
Development and validation of predictive nomograms for survival in early-onset colon cancer patients with II–III stage across various tumor sites
Highlight box
Key findings
• The developed nomograms are capable to predict overall survival (OS) and cancer-specific survival (CSS) of early-onset colon cancer (EOCC) patients. The right-sided groups derived greater benefits from adjuvant chemotherapy compared to the left-sided groups, whereas radiation therapy provided no discernible benefits across all subgroups.
What is known and what is new?
• The incidence of EOCC has been escalating with few relative prognostic studies.
• Based on a large population-level data, we developed nomograms to predict OS and CSS after dividing patients into two risk groups. Then we assessed the potential benefits of various therapies across subgroups after propensity score matching.
What is the implication, and what should change now?
• Identifying high-risk EOCC patients and providing them with targeted medical interventions can potentially improve their prognosis.
Introduction
Colorectal cancer (CRC) is currently the third most prevalent malignancy and the second leading cause of cancer-related mortality worldwide (1). Fortunately, the implementation of colonoscopy screening and therapeutic advancements have contributed to a reduction in both the incidence and mortality rates of CRC in recent years (2). Nonetheless, there is a troubling upward trend in CRC cases among individuals under 50, underscoring the rise of early-onset colon cancer (EOCC) as a significant factor in this increase, with the majority of cases presenting at II and III stage (3). Evidence available suggests that EOCC is currently linked to worse tumor differentiation and more advanced illness upon diagnosis in comparison to late-onset CRC (4). However, there is still a clinical need for the development of precise diagnostic and treatment regimens for EOCC. When the colon differentiates from the midgut and hindgut, it exhibits unique right- and left-sided characteristics from an embryological standpoint (5). According to recent researches, the prevalence of BRAF mutations, microsatellite instability-high (MSI-H), and CpG island methylator phenotype (CIMP) gradually increases from the rectum to the ascending colon across different colorectal subsites (6). Depending on its anatomical location, colon cancer (CC) might exhibit different patterns of disease progression, survival rates, and treatment effects (7,8). Additionally, some evidence suggests that various tumor sites may have different best treatment approaches for different risk groupings (9,10). A previous study has revealed a number of risk variables for CC, such as chemotherapy, surgical intervention, and pathological grade (11). However, specific risk factors associated with various tumor sites in the II-III stage EOCC are still not clear and it needs more studies.
The aim of this study is to discover predictive markers and create nomograms for predicting overall survival (OS) and cancer-specific survival (CSS) in patients with stage II–III EOCC using data from the Surveillance, Epidemiology, and End Results (SEER) database (seer.cancer.gov). We also contribute to comparing treatment alternatives among subgroups in order to determine the best therapy strategies for this population. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2290/rc).
Methods
Study population and inclusion and exclusion criteria
In this retrospective analysis, we investigated clinicopathological profiles of patients diagnosed with stage II-III EOCC utilizing data extracted from the SEER database, encompassing the period from January 1, 2010, through December 31, 2017. Data manipulation and statistical analysis were performed using SEER*Stat software version 8.4.0.1. The dataset coverd an array of variables, including age, race, sex, tumor and histologic grades, American Joint Committee on Cancer (AJCC) 7th edition staging (T, N, M) (12), tumor size, lymph node count, treatment methods (surgery, chemoradiotherapy), preoperative carcinoembryonic antigen (CEA) levels, perineural invasion, and tumor deposits. The grade is assessed through pathologic examination or the tumor’s differentiation level, with a lower grade indicating a higher differentiation degree. Tumor deposits are defined as microscopic or macroscopic tumor nodules found in the lymphatic drainage area of the primary tumor. The absence of tumor deposits is documented as negative, and similarly, the lack of perineural invasion by the tumor is also recorded as negative. CEA positive is defined as when the serum CEA level exceeds 5 ng/mL.
The inclusion criteria were: (I) individuals with a primary diagnosis of stage II or III EOCC; (II) patients with CC as their sole primary malignancy; (III) patients who underwent surgical treatment and subsequent pathological evaluation. Exclusion criteria included: (I) patients with multiple primary cancers; (II) cases documented solely through autopsy or death certificate information; (III) incomplete or missing data; (IV) individuals aged 50 years or older; (V) patients with a survival duration of zero months.
