
CDKN2A as a prognostic and diagnostic marker is a possible key contributor in hepatocellular carcinoma
Highlight box
Key findings
• Cyclin-dependent kinase inhibitor 2A (CDKN2A) has emerged as a promising prognostic and diagnostic marker for hepatocellular carcinoma (HCC) and may play crucial roles in HCC development and metastasis by regulating the tumor cell cycle.
What is known and what is new?
• Despite ongoing efforts, the incidence and mortality rates of HCC continue to rise annually. A primary contributor to this trend is the lack of effective early diagnostic markers and therapeutic targets.
• This study suggests that CDKN2A may serve as a novel and promising prognostic and diagnostic marker, as well as an important therapeutic target in HCC.
What is the implication, and what should change now?
• This study offers a promising potential intervention target for the (early) diagnosis and effective treatment of HCC patients. However, some of the current findings require large-scale validation using expanded experimental samples. Additionally, the proposed mechanisms, which have been predicted using computer algorithms, will need to be confirmed through a series of subsequent functional experiments.
Introduction
Liver cancer mainly includes hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (ICC), of which HCC accounts for 85–90% (1). Early diagnosis of HCC is difficult, and most patients with HCC (about 90%) are already in the middle and late stages when diagnosed, resulting in the survival of HCC patients being generally less than 6 months and the survival rate of patients with more than 5 years being less than 10% (2). Over the years, some breakthroughs have been made in the treatment of HCC, such as surgery, liver transplantation, targeting and immunotherapy, which have improved the treatment effect of HCC and extended the survival time of patients. However, the incidence and mortality of HCC are not decreased, but still increased year by year. The main cause for this issue is the lack of early diagnostic markers and effective therapeutic targets. Therefore, there is an urgent need to identify new molecular markers for early diagnosis, pinpoint key regulatory factors, and elucidate the specific molecular mechanisms underlying HCC pathogenesis. Developing effective diagnostic and treatment methods based on these findings is crucial for the prevention and ultimate cure of HCC. This approach represents the most promising strategy to comprehensively address and combat the disease. In general, the molecules with potential to serve as diagnostics and key players in HCC are bound to undergo the most pronounced changes. Following extensive screening and analysis using big data genomics and comprehensive literature reviews, cyclin-dependent kinase inhibitor 2A (CDKN2A) has emerged as one of the most aberrantly expressed yet under-investigated molecules in HCC. Human CDKN2A is localized on chromosome 9 (9p21.3), and has been reported to be aberrantly expressed in a variety of tumors, including lung, oesophageal, gastric, colorectal, ovarian, leukaemia and renal cancers (3-6). Previous study has revealed that CDKN2A plays a key role in the cell cycle as an oncogene and prognostic marker in acute lymphoblastic leukemia (4). CDKN2A specifically binds to cyclin-dependent kinase 4/6 (CDK4/6), inhibiting the formation of the CDK4/6-Cyclin D complex. This leads to the suppression of the Rb/E2F transcriptional complex activity, thereby inducing G1 phase cell cycle arrest (3). Recent studies have identified CDKN2A as an important tumor suppressor in cervical and lung cancers (5,6). These findings suggest that CDKN2A may act an important role and has a potential diagnostic value in HCC. But the expression regulation, clinical values, main function, and possible mechanism of CDKN2A in HCC, are still largely unclear.
In this study, we found that CDKN2A may be an important candidate for (early) clinical diagnosis of HCC and may play key roles in the process of HCC development and metastasis. We present this article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1845/rc).
Methods
Patient samples
Tissue microarray (TMA) containing cancerous and paracancerous tissues from 89 HCC patients who underwent surgical resection between 2007 and 2008 were purchased from Shanghai Outdo Biotech Co., Ltd. (Shanghai, China). The clinicopathological information was retrieved from the patient’s electronic medical record, including age, gender, lymph nodes (negative or positive), tumor size, histological grade and clinical stage defined according to the 7th edition of the American Joint Committee on Cancer (AJCC) and the new tumor node metastasis (TNM) classification, as well as follow-up information (5–8 years) for overall survival (OS) rate and disease-free survival (DFS) rate. The patients were followed for a maximum of 104 months and a median follow-up of 49 months. OS was defined as the time from surgery to death or last observation. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Shanghai Outdo Biotech Company (No. YB M-05-02) and the Ethics Committee of Third Affiliated Hospital of Chongqing Medical University (CQMU, No. 2021-95). All patients signed an informed consent form.
