancer reveals an innovative immunotherapy combination strategy. Cell Metab 2023;35:84-100.e8. [Crossref] [PubMed]
- Hong Y, Ren T, Wang X, et al. APR-246 triggers ferritinophagy and ferroptosis of diffuse large B-cell lymphoma cells with distinct TP53 mutations. Leukemia 2022;36:2269-80. [Crossref] [PubMed]
- Zhang C, Liu N. Ferroptosis, necroptosis, and pyroptosis in the occurrence and development of ovarian cancer. Front Immunol 2022;13:920059. [Crossref] [PubMed]
- Guo J, Xu B, Han Q, et al. Ferroptosis: A Novel Anti-tumor Action for Cisplatin. Cancer Res Treat 2018;50:445-60. [Crossref] [PubMed]
- Lau A, Rahn JJ, Chappellaz M, et al. Dipeptidase-1 governs renal inflammation during ischemia reperfusion injury. Sci Adv 2022;8:eabm0142. [Crossref] [PubMed]
- Zhang Z, Mei Y, Feng M, et al. The relationship between common variants in the DPEP1 gene and the susceptibility and clinical severity of osteoarthritis. Int J Rheum Dis 2021;24:1192-9. [Crossref] [PubMed]
- Miller RE, Lu Y, Tortorella MD, et al. Genetically Engineered Mouse Models Reveal the Importance of Proteases as Osteoarthritis Drug Targets. Curr Rheumatol Rep 2013;15:350. [Crossref] [PubMed]
- Shayan M, Elyasi S. Cilastatin as a protective agent against drug-induced nephrotoxicity: a literature review. Expert Opin Drug Saf 2020;19:999-1010. [Crossref] [PubMed]
- Nauseef WM, Borregaard N. Neutrophils at work. Nat Immunol 2014;15:602-11. [Crossref] [PubMed]
- Genga KR, Russell JA. Update of Sepsis in the Intensive Care Unit. J Innate Immun 2017;9:441-55. [Crossref] [PubMed]
- Wang Q, Wang Z, Zhang Z, et al. Landscape of cell heterogeneity and evolutionary trajectory in ulcerative colitis-associated colon cancer revealed by single-cell RNA sequencing. Chin J Cancer Res 2021;33:271-88. [Crossref] [PubMed]
- Zhang Y, Cai J, Lu W, et al. Comprehensive Network-Based Analyses Reveal Novel Renal Function-Related Targets in Acute Kidney Injury. Front Genet 2022;13:907145. [Crossref] [PubMed]
- Toiyama Y, Inoue Y, Yasuda H, et al. DPEP1, expressed in the early stages of colon carcinogenesis, affects cancer cell invasiveness. J Gastroenterol 2011;46:153-63. [Crossref] [PubMed]
- Zeng C, Qi G, Shen Y, et al. DPEP1 promotes drug resistance in colon cancer cells by forming a positive feedback loop with ASCL2. Cancer Med 2023;12:412-24. [Crossref] [PubMed]
- Green AR, Krivinskas S, Young P, et al. Loss of expression of chromosome 16q genes DPEP1 and CTCF in lobular carcinoma in situ of the breast. Breast Cancer Res Treat 2009;113:59-66. [Crossref] [PubMed]
- Llombart V, Mansour MR. Therapeutic targeting of "undruggable" MYC. EBioMedicine 2022;75:103756. [Crossref] [PubMed]
- Wang S, Diao YJ, Zhu BB. MiR-193a-5p suppresses cell proliferation and induces cell apoptosis by regulating HOXA7 in human ovarian cancer. Neoplasma 2020;67:825-33. [Crossref] [PubMed]
- Salim T, Sand-Dejmek J, Sjölander A. The inflammatory mediator leukotriene D4 induces subcellular β-catenin translocation and migration of colon cancer cells. Exp Cell Res 2014;321:255-66. [Crossref] [PubMed]
- Wu YF, Wang CY, Tang WC, et al. Expression Profile and Prognostic Value of Wnt Signaling Pathway Molecules in Colorectal Cancer. Biomedicines 2021;9:1331. [Crossref] [PubMed]
- Liu J, Xiao Q, Xiao J, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther 2022;7:3. [Crossref] [PubMed]
- Stange DE, Engel F, Longerich T, et al. Expression of an ASCL2 related stem cell signature and IGF2 in colorectal cancer liver metastases with 11p15.5 gain. Gut 2010;59:1236-44. [Crossref] [PubMed]
- Orouji E, Raman AT, Singh AK, et al. Chromatin state dynamics confers specific therapeutic strategies in enhancer subtypes of colorectal cancer. Gut 2022;71:938-49. [Crossref] [PubMed]
- Jubb AM, Chalasani S, Frantz GD, et al. Achaete-scute like 2 (ascl2) is a target of Wnt signalling and is upregulated in intestinal neoplasia. Oncogene 2006;25:3445-57. [Crossref] [PubMed]
- Song D, Lin Z, Yuan Y, et al. DPEP1 Balance GSH Involve in Cadmium Stress Response in Blood Clam Tegillarca granosa. Front Physiol 2018;9:964. [Crossref] [PubMed]
- Moine L, Rivoira M, Díaz de Barboza G, et al. Glutathione depleting drugs, antioxidants and intestinal calcium absorption. World J Gastroenterol 2018;24:4979-88. [Crossref] [PubMed]
- Heiser CN, Simmons AJ, Revetta F, et al. Molecular cartography uncovers evolutionary and microenvironmental dynamics in sporadic colorectal tumors. Cell 2023;186:5620-5637.e16. [Crossref] [PubMed]
- Eisenach PA, Soeth E, Röder C, et al. Dipeptidase 1 (DPEP1) is a marker for the transition from low-grade to high-grade intraepithelial neoplasia and an adverse prognostic factor in colorectal cancer. Br J Cancer 2013;109:694-703. [Crossref] [PubMed]
- Cui X, Liu X, Han Q, et al. DPEP1 is a direct target of miR-193a-5p and promotes hepatoblastoma progression by PI3K/Akt/mTOR pathway. Cell Death Dis 2019;10:701. [Crossref] [PubMed]
- Tang L. Detecting protein glycosylation. Nat Methods 2019;16:26. [Crossref] [PubMed]
- Xu C, Zhang M, Bian L, et al. N-glycosylated SGK196 suppresses the metastasis of basal-like breast cancer cells. Oncogenesis 2020;9:4. [Crossref] [PubMed]
- Radovani B, Gudelj I. N-Glycosylation and Inflammation; the Not-So-Sweet Relation. Front Immunol 2022;13:893365. [Crossref] [PubMed]
- Polmear J, Good-Jacobson KL. Antibody glycosylation directs innate and adaptive immune collaboration. Curr Opin Immunol 2022;74:125-32. [Crossref] [PubMed]