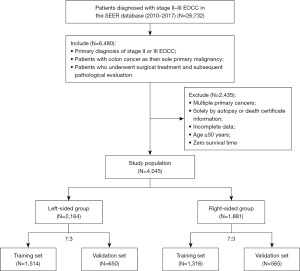
A total of 4,045 patients met the study’s criteria, with 2,164 classified in the left-sided group and 1,881 in the right-sided group ultimately (Figure 1). These cohorts were stratified in a 7:3 ratio to form training and test datasets, respectively, with the latter utilized for internal validation of the predictive nomogram. The allocating process was realized by random allocation of database by the instructions of R software. The study’s primary endpoints were OS and CSS, with the median follow-up period representing the observed survival duration. OS was defined as the interval from the date of diagnosis to death due to any cause or the date of last contact, while CSS was calculated from the date of diagnosis to death specifically attributed to cancer or the date of last follow-up. Ethics committee approval and patient consent were not required as we utilized publicly accessible, de-identified data. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
The Mann-Whitney U test was utilized to analyze continuous variables, and the Chi-squared test was employed for categorical data. To ascertain the relationship between variables and survival outcomes, both univariate and multivariate Cox proportional hazards models were conducted, yielding estimates of hazard ratios (HRs) alongside their 95% confidence intervals (CIs). Variables exhibiting a statistical significance level of P<0.05 in the multivariate analysis were integrated into a graphical predictive tool, namely a nomogram, to depict the probabilities of 1-, 3-, and 5-year survival. The analysis also included the computation of HRs with their respective 95% CIs. By entering the data of tumor size and OS into the X-tile software (version 3.6.1), a computer program was used to assess the most effective threshold of the tumor size. The X-tile worked by finding the best cutoff value that maximizes differences between groups, such as survival differences, by exhausting all possible values of a continuous variable. The predictive accuracy of the model was gauged using the concordance index (C-index) and receiver operating characteristic (ROC) curve analysis, which encompassed the calculation of the area under the curve (AUC). The C-index served as an equivalent variable to the AUC of the ROC curve when analyzing censored data. Additionally, calibration plots were constructed to assess the agreement between the predicted survival outcomes and the observed outcomes at various time points. The clinical utility of the model was further evaluated through decision curve analysis (DCA). Patients within the development cohort were stratified into distinct risk categories based on cumulative scores, and the divergence in survival was scrutinized using the Kaplan-Meier estimator coupled with log-rank tests. The core principle of the log-rank test is to determine whether there is a statistically significant difference in the survival curve by comparing the difference between the observed number of death events and the number of death events that would be expected under the invalid hypothesis. To mitigate potential biases and confounding effects, propensity score matching (PSM) was implemented in the subgroup analyses, taking into consideration a comprehensive set of covariates including age, sex, race, T stage, N stage, tumor grade, tumor size, CEA levels, perineural invasion, and the presence of tumor deposits. PSM analysis was executed utilizing the matching package of R software, employing the 1:1 nearest neighbor matching approach to ensure balanced comparison groups. All statistical computations were executed utilizing SPSS software (version 22.0) and R (version 4.2.0).
Results
Characteristics of II-III stage EOCC patients
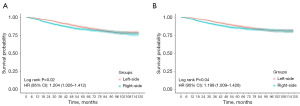
A cohort of 4,045 cases with stage II–III EOCC was examined, which included 2,164 individuals with left-sided group and 1,881 with right-sided group (Table 1). The median age was 42 years, and the median OS was 55 months. Surgical intervention was administered to all participants, with 97.65% undergoing adequate lymph node dissection. The demographic analysis indicated a higher prevalence of female and white patients in the left-sided group, whereas the right-sided group was characterized by inferior tumor differentiation and histological types. It is noteworthy that the right-sided group had advanced T stage but better N stage characteristics. A greater proportion of the left-sided group underwent chemoradiotherapy. However, this group also exhibited a higher incidence of perineural invasion and tumor deposits. Kaplan-Meier survival analysis demonstrated a significantly superior OS and CSS in the left-sided group when compared to the right-sided group (Figure 2). The cutoffs of the tumor size of the left-sided EOCC and the right-sided EOCC were 79 and 39 mm, respectively (Figure S1). The left-sided group also showed better survival outcomes and smaller tumor sizes. For further analysis, both groups were stratified into a training subset, encompassing 1,514 left-sided and 1,316 right-sided patients, and a validation subset, consisting of 650 left-sided and 565 right-sided patients. Statistical evaluation using the Mann-Whitney U and Chi-squared tests indicated no significant differences in the distribution of features between the training and validation cohorts (Tables S1,S2).
Table 1
| Characteristics | Total (n=4,045) | Left-side (n=2,164) | Right-side (n=1,881) | P |