TMA generation
The samples of 89 HCC patients were histologically examined using hematoxylin and eosin staining. To construct TMA sections, two cores were taken from each representative tumor and adjacent non-cancerous tissue (within distance of 20 mm). Adjacent non-cancerous tissue was compared with normal tissue, stained with hematoxylin-eosin and examined histologically by at least two pathologists.
Immunohistochemical (IHC) analysis
IHC analysis was performed using CDKN2A antibody (specifically recognizes p16/INK4A, GB111143-100, Servicebio, Wuhan, China). IHC staining results were assessed, evaluated, and quantified under a microscope by two pathologists. The percentage of positive cells was calculated based on the ratio of positive cells to the total number of cells. Staining was considered positive when the percentage of tumor cells staining was ≥10%. The percentage of positive cells was scored into five categories: 0 (<10%), 1 (10–25%), 2 (26–50%), 3 (51–75%), and 4 (>75%). Staining intensity was scored in four categories: 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). Protein expression levels were determined by multiplying the scores of positive cell percentage and staining intensity (the range of CDKN2A protein expression: 0–12). Samples were then grouped into high and low expression categories based on the average value.
Analysis of available datasets
Screening for important changed molecules in HCC using the Gene Expression Profiling Interactive Analysis (GEPIA) 2.0 website
Whole transcriptome sequencing data from tumours and normal tissues in the HCC dataset (|Log2 fold change| ≥1, P≤0.01) were analyzed via the GEPIA 2.0 (http://gepia2.cancer-pku.cn/) website to identify molecules that underwent significant changes. The GEPIA 2.0 website was further used to analyze the differences of the top 10 expression changed molecules, and determine the correlations of the changed molecules with clinical stage, OS, and DFS (using median grouping). TIMER2.0 database (http://timer.cistrome.org/) and Oncomine database (https://www.oncomine.org) were used to further confirm CDKN2A expression in HCC.
HCC clinical sample data sources and processing
The University of California Santa Cruz (UCSC) Xena database (http://xena.ucsc.edu/) was used to download the CDKN2A gene expression matrix, clinical correlation data, gene promoter region methylation data, and copy number variant data from The Cancer Genome Atlas (TCGA) cohort. Integration of clinical and pathological information via the VLOOKUP function identified 374 HCC tissue samples and 50 para-cancerous tissues for which expression and clinical parameter data were available. These data were used for differential expression analysis of CDKN2A, correlation analysis of clinicopathological indicators, receiver operating characteristic (ROC) curve analysis and expression regulation analysis. The high and low expression groups of CDKN2A were classified according to the median expression.
Analysis of the correlation between CDKN2A expression and clinicopathological indicators of HCC
The downloaded CDKN2A expression matrix data were matched with clinical data using VLOOKUP function. Patients were grouped using clinical parameters to further compare the differences in CDKN2A expression among different patients, and the results were visualised using Graphpad 8.0, and the P value was calculated using the Mann-Whitney test. The P values less than 0.05 were considered statistically significant.
Kaplan-Meier survival analysis
The Kaplan-Meier plotter (https://kmplot.com/analysis/) database was used to analyze the prognostic value of CDKN2A in HCC patients, and the same analysis was performed for the patients with different clinical stages. The boundaries of high and low expression of CDKN2A were determined using the median of expression, and the P value was calculated using the Logrank test. A P value that less than 0.05 considered to be statistically significant.
ROC curve analysis
The above data were analyzed by ROC curve using Graphpad 8.0 software, with the horizontal X-axis as specificity (also known as false positive rate) and the vertical Y-axis as sensitivity (also known as true positive rate), and the area under the ROC curve was calculated to assess the diagnostic value of CDKN2A expression for HCC.
Analysis of copy number variation (CNV) for CDKN2A gene
The UCSC Xena database was used to download the CNV data of the CDKN2A gene from the TCGA cohort of HCC patients, and the VLOOKUP function was used to match the CNV and clinical parameter data of 364 HCC tissue samples. CNV was determined by the copy number threshold, with −1 or −2 defined as a copy number deletion (del), 0 as a copy number normal [wild type (wt)], and 1 or 2 as a copy number amplification (amp). The distribution of copy number variants in the patients with different clinical parameters was assessed by chi-square test. The Mann-Whitney test was used for analysis of variance (ANOVA) and P values were calculated. All results were visualised using Graphpad 8.0, and P values less than 0.05 were considered statistically significant.
Methylation analysis of the promoter region of CDKN2A gene
The UCSC Xena website was used to download CDKN2A methylation data from the TCGA cohort of HCC patients. Gene methylation β-value and gene expression of 370 HCC samples were obtained after matching from UCSC Xena website. The promoter region of CDKN2A gene (the promoter region is generally located 2,000 bp before the transcription start site) was located through the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/), and the promoter methylation β-value was obtained by calculating the average of methylation β-values of all probes located at the promoter region. The median was used for the boundaries of the high and low methylation groups, the differences were analyzed using the Mann-Whitney test, and all results were visualised using Graphpad 8.0. The P values less than 0.05 were considered statistically significant.
Correlation analyses of CDKN2A expression with tumour microenvironment
The stromal score, immune score and ESTIMATE score in HCC were calculated by the ESTIMATE algorithm and used to predict the infiltration of non-tumour cells. To analyze whether the above three metrics differed in the patients with different CDKN2A expression levels, the data were obtained from the ESTIMATE data (https://bioinformatics.mdanderson.org/estimate/). The TIMER2.0 database provides six types of immune cell infiltration (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells), which was used to further validate the correlation between CDKN2A expression and immune cell infiltration.
CDKN2A-related gene enrichment analysis
Genes co-expressed with CDKN2A in HCC were obtained from the LinkedOmics database (http://www.linkedomics.org/) and entered into the DAVID database for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichments and functional annotation.
CDKN2A interaction protein network and enrichment analysis
STRING (https://cn.string-db.org/) and GeneMANIA (http://genemania.org/) were used to construct the CDKN2A protein-protein interaction (PPI) network, and the interacting proteins were subjected to GO and KEGG enrichments and functional annotation.
Statistical analysis
Statistical analyses were performed using SPSS 26.0 software (SPSS, Inc., Chicago, IL, USA). Spearman and Pearson tests were used for correlation analysis, t-test was selected for comparison between two groups, and one-way ANOVA test was used for comparison between multiple groups. The OS and DFS were calculated using the Kaplan-Meier method and evaluated using the log-rank test. CDKN2A expression was classified as high and low groups using the median score. The P values less than 0.05 were considered statistically significant difference.
Results
The screening for candidate molecules of diagnosis and key participant in HCC
By analyzing the transcriptome sequencing data of HCC and normal tissues, the expression levels of 2,207 genes were significantly changed (|Log2 fold change| ≥1, P≤0.01) in HCC compared with normal tissues. Among these altered genes, 1,482 were up-regulated and 725 were down-regulated (Figure 1A). The distribution of these altered genes on chromosomes revealed that the up-regulated genes were preferentially located on chromosomes 1, 6, 11, 17, 19 and 20, while the down-regulated genes were preferentially located on chromosomes 1, 11, 16 and 19 (Figure 1A). The screened 2,207 molecules were subjected to GO and KEGG enrichment analyses. The analyzed GO processes were sorted according to the number of aggregated molecules, and the top 20 GO processes were displayed (Figure S1). These altered genes mainly involved in ribosome structure and protein localization from the top 20 GO processes (Figure S1). The KEGG enrichments were also analyzed, and the top 10 positive and negative signaling pathways were displayed (Figure S1). The positive genes primarily involved in disease-related pathways, and the negative genes majorly involved in metabolic pathways from the top 10 signaling pathways (Figure S1).