|---|---|---|---|---|
| Sex, n (%) | <0.001* | |||
| Female | 1,940 (47.96) | 1,108 (51.20) | 832 (44.23) | |
| Male | 2,105 (52.04) | 1,056 (48.80) | 1,049 (55.77) | |
| Race, n (%) | <0.001* | |||
| Others | 1,140 (28.18) | 552 (25.51) | 588 (31.26) | |
| White | 2,905 (71.82) | 1,612 (74.49) | 1,293 (68.74) | |
| Grade, n (%) | <0.001* | |||
| I/II | 3,208 (79.31) | 1,827 (84.43) | 1,381 (73.42) | |
| III/IV | 837 (20.69) | 337 (15.57) | 500 (26.58) | |
| Histology, n (%) | <0.001* | |||
| Adenocarcinoma | 3,619 (89.47) | 2,019 (93.30) | 1,600 (85.06) | |
| Others | 426 (10.53) | 145 (6.70) | 281 (14.94) | |
| T stage, n (%) | <0.001* | |||
| I–II | 229 (5.66) | 155 (7.16) | 74 (3.93) | |
| III–IV | 3,816 (94.34) | 2,009 (92.84) | 1,807 (96.07) | |
| N stage, n (%) | <0.001* | |||
| N0 | 1,675 (41.41) | 800 (36.97) | 875 (46.52) | |
| N1 | 1,443 (35.67) | 821 (37.94) | 622 (33.07) | |
| N2 | 927 (22.92) | 543 (25.09) | 384 (20.41) | |
| Number of nodes examined, n (%) | 0.28 | |||
| <12 | 95 (2.35) | 56 (2.59) | 39 (2.07) | |
| ≥12 | 3,950 (97.65) | 2,108 (97.41) | 1,842 (97.93) | |
| Radiation, n (%) | <0.001* | |||
| No/unknown | 3,946 (97.55) | 2,094 (96.77) | 1,852 (98.46) | |
| Yes | 99 (2.45) | 70 (3.23) | 29 (1.54) | |
| Chemotherapy, n (%) | <0.001* | |||
| No/unknown | 1,277 (31.57) | 598 (27.63) | 679 (36.10) | |
| Yes | 2,768 (68.43) | 1,566 (72.37) | 1,202 (63.90) | |
| Systemic sequence, n (%) | <0.001* | |||
| Adjuvant therapy with surgery | 2,737 (67.66) | 1,549 (71.58) | 1,188 (63.16) | |
| Neoadjuvant with surgery | 34 (0.84) | 18 (0.83) | 16 (0.85) | |
| Only surgery | 1,274 (31.50) | 597 (27.59) | 677 (35.99) | |
| Carcinoembryonic antigen, n (%) | 0.50 | |||
| Negative | 2,660 (65.76) | 1,413 (65.30) | 1,247 (66.29) | |
| Positive | 1,385 (34.24) | 751 (34.70) | 634 (33.71) | |
| Perineural invasion, n (%) | <0.001* | |||
| No | 3,413 (84.38) | 1,785 (82.49) | 1,628 (86.55) | |
| Yes | 632 (15.62) | 379 (17.51) | 253 (13.45) | |
| Tumor deposits, n (%) | 0.02* | |||
| No | 3,414 (84.40) | 1,799 (83.13) | 1,615 (85.86) | |
| Yes | 631 (15.60) | 365 (16.87) | 266 (14.14) | |
| Survival time, months, mean (SD) | 58.72 (30.29) | 59.13 (29.92) | 58.25 (30.71) | 0.36 |
| Tumor size, mm, mean (SD) | 56.36 (28.64) | 51.24 (23.78) | 62.26 (32.37) | <0.001* |
*, statistical significance. SD, standard deviation.
Significance of variable features in survival prediction
To identify prognostic factors affecting OS in the left-sided group, we performed both univariate and multivariate Cox regression analyses on the training set (Table 2). These analyses revealed several significant risk factors associated with OS outcomes, including tumor histology, grade, size, N stage, CEA levels, perineural invasion, tumor deposits, and race. Notably, N1 stage (HR: 2.09; 95% CI: 1.36–3.20) and N2 stage (HR: 2.46; 95% CI: 1.60–3.76) were linked to worse OS, alongside poor tumor grade (HR: 1.58; 95% CI: 1.14–2.17), unfavorable histology (HR: 2.03; 95% CI: 1.32–3.12), larger tumor size (HR: 1.74; 95% CI: 1.18–2.58), race (HR: 1.42; 95% CI: 1.06–1.91), elevated CEA levels (HR: 1.51; 95% CI: 1.14–2.01), perineural invasion (HR:1.62; 95% CI: 1.18–2.22), and tumor deposits (HR: 1.51; 95% CI: 1.09–2.10). Subsequently, we conducted similar univariate and multivariate Cox regression analyses in the training set to evaluate prognostic factors for OS in the right-sided group (Table S3). Here, we found that N1 stage (HR: 2.51; 95% CI: 1.62–3.90) and N2 stage (HR: 6.17; 95% CI: 3.96–9.61) were again associated with poorer OS. Additionally, poor tumor grade (HR: 1.40; 95% CI: 1.05–1.87), elevated CEA levels (HR: 1.36; 95% CI: 1.02–1.81), perineural invasion (HR: 1.44; 95% CI: 1.05–2.00), and tumor deposits (HR: 2.14; 95% CI: 1.56–2.94) were also identified as negative prognostic indicators. Interestingly, while radiation (HR: 3.27; 95% CI: 1.79–5.97) correlated with worse OS, chemotherapy (HR: 0.56; 95% CI: 0.39–0.80) was associated with improved OS outcomes.
Table 2
| Characteristics | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Sex (vs. female) | |||||
| Male | 1.18 (0.90–1.55) | 0.24 | |||
| Histology (vs. adenocarcinoma) | |||||
| Others | 2.66 (1.79–3.94) | <0.001* | 2.03 (1.32–3.12) | 0.001* | |
| Lymph nodes examined (vs. <12) | |||||
| ≥12 | 1.07 (0.44–2.60) | 0.88 | |||
| Radiation (vs. no) | |||||
| Yes | 1.68 (0.96–2.95) | 0.07 | |||
| Chemotherapy (vs. no/unknown) | |||||
| Yes | 1.26 (0.91–1.75) | 0.16 | |||
| Grade (vs. I–II) | |||||
| III–IV | 2.17 (1.60–2.94) | <0.001* | 1.58 (1.14–2.17) | 0.005* | |
| Carcinoembryonic antigen level (vs. negative) | |||||
| Positive | 1.87 (1.42–2.46) | <0.001* | 1.51 (1.14–2.01) | 0.004* | |
| Perineural invasion (vs. no) | |||||
| Yes | 2.10 (1.57–2.82) | <0.001* | 1.62 (1.18–2.22) | 0.003* | |
| Tumor deposits (vs. no) | |||||
| Yes | 2.05 (1.51–2.80) | <0.001* | 1.51 (1.09–2.10) | 0.01* | |
| Tumor size (mm) (vs. <79) | |||||
| ≥79 | 1.89 (1.32–2.70) | <0.001* | 1.74 (1.18–2.58) | 0.006* | |
| Race (vs. White) | |||||
| Others | 1.43 (1.07–1.92) | 0.02* | 1.42 (1.06–1.91) | 0.02* | |
| N stage (vs. N0) | |||||
| N1 | 1.80 (1.25–2.61) | 0.002* | 2.09 (1.36–3.20) | <0.001* | |
| N2 | 2.80 (1.94–4.05) | <0.001* | 2.46 (1.60–3.76) | <0.001* | |
| T stage (vs. T1–T2) | |||||
| T3–T4 | 2.14 (1.06–4.34) | 0.04* | 1.63 (0.78–3.39) | 0.19 | |
*, statistical significance. CI, confidence interval.