To search for the candidate molecules of diagnosis and key participant in HCC, the most altered molecules (top 20) were selected and were then confirmed in independent HCC samples. The most significantly altered candidates among the expression-changed genes included RP11-40C6.2, glypican 3 (GPC3), aldo-keto reductase family 1 member B10 (AKR1B10), ubiquitin D (UBD), midkine (MDK), phosphatidylinositol glycan anchor biosynthesis class Y (PIGY), C-C motif chemokine ligand 20 (CCL20), CDKN2A, ubiquitin conjugating enzyme E2 C (UBE2C), and cyclin B1 (CCNB1) (Figure 1B). The results demonstrate that RP11-40C6.2, GPC3, AKR1B10, UBD, MDK, PIGY, CCL20, CDKN2A, UBE2C and CCNB1 are likely the candidates of diagnosis and key participant in HCC.
The clinical relevance of the candidates with patient’s stage and prognosis
The expression differences of these candidates (RP11-40C6.2, GPC3, AKR1B10, UBD, MDK, PIGY, CCL20, CDKN2A, UBE2C, and CCNB1) were then analyzed in the HCC patients at different clinical stages. The expression of CDKN2A, UBE2C, and CCNB1 was closely associated with clinical stages, whereas the expression of other candidates was not correlated with clinical stages of HCC patients (Figure 2A; Figure S2). For prognostic values of these candidates, the expression of CDKN2A and CCNB1 was strongly associated with the OS and DFS of HCC patients, the expression of AKR1B10 was associated with OS of HCC patients, and the expression of UBE2C was only associated with DFS of HCC patients (Figure 2B). However, the expression of other candidates was not associated with OS and DFS of HCC patients (Figure S3). These data suggest that CDKN2A and CCNB1 probably acts important roles in diagnosis, prognosis, and metastasis of HCC. After a literature searching, the roles and potential clinical significance of CCNB1 had been well studied in many types of tumors, including HCC. However, CDKN2A has been less studied in cancers specially in HCC and deserves further in-depth research.

The validation of CDKN2A at different expression levels in HCC
To further demonstrate the expression of CDKN2A in cancers, a pan-cancer analysis was performed using multiple datasets from different databases. The transcriptional expression level of CDKN2A were clearly up-regulated in the most tumor types especially HCC (Figure 3A,3B). To further determine the changed expression of CDKN2A at protein level in HCC, the IHC using CDKN2A antibody was performed in HCC tissues on TMA. The protein expression of CDKN2A was significantly higher in HCC tissues than that in adjacent normal tissues (Figure 3C). In addition, CDKN2A protein appeared to be primarily expressed at membrane, cytoplasm and nucleus of tumor cells (Figure 3C).

The associations of CDKN2A with clinical indexes and survival prognosis
To further explore the relationship between CDKN2A expression and clinical data, as well as the prognostic value of HCC, we analyzed the associations of CDKN2A expression with clinical indexes and survival prognosis in HCC using the TCGA data downloaded from UCSC database. The up-regulation of CDKN2A mRNA was confirmed in HCC compared to normal tissues (P<0.001, Figure 4A). To assess the clinical significance of this changes in CDKN2A expression, we used the VLOOKUP function to match CDKN2A expression with clinical data of HCC patients, and compared the expression levels of CDKN2A mRNA in patients with different clinical indicators. The expression of CDKN2A mRNA was closely associated with tumor sizes (T) and clinical stages of HCC patients (P<0.001, Figure 4A). These results suggest that CDKN2A may be an important molecule in promoting HCC growth and progression. To analyze the prognostic value of CDKN2A expression changes in HCC, survival analysis was performed using Kaplan-Meier Plotter database. The HCC patients with high CDKN2A expression had a poorer prognosis compared to those with low CDKN2A expression (Logrank P=0.005, Figure 4B). Further analysis revealed that high CDKN2A expression was associated with poorer prognosis of the patients at stage II (Logrank P=0.045, Figure 4C), whereas CDKN2A expression was not associated with prognosis of the patients at stage I (Logrank P=0.052) and III (Logrank P=0.21, Figure 4C). The results suggest that CDKN2A may be a prognostic marker for HCC patients.
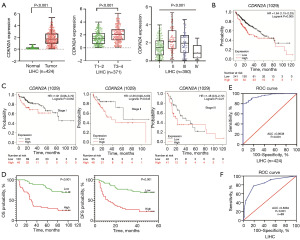
To further investigate the clinical value of CDKN2A expression at protein level in HCC, IHC staining of CDKN2A antibody was performed on TMAs containing 89 HCC cases. The expression score was the product of staining intensity and positivity of CDKN2A, and the patients were divided into two groups based on the average of the score: low expression group (score ≤4) and high expression group (4< score ≤12). Analyses of the relationship between CDKN2A protein expression and the prognosis of HCC patients showed that the OS of the patients with high CDKN2A protein expression was significantly shorter than that of the patients with low CDKN2A protein expression, and the DFS of the patients with high CDKN2A expression was also clearly shorter than that of the patients with low expression (P<0.001, Figure 4D).
The diagnostic value of altered CDKN2A expression in HCC
To evaluate the diagnostic value of CDKN2A expression in HCC, ROC curve analysis was performed using CDKN2A expression and patient data of HCC from the TCGA database. The area under the curve (AUC) of CDKN2A in HCC was 0.9538 [95% confidence interval (CI): 0.9332–0.9744; P<0.001, Figure 4E], suggesting that the altered CDKN2A expression possess a good diagnostic value. To further confirm the diagnostic value of CDKN2A, the changed protein expression was analyzed to determine the diagnostic significance in the HCC cases. ROC curve analysis showed that the AUC of CDKN2A was 0.8384 (95% CI: 0.7793–0.8975; P<0.001, Figure 4F). These data indicate that the altered CDKN2A expression is important for disease diagnosis of HCC both at mRNA and protein levels, suggesting that CDKN2A may be a promising marker for diagnosis of HCC.
CDKN2A promoter methylation is associated with its expression with clear diagnostic value
To further explore the cause of CDKN2A changed expression in HCC, we analyzed the associations of CDKN2A expression with its CNV and DNA methylation using the HCC data from public TCGA database. The deletions (del) of CDKN2A predominantly existed in HCC patients accounting for 40.93%, whereas there was no significant difference of deletions and amplifications (amp) with different tumor sizes (P=0.26, Figure 5A) and clinical stages (P=0.06, Figure 5A) in the HCC patients. The relationship between CDKN2A expression and its CNV was next analyzed in HCC. CDKN2A expression was lower in the patients with CDKN2A deletions (P=0.009, Figure 5B). Meanwhile, the methylation β-value of CDKN2A was positively correlated with its expression, and the methylation degree of CDKN2A promoter region was significantly higher in HCC than normal tissues (P<0.001, Figure 5B). ROC analysis showed that the AUC of the methylation β value of CDKN2A promoter region was 0.9538 (95% CI: 0.9342–0.9734; P<0.001) in HCC (Figure 5C), suggesting the methylation of CDKN2A promoter as a valuable diagnostic marker for HCC. These results demonstrate that the up-regulation of CDKN2A expression in HCC may contribute to the methylation of its promoter region, and this methylation pattern has a great diagnostic value for HCC.

CDKN2A expression is associated with tumor microenvironment
The stromal score, immune score, and ESTIMATE score were calculated using the ESTIMATE algorithm. These scores were then analyzed to determine whether they differ in patients with varying levels of CDKN2A expression. The HCC patients with high CDKN2A expression had lower stromal scores compared to the patients with low CDKN2A expression (P<0.001), whereas there was no significant difference in immune score and ESTIMATE score between the patients with high CDKN2A expression and that with low CDKN2A expression (Figure 6A). Immune cells are non-tumor components that play important roles in the body’s fight against tumors. To analyze the correlation of CDKN2A expression with immune cell infiltration, the immune cell infiltration data and expression data of six types of immune cells from the TIMER database were utilized. The CDKN2A expression levels were positively correlated with B cells, CD8+ T cells, neutrophils, macrophages and dendritic cell (Figure 6B). These data reveal that CDKN2A expression is closely associated with tumor microenvironment in HCC.