We further evaluated prognostic factors for CSS in the left-sided group by the same way (Table 3). The results indicated several possible risk factors, including poor tumor grade (HR: 1.74; 95% CI: 1.25–2.43), elevated CEA levels (HR: 1.63; 95% CI: 1.20–2.20), perineural invasion (HR: 1.58; 95% CI: 1.14–2.19), tumor deposits (HR: 1.41; 95% CI: 1.01–1.98), larger tumor size (HR: 1.98; 95% CI: 1.33–2.94), race (HR: 1.41; 95% CI: 1.02–1.93), N1 stage (HR: 2.63; 95% CI: 1.62–4.27), and N2 stage (HR: 3.46; 95% CI: 2.13–5.60). Notably, T3/4 stage was also identified as a risk factor for CSS (HR: 2.84; 95% CI: 1.03–7.80). Subsequently, we performed similar univariate and multivariate Cox regression analyses to assess prognostic factors for CSS in the right-sided group (Table S4). It showed that radiation (HR: 2.62; 95% CI: 1.38–4.98), poor tumor grade (HR: 1.42; 95% CI: 1.05–1.93), elevated CEA levels (HR: 1.39; 95% CI: 1.03–1.87), perineural invasion (HR: 1.47; 95% CI: 1.04–2.08), tumor deposits (HR: 1.85; 95% CI: 1.32–2.60), unfavorable histology (HR: 1.58; 95% CI: 1.11–2.25), N1 stage (HR: 2.84; 95% CI: 1.76–4.60), and N2 stage (HR: 7.17; 95% CI: 4.44–11.56) were associated with worse OS outcomes. Conversely, chemotherapy was linked to improved OS (HR: 0.60; 95% CI: 0.41–0.89).
Table 3
| Characteristics | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Sex (vs. female) | |||||
| Male | 1.18 (0.88–1.58) | 0.27 | |||
| Histology (vs. adenocarcinoma) | |||||
| Others | 2.67 (1.76–4.05) | <0.001* | 1.57 (1.00–2.47) | 0.051 | |
| Lymph nodes examined (vs. <12) | |||||
| ≥12 | 1.61 (0.52–5.05) | 0.41 | |||
| Radiation (vs. no) | |||||
| Yes | 1.70 (0.90–3.22) | 0.10 | |||
| Chemotherapy (vs. no/unknown) | |||||
| Yes | 1.56 (1.08–2.25) | 0.02* | 0.83 (0.55–1.25) | 0.37 | |
| Grade (vs. I/II) | |||||
| III/IV | 2.36 (1.72–3.23) | <0.001* | 1.74 (1.25–2.43) | 0.001* | |
| Carcinoembryonic antigen level (vs. negative) | |||||
| Positive | 1.95 (1.45–2.61) | <0.001* | 1.63 (1.20–2.20) | 0.002* | |
| Perineural invasion (vs. no) | |||||
| Yes | 2.37 (1.73–3.23) | <0.001* | 1.58 (1.14–2.19) | 0.006* | |
| Tumor deposits (vs. no) | |||||
| Yes | 2.29 (1.66–3.16) | <0.001* | 1.41 (1.01–1.98) | 0.045* | |
| Tumor size (mm) (vs. <79) | |||||
| ≥79 | 2.11 (1.46–3.06) | <0.001* | 1.98 (1.33–2.94) | <0.001* | |
| Race (vs. White) | |||||
| Others | 1.48 (1.08–2.02) | 0.02* | 1.41 (1.02–1.93) | 0.04* | |
| N stage (vs. N0) | |||||
| N1 | 2.44 (1.58–3.76) | <0.001* | 2.63 (1.62–4.27) | <0.001* | |
| N2 | 4.20 (2.75–6.43) | <0.001* | 3.46 (2.13–5.60) | <0.001* | |
| T stage (vs. T1–T2) | |||||
| T3–T4 | 3.62 (1.35–9.76) | 0.01* | 2.84 (1.03–7.80) | 0.043* | |
*, statistical significance. CI, confidence interval.
Nomogram construction
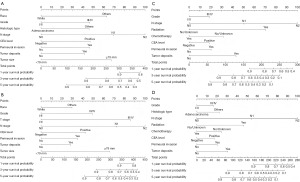
Based on the above analysis from the training set, we created nomogram models to predict OS and CSS for the two groups (Figure 3). Each of the identified prognostic factors was assigned a score ranging from 0 to 100, which quantitatively represents its relative contribution to the predictive accuracy of the model. By aggregating the scores for each individual patient, we were able to compute a total point value. This value was subsequently employed to estimate the probabilities of 1-, 3-, and 5-year survival for both OS and CSS. Notably, higher total scores were linked to poorer patient prognosis.