CDKN2A likely involves in HCC progression by major regulating cell cycle
To determine the major potential function and the possible mechanism of CDKN2A in HCC, the co-expressed genes of CDKN2A in HCC were downloaded from the LinkedOmics database. There are 3,946 genes clearly associated with CDKN2A expression in HCC [false discovery rate (FDR) <0.01], of which 2,259 were positively associated genes and 1,687 were negatively associated genes (FDR <0.01, Figure 7A). Enrichment analyses were conducted on the top 500 genes most positively and negatively associated with CDKN2A in HCC. GO enrichment analysis showed that the total 1,000 co-expressed genes of CDKN2A in HCC were mainly involved in the biological processes (BPs) such as cell division, mitotic spindle organization, DNA repair, and DNA replication; cellular components (CCs) such as nucleoplasm, nucleus, cytosol, and spindle; molecular functions (MFs) such as protein binding, adenosine triphosphate (ATP) binding, microtubule binding, and DNA helicase activity (Figure 7B, Figure S4A). The top 500 positively co-expressed genes were primarily involved in the BPs such as cell division, DNA repair, DNA replication, and cell cycle; CCs such as nucleoplasm, nucleus, kinetochore, and cytosol; MFs such as protein binding, DNA binding, DNA helicase activity, and microtubule binding (Figure 7B, Figure S4B). The top 500 negatively co-expressed genes were mainly involved in the BPs such as cell adhesion, actin cytoskeleton organization, vasculature development, and vasculogenesis; CCs such as extracellular matrix, mitochondrial matrix, mitochondrion, and endosome; MFs such as extracellular matrix structural constituent, calcium ion binding, extracellular matrix constituent conferring elasticity, and growth factor binding (Figure 7B, Figure S4C). KEGG enrichment analysis showed that the total 1,000 associated genes of CDKN2A were primarily involved in the signaling pathways such as cell cycle, DNA replication, Fanconi anemia pathway, and cellular senescence; the top 500 positively co-expressed genes were mainly involved in the signaling pathways such as cell cycle, DNA replication, Fanconi anemia pathway, and homologous recombination; the top 500 negatively co-expressed genes were primarily involved in the signaling pathways such as citrate cycle, valine/leucine/isoleucine degradation, and proximal tubule bicarbonate reclamation in HCC (Figure 7B).
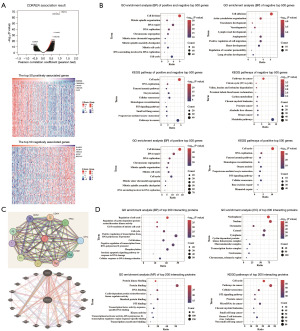
Next, PPI network and interacting proteins of CDKN2A were analyzed using STRING and Gene MINIA databases. The PPI network results showed that the main proteins interacting with CDKN2A were regulatory proteins of cell cycle, such as CDK2, CDK4, CDK6, TP53, CCND1, CCND2 and MDM2 (Figure 7C). A medium confidence level of 0.400 was used as the minimum required interaction score, and a total of 200 interacting proteins of CDKN2A were selected for enrichment analysis using the STRING database. The GO enrichment analysis of these interacting proteins showed that CDKN2A interacting proteins were involved in the BPs such as cell cycle, cyclin-dependent protein serine/threonine kinase activity, G1/S transition of mitotic cell cycle, and cell cycle; CCs such as nucleoplasm, nucleus, chromatin, and cytosol; MFs such as protein kinase binding, protein binding, DNA binding, and cyclin-dependent protein serine/threonine kinase regulator activity (Figure 7D). KEGG enrichment analysis of these interacting proteins revealed that CDKN2A interacting proteins were involved in cell cycle, cancer-related pathways, cellular senescence, and P53 signaling pathway (Figure 7D). These findings suggest that CDKN2A may play a significant role in promoting the development of HCC.