Model validation
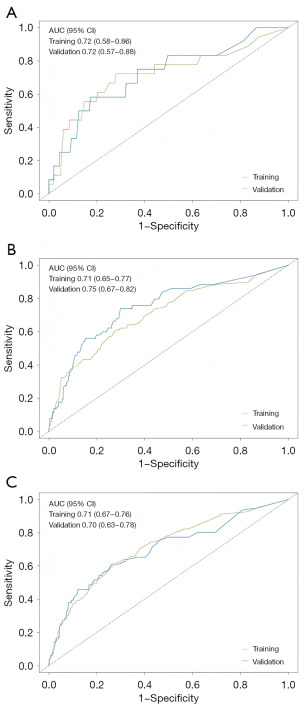
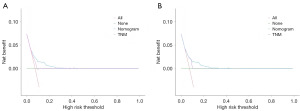
In the training cohort for the left-sided group, our nomogram for OS demonstrated a C-index of 0.70 (95% CI: 0.66–0.74), which was significantly higher than that of 0.61 (95% CI: 0.54–0.67) observed for the 7th edition of AJCC staging system. The predictive accuracy of our model was further substantiated by the ROC curves, which yielded AUC values of 0.72, 0.71, and 0.71 for 1-, 3-, and 5-year OS predictions respectively (Figure 4). Calibration plots revealed a strong concordance between the predicted probabilities and observed outcomes (Figure 5), and DCA highlighted the superior clinical utility of our nomogram over the AJCC system (Figure 6). In the validation set, the C-index further improved to 0.75 (95% CI: 0.73–0.77), with corresponding AUC values of 0.72, 0.75, and 0.70 for the same survival time points (Figure 4A-4C). Calibration curves consistently confirmed the predictive accuracy (Figure 5D-5F), and DCA further underscored the enhanced clinical utility of our nomogram relative to the AJCC system (Figure 6B).
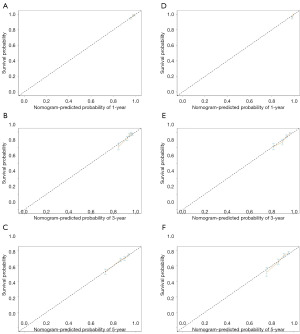
Similarly, the CSS nomogram for the left-sided group exhibited robust validation. For the training set, it achieved a C-index of 0.75 (95% CI: 0.72–0.79), surpassing the AJCC’s C-index of 0.61 (95% CI: 0.55–0.66). The ROC curves generated AUC values of 0.92, 0.77, and 0.75 for 1-, 3-, and 5-year CSS, respectively (Figure S2A-S2C). Calibration plots indicated a strong correlation between predicted and observed outcomes (Figure S2D-S2F), and DCA confirmed the model’s superior clinical efficacy (Figure S2G). In the validation cohort, the C-index was 0.64 (95% CI: 0.57–0.70), with AUC values of 0.71, 0.69, and 0.65 for the respective time points (Figure S2A-S2C). Consistent results were noted in calibration curves (Figure S2H-S2J), and DCA conformed the model’s enhanced clinical utility compared to the AJCC system (Figure S2K).
For the nomograms associated with the right-sided group, a similar analysis was conducted. In the training set, the nomogram for OS yielded a C-index of 0.77 (95% CI: 0.73–0.80), markedly higher than the 7th edition of the AJCC staging system’s C-index of 0.59 (95% CI: 0.53–0.66). The model’s performance was further evaluated through ROC curves, resulting in AUC values of 0.75, 0.79, and 0.78 for 1-, 3-, and 5-year OS, respectively (Figure S3A-S3C). Calibration plots demonstrated a strong alignment between predicted and actual outcomes (Figure S3D-S3F), and DCA underscored the nomogram’s superior clinical utility compared to the AJCC system (Figure S3G). In the validation cohort, the C-index was 0.75 (95% CI: 0.70–0.80), with AUC values of 0.84, 0.77, and 0.76 at the same intervals (Figure S3A-S3C). Calibration curves reaffirmed the correlation between predictions and observed results (Figure S3H-S3J), while DCA further illustrated the nomogram’s enhanced clinical relevance over the AJCC system (Figure S3K). Additionally, the CSS nomogram for the right-sided group showed strong validation. In the training set, it achieved a C-index of 0.77 (95% CI: 0.74–0.81), again exceeding the AJCC’s C-index of 0.58 (95% CI: 0.52–0.65). The ROC curves produced AUC values of 0.81, 0.80, and 0.78 for 1-, 3-, and 5-year CSS, respectively (Figure S4A-S4C). Calibration plots indicated a strong correlation between predicted and observed outcomes (Figure S4D-S4F), and DCA confirmed the model’s superior clinical efficacy (Figure S4G). In the validation cohort, the C-index remained at 0.77 (95% CI: 0.71–0.82), with AUC values of 0.80, 0.78, and 0.78 for the respective time points (Figure S4A-S4C). Consistent results were observed in calibration curves (Figure S4H-S4J), and DCA reiterated the model’s enhanced clinical utility compared to the AJCC system (Figure S4K).