Discussion
Previous studies have shown that CDKN2A is aberrantly expressed in a variety of tumor types including lung, oesophageal, gastric, colorectal, ovarian, leukaemia and renal cancers (3-6). However, little is known about its clinical relevance and value in these cancers. In this study, we comprehensively investigate the clinical relevance and significance of CDKN2A in HCC. Our findings indicate that CDKN2A expression is generally elevated in HCC and exhibits a strong correlation with tumor size and clinical stage in patients. The patients with low CDKN2A expression had correspondingly prolonged OS and DFS compared to the patients with high CDKN2A expression. Moreover, ROC curve analysis shows a good diagnostic value of CDKN2A expression specially the methylation of CDKN2A promoter for HCC. These data suggest that CDKN2A is a promising biomarker for HCC prognosis and diagnosis.
Previous evidence suggests that CDKN2A is a susceptibility gene for melanoma and pancreatic cancer (7). Recent studies have identified CDKN2A as an important tumor suppressor in cervical and lung cancers (5,6). However, there are few studies on CDKN2A in HCC. In the present study, CDKN2A expression is significantly elevated and closely associated with tumor size and clinical stage of HCC patients, suggesting CDKN2A likely an important participant in HCC growth and progression. The GO enrichments of co-expressed genes and interacting proteins of CDKN2A in HCC reveal that CDKN2A may act crucial roles on regulation of tumor cell cycle, which further confirm the important roles of CDKN2A in HCC growth and progression.
Few studies have demonstrated the mechanisms of CDKN2A expression regulation and tumor progression. A recent study has revealed that CDKN2A inhibits the proliferation and invasion of cervical cancer cells through the LDHA-mediated AKT-mTOR pathway (5). In the present study, KEGG enrichments of co-expressed genes and interacting proteins of CDKN2A indicate that CDKN2A likely acts important roles in HCC by regulation of the signaling pathways related to cell cycle, cellular senescence and apoptosis (e.g., P53 signaling pathway). These data suggest that CDKN2A probably participates in HCC progression by primarily regulating tumor cell cycle. Moreover, CDKN2A as a cyclin-dependent kinase inhibitor should play key roles in cell cycle regulation. We noted that the cell cycle, cellular senescence, and P53 signaling pathway were enriched in the interacting proteins with CDKN2A. We speculate that CDKN2A may promote HCC progression by regulating the P53 signaling pathway. However, further experiments are required to confirm this speculation, and more studies are needed to understand how CDKN2A regulates P53, which induces changes in downstream molecules.
The immune cells are non-tumor cells in tumor microenvironment. The relationship between immune cells and tumor development is well established. Immune cells are able to sense various signals in the microenvironment, and initiate specific immune functions in response. There is growing evidence that the immune response is associated with dramatic changes in tissue metabolism, including nutrient depletion, increased oxygen consumption, and the production of reactive nitrogen and oxygen intermediates (8-10). Similarly, many metabolites in the tumor microenvironment can in turn affect immune cell differentiation and effector functions (11,12). Emerging evidences suggest that cancer cells are able to suppress anti-tumor immune responses by competing for and consuming essential nutrients or otherwise reducing the metabolic capacity of tumour-infiltrating immune cells (13,14). In this study, we analyzed the correlation between CDKN2A and immune cell infiltration, and found that in HCC, CDKN2A expression was closely correlated with the infiltration of various immune cells. These data suggest that CDKN2A may play a role in HCC by altering the tumour microenvironment. Further studies are required to address this issue.