Potential benefits of different treatment options across varying risk groups
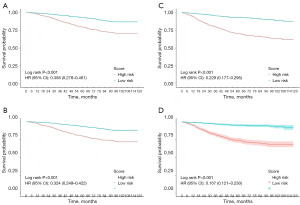
The left-sided group was divided into two subgroups according to prediction model scores: the low-risk group (scores <14 for OS and <185 for CSS) and the high-risk group (scores ≥114 for OS and ≥185 for CSS). Similarly, the right-sided group was classified into low-risk (scores <50 for OS and <54 for CSS) and high-risk (scores ≥50 for OS and ≥54 for CSS) subgroups. Kaplan-Meier survival analysis revealed significant differences in both OS and CSS between these subgroups (Figure 7). Notably, patients with higher risk scores demonstrated worse OS and CSS, while those in the low-risk group experienced better outcomes.
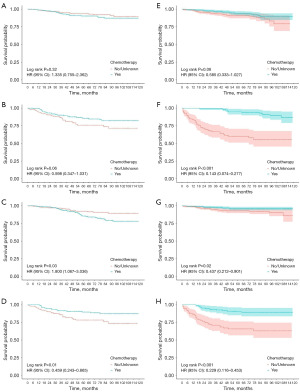
We further examined the impact of various treatment options on OS and CSS in these subgroups after PSM (Figure 8 and Figure S5). After each process of PSM, the baseline information of the variables was balanced and comparable (Appendix 1). In the left-sided cohort, our analysis indicated that chemotherapy conferred a significant improvement in CSS among patients classified as high risk. In contrast, within the right-sided cohort, chemotherapy was observed to significantly enhance both OS and CSS for high-risk patients, as well as CSS for those in the low-risk subgroup. Conversely, radiotherapy did not confer any significant survival advantages and was correlated with poorer outcomes in the low-risk subgroup. Furthermore, we compared the efficacy of adjuvant versus neoadjuvant chemotherapy within selected subgroups and observed no significant superiority of neoadjuvant chemotherapy (Figure S6). However, the limited sample size of patients receiving neoadjuvant chemotherapy may be a contributing factor to the observed outcomes.
Discussion
There has been a significant escalation in the incidence of EOCC over the past three decades, while the incidence of CRC among the elderly has either stabilized or declined in many countries. However, there is a lack of explanations for the rising incidence rate (13,14). EOCC is frequently diagnosed at advanced stages, suggesting a more aggressive tumorigenesis compared to late-onset colon cancer (LOCC) (15). Furthermore, the molecular signatures and the microbiome-metabolome profiles of EOCC are significantly different from those observed in late-onset cases (16). Moreover, emerging evidence implicates the younger tissue microenvironment as a contributing factor to the accelerated progression of EOCC (17).
Tumor location plays a critical role in shaping the biological features and prognosis of CRC (18). A recent study reported that there were differences between left-sided and right-sided CRC in the gut microbiome, metabolites, and host genomics (19). Additionally, research indicated that the infiltration of M0 macrophages was significantly higher in left-sided CC, whereas right-sided groups showed increased levels of differentiated M1 and M2 macrophages, as well as elevated CD4+ and CD8+ T cell infiltration (20). Furthermore, right-sided CC were associated with higher levels of myeloid-derived suppressor cells (MDSCs) compared to their left-sided counterparts (21). Adjuvant chemotherapy is the standard treatment for stage II–III CC; however, treatment responses vary among different subgroups (22). Neoadjuvant therapy has also shown benefits in selected patients with CC (23). These differences in the distribution of immune-related genes and biological characteristics may clarify the divergent responses to various therapies observed between left-sided and right-sided CC groups (24,25). Nevertheless, researches aimed at survival predictions and optimal treatment strategies for patients with stage II–III EOCC remain rare.
Our study investigated prognostic factors and developed prediction nomograms for OS and CSS in patients with stage II-III EOCC. For the left-sided cohort, multivariate Cox regression analysis identified independent risk factors for both OS and CSS, which included race, N stage, tumor grade, tumor size, histology, elevated CEA levels, presence of tumor deposits, and perineural invasion. Notably, T stage emerged as a significant risk factor exclusively for CSS. In the right-sided one, predictors for OS and CSS were N stage, tumor grade, histology, elevated CEA levels, tumor deposits, perineural invasion, and radiation and chemotherapy. We developed and assessed the accuracy of nomograms through C-index values, AUC in ROC analysis, and calibration curves. To evaluate the clinical utility and potential benefits of our model, DCA was conducted (26). The predictive nomogram effectively categorized patients into low-risk and high-risk subgroups. Kaplan-Meier analysis revealed significant differences in OS and CSS between these two groups. Furthermore, we evaluated the benefits of various treatment strategies after PSM in the training cohort.
In prior nomograms predicting outcomes for EOCC, the impact of tumor location was seldom analyzed (27). Our study revealed that the right-sided group correlated with poorer survival outcomes, aligning with findings in CC (28). A Recent study has identified N stage, tumor deposits, elevated CEA levels, and perineural invasion as crucial risk factors of survival prognosis in CC patients, consistent with our own findings (29). Contrary to an earlier study that locally advanced CC patients may benefit from chemotherapy, our research indicated that chemotherapy could improve OS and CSS in the right-sided EOCC cohort, but show benefits only for CSS in the high-risk group of left-sided EOCC cohort (30). One possible explanation is that the right-sided EOCC are often associated with hypermethylation, microsatellite instability and hypermutation, whereas left-sided EOCC tends to be more frequently characterized by chromosomal instability that reduces the effectiveness of adjuvant therapy (31).