CDKN2A expression is clearly elevated in HCC, to clarify the possible regulatory mechanism of this elevated expression, we analyzed the contributions of CNV and methylation of CDKN2A gene to its expression. The association analysis shows that CDKN2A expression in HCC may be mainly regulated by the methylation of its promoter region. Moreover, the methylation of CDKN2A promoter region possesses a good diagnostic value for HCC, suggesting that detecting CDKN2A methylation can be used as an important mean for diagnosis of HCC.
Conclusions
CDKN2A has been identified as a promising prognostic and diagnostic candidate for HCC, and may have crucial roles on the process of HCC development and metastasis by regulating tumor cell cycle, providing an important therapeutic target in HCC. However, there are shortcomings in this study. Some of the current results need to be validated on a large scale in our experimental sample. The possible mechanisms are predicted by computer algorithms and will require a series of subsequent functional experiments for further validation.
Acknowledgments
We would like to thank the patients involved in this study for their support, and the public availability of the various databases.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1845/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1845/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1845/prf
Funding: This work was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1845/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Shanghai Outdo Biotech Company (No. YB M-05-02) and the Ethics Committee of Third Affiliated Hospital of Chongqing Medical University (CQMU, No. 2021-95). All patients signed an informed consent form.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. [Crossref] [PubMed]
- Vogel A, Meyer T, Sapisochin G, et al. Hepatocellular carcinoma. Lancet 2022;400:1345-62. [Crossref] [PubMed]
- Zhao R, Choi BY, Lee MH, et al. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer. EBioMedicine 2016;8:30-9. [Crossref] [PubMed]
- Agarwal M, Bakhshi S, Dwivedi SN, et al. Cyclin dependent kinase inhibitor 2A/B gene deletions are markers of poor prognosis in Indian children with acute lymphoblastic leukemia. Pediatr Blood Cancer 2018;65:e27001. [Crossref] [PubMed]
- Luan Y, Zhang W, Xie J, et al. CDKN2A inhibits cell proliferation and invasion in cervical cancer through LDHA-mediated AKT/mTOR pathway. Clin Transl Oncol 2021;23:222-8. [Crossref] [PubMed]
- Liu W, Zhuang C, Huang T, et al. Loss of CDKN2A at chromosome 9 has a poor clinical prognosis and promotes lung cancer progression. Mol Genet Genomic Med 2020;8:e1521. [Crossref] [PubMed]
- Chan SH, Chiang J, Ngeow J. CDKN2A germline alterations and the relevance of genotype-phenotype associations in cancer predisposition. Hered Cancer Clin Pract 2021;19:21. [Crossref] [PubMed]
- Terry S, Engelsen AST, Buart S, et al. Hypoxia-driven intratumor heterogeneity and immune evasion. Cancer Lett 2020;492:1-10. [Crossref] [PubMed]
- Huang B, Song BL, Xu C. Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat Metab 2020;2:132-41. [Crossref] [PubMed]
- Chen B, Gao A, Tu B, et al. Metabolic modulation via mTOR pathway and anti-angiogenesis remodels tumor microenvironment using PD-L1-targeting codelivery. Biomaterials 2020;255:120187. [Crossref] [PubMed]
- Karayama M, Masuda J, Mori K, et al. Comprehensive assessment of multiple tryptophan metabolites as potential biomarkers for immune checkpoint inhibitors in patients with non-small cell lung cancer. Clin Transl Oncol 2021;23:418-23. [Crossref] [PubMed]
- Yan Y, Chang L, Tian H, et al. 1-Pyrroline-5-carboxylate released by prostate Cancer cell inhibit T cell proliferation and function by targeting SHP1/cytochrome c oxidoreductase/ROS Axis. J Immunother Cancer 2018;6:148. [Crossref] [PubMed]
- Hurley HJ, Dewald H, Rothkopf ZS, et al. Frontline Science: AMPK regulates metabolic reprogramming necessary for interferon production in human plasmacytoid dendritic cells. J Leukoc Biol 2021;109:299-308. [Crossref] [PubMed]
- Guerra L, Bonetti L, Brenner D. Metabolic Modulation of Immunity: A New Concept in Cancer Immunotherapy. Cell Rep 2020;32:107848. [Crossref] [PubMed]