Researches have reported considerable benefits of neoadjuvant chemotherapy for patients with locally advanced CC (32). In contrast, our study found no significant improvements associated with this treatment. A possible explanation is that our cohort had a deficient DNA mismatch repair (dMMR), which may contribute to a suboptimal response to neoadjuvant chemotherapy (33). For stage II-III CC, indications for neoadjuvant chemotherapy are focused on locally advanced (such as T4 or N+) or potentially resectable metastatic cases. Its application should be combined with the specific situation of patients, the biological characteristics of the tumor, and the evaluation of the multidisciplinary team. In the future, more high-quality studies are needed to clarify the value and optimal application of neoadjuvant chemotherapy in CC, especially in the group of EOCC. Radiotherapy is a standard treatment for rectal cancer, yet it is rarely used in EOCC (34). Our study found no significant survival benefits linked to radiation therapy in EOCC patients, aligning with prior research (35). However, the small sample size of patients receiving neoadjuvant therapy and radiation may impact the analysis. Furthermore, due to the lack of detailed drug and dosage of chemotherapy in the SEER database, the universality of our conclusions in clinical application is limited. Therefore, larger detailed clinical trials are necessary to validate these findings.
Comprehending OS and CSS is crucial for alleviating patient anxiety and enhancing quality of life, especially for those with initially poor prognosis. OS and CSS nomograms enable clinicians to evaluate mortality risk and develop personalized follow-up and monitoring strategies. This framework provides essential insights into the changing landscape of postoperative survival, empowering both patients and healthcare providers to make more informed treatment choices. However, the nomogram we developed is only applicable to stage II–III EOCC. The stage IV EOCC and any stage of LOCC are not applicable. Therefore, future studies with large sample sizes of more levels are needed before the model is widely used in clinical practice.
There are some limitations in this study. First, the SEER database lacked key biomarker data, such as molecular subtypes, MSI and dMMR status, both of which are vital prognostic markers. Additionally, it provided only basic therapeutic records without details on surgical techniques, chemotherapy regimens (including specific drug combinations and doses), radiation doses, patient health conditions, or socio-economic factors that may influence survival outcomes. Furthermore, the reasons of for not receiving specific treatments were missing. These omissions limited the scope of our analysis, and future researches should incorporate these variables to better assess their impact and generalize our findings. Furthermore, the retrospective nature of the study introduced potential selection bias. To validate our findings and minimize bias, prospective cohort studies or randomized controlled trials are necessary.
Conclusions
Our study offered a detailed analysis of prognostic factors that may affect OS and CSS in EOCC patients by analyzing data from the SEER database. We developed and validated predictive nomograms for OS and CSS and assessed the impact of various treatment options across different subgroups. Although our model demonstrated encouraging performance in predicting survival outcomes for EOCC patients, further multi-center studies are necessary to confirm its clinical applicability.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2290/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2290/prf
Funding: The research was funded by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2290/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin 2024;74:12-49. [Crossref] [PubMed]
- Benson AB, Venook AP, Adam M, et al. Colon Cancer, Version 3.2024, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2024;22:e240029. [Crossref] [PubMed]
- Takada K, Hotta K, Kishida Y, et al. Comprehensive Analysis of Early-onset Colorectal Cancer: A Review. J Anus Rectum Colon 2023;7:241-9. [Crossref] [PubMed]
- Liu H, Xu H, Liu Y, et al. Comparative characteristics of early-onset vs. late-onset advanced colorectal cancer: a nationwide study in China. BMC Cancer 2024;24:503. [Crossref] [PubMed]
- Kuliavas J, Marcinkevičiūtė K, Baušys A, et al. Short- and long-term outcome differences between patients undergoing left and right colon cancer surgery: cohort study. Int J Colorectal Dis 2024;39:66. [Crossref] [PubMed]
- Zhu Q, Zhu C, Zhang X, et al. Comprehension of rectosigmoid junction cancer molecular features by comparison to the rectum or sigmoid colon cancer. J Gastrointest Oncol 2023;14:1307-19. [Crossref] [PubMed]
- Ulanja MB, Asafo-Agyei KO, Neelam V, et al. Survival trends for left and right sided colon cancer using population-based SEER database: A forty-five-year analysis from 1975 to 2019. Cancer Med 2024;13:e7145. [Crossref] [PubMed]
- Liu B, Li S, Cheng Y, et al. Distinctive multicellular immunosuppressive hubs confer different intervention strategies for left- and right-sided colon cancers. Cell Rep Med 2024;5:101589. [Crossref] [PubMed]
- Tan C, Wang Q, Yao S. Effects of Adjuvant Chemotherapy on Early-onset Stage II Colon Cancer at Different Tumor Sites. Am J Clin Oncol 2024;47:253-8. [Crossref] [PubMed]
- Santos FA, Reis RM, Barroti LC, et al. Overall Survival, BRAF, RAS, and MSI Status in Patients Who Underwent Cetuximab After Refractory Chemotherapy for Metastatic Colorectal Cancer. J Gastrointest Cancer 2024;55:344-54. [Crossref] [PubMed]
- Gao J, Zhuang L, He C, et al. Risk and prognostic factors in patients with colon cancer with liver metastasis. J Int Med Res 2023;51:3000605231191580. [Crossref] [PubMed]
- Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 2010;17:1471-4.
- Siegel RL, Wagle NS, Cercek A, et al. Colorectal cancer statistics, 2023. CA Cancer J Clin 2023;73:233-54. [Crossref] [PubMed]
- Laskar RS, Qu C, Huyghe JR, et al. Genome-wide association studies and Mendelian randomization analyses provide insights into the causes of early-onset colorectal cancer. Ann Oncol 2024;35:523-36. [Crossref] [PubMed]
- Zhang R, Boakye D, Yang N, et al. Field Synopsis of Environmental and Genetic Risk Factors of Sporadic Early-Onset Colorectal Cancer and Advanced Adenoma. Cancer Epidemiol Biomarkers Prev 2023;32:1048-60. [Crossref] [PubMed]
- Kong C, Liang L, Liu G, et al. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut 2023;72:1129-42. [Crossref] [PubMed]
- Giardina C, Kuo A, Nito K, et al. Early onset colorectal cancer: Cancer promotion in young tissue. Biochem Pharmacol 2024;226:116393. [Crossref] [PubMed]
- Ugai T, Akimoto N, Haruki K, et al. Prognostic role of detailed colorectal location and tumor molecular features: analyses of 13,101 colorectal cancer patients including 2994 early-onset cases. J Gastroenterol 2023;58:229-45. [Crossref] [PubMed]
- Liang L, Kong C, Li J, et al. Distinct microbes, metabolites, and the host genome define the multi-omics profiles in right-sided and left-sided colon cancer. Microbiome 2024;12:274. [Crossref] [PubMed]
- Hu Y, Ding J, Wu C, et al. Differential Expression and Prognostic Correlation of Immune Related Factors Between Right and Left Side Colorectal Cancer. Front Oncol 2022;12:845765. [Crossref] [PubMed]
- Su C, Lin Z, Cui Y, et al. Identification of Essential Tumor-Infiltrating Immune Cells and Relevant Genes in Left-Sided and Right-Sided Colon Cancers. Cancers (Basel) 2022;14:4713. [Crossref] [PubMed]
- Kesireddy M, Tenner L. Colon Cancer Survivorship in Patients Who Have Received Adjuvant Chemotherapy. Clin Colorectal Cancer 2023;22:361-74. [Crossref] [PubMed]
- Morton D, Seymour M, Magill L, et al. Preoperative Chemotherapy for Operable Colon Cancer: Mature Results of an International Randomized Controlled Trial. J Clin Oncol 2023;41:1541-52. [Crossref] [PubMed]
- Gholamalizadeh H, Zafari N, Velayati M, et al. Prognostic value of primary tumor location in colorectal cancer: an updated meta-analysis. Clin Exp Med 2023;23:4369-83. [Crossref] [PubMed]
- Luo S, Cai S, Zhao R, et al. Comparison of left- and right-sided colorectal cancer to explore prognostic signatures related to pyroptosis. Heliyon 2024;10:e28091. [Crossref] [PubMed]
- Zhao L, Leng Y, Hu Y, et al. Understanding decision curve analysis in clinical prediction model research. Postgrad Med J 2024;100:512-5. [Crossref] [PubMed]
- Chen B, Ma Y, Zhou J, et al. Predicting survival and prognosis in early-onset locally advanced colon cancer: a retrospective observational study. Int J Colorectal Dis 2023;38:250. [Crossref] [PubMed]
- Asghari-Jafarabadi M, Wilkins S, Plazzer JP, et al. Prognostic factors and survival disparities in right-sided versus left-sided colon cancer. Sci Rep 2024;14:12306. [Crossref] [PubMed]
- Shi Y, Wu X, Qu W, et al. Construction and validation of a prognostic nomogram for predicting cancer-specific survival in patients with intermediate and advanced colon cancer after receiving surgery and chemotherapy. J Cancer Res Clin Oncol 2023;149:12821-34. [Crossref] [PubMed]
- Arjona-Sánchez A, Espinosa-Redondo E, Gutiérrez-Calvo A, et al. Efficacy and Safety of Intraoperative Hyperthermic Intraperitoneal Chemotherapy for Locally Advanced Colon Cancer: A Phase 3 Randomized Clinical Trial. JAMA Surg 2023;158:683-91. [Crossref] [PubMed]
- Aljama S, Lago EP, Zafra O, et al. Dichotomous colorectal cancer behaviour. Crit Rev Oncol Hematol 2023;189:104067. [Crossref] [PubMed]
- Gupta K, Elfiky A, Patel E. Clinical Evidence for the Benefits of Neoadjuvant Chemotherapy and Immunotherapy in Colon Cancer: A Concise Review. Discov Med 2023;35:928-35. [Crossref] [PubMed]
- Jin Z, Sinicrope FA. Mismatch Repair-Deficient Colorectal Cancer: Building on Checkpoint Blockade. J Clin Oncol 2022;40:2735-50. [Crossref] [PubMed]
- Hitchcock KE, Miller ED, Shi Q, et al. Alliance for clinical trials in Oncology (Alliance) trial A022101/NRG-GI009: a pragmatic randomized phase III trial evaluating total ablative therapy for patients with limited metastatic colorectal cancer: evaluating radiation, ablation, and surgery (ERASur). BMC Cancer 2024;24:201. [Crossref] [PubMed]
- Zhu S, Tu J, Pei W, et al. Development and validation of prognostic nomograms for early-onset colon cancer in different tumor locations: a population-based study. BMC Gastroenterol 2023;23:362. [Crossref] [PubMed]


