
Ubiquitin-specific peptidase 20 affects immune cell infiltration by regulating prostaglandin E2 on intraperitoneal metastasis model of colorectal cancer
Highlight box
Key findings
• Ubiquitin-specific peptidase 20 (USP20) expression is significantly upregulated in colorectal cancer (CRC), and can promote tumor metastasis by affecting immune cell infiltration.
What is known and what is new?
• Previously, there are only a few reports about the effect of USP20 on CRC. In terms of tumor-promoting mechanisms, USP20 is focused on regulating stabilities of β-catenin and SOX4 in colon cancer cells.
• In this study, it is found that USP20 regulates prostaglandin E2 through the protein degradation of microsomal prostaglandin E synthase-1 and 15-hydroxyprostaglandin dehydrogenase mediated by proteasome to influence immune cell infiltration.
What is the implication, and what should change now?
• Our findings provide insights on new molecular mechanisms of USP20 influencing immune cell infiltration in CRC.
Introduction
Ubiquitin-specific peptidase 20 (USP20) is a specific member of the ubiquitin-specific proteolytic enzyme family. It was first identified as a von Hippel-Lindau (VHL) syndrome-related deubiquitinase (1). Human USP20 comprises 914 amino acids, which include an N-terminal zinc-finger ubiquitin binding domain (ZnF-UBP), a USP catalytic domain, and two tandem ubiquitin-specific protease domains (DUSP) (2,3). USP20 can regulate the abundance of several proteins by removing ubiquitin molecules from the labeled proteins (2,3).
Several studies have revealed that USP20 plays key roles in the managements of virous cancers, including the cancer of colon, breast, gastric, bladder, etc., by stabilizing tumorigenic or antitumor proteins (4-8). USP20 is commonly considered as an oncogene, and it affects cell proliferation, migration, tumor growth, and glucose metabolism by regulating different signaling pathways (9-12). However, it also acts as a tumor suppressor in a few types of cancer (7,13). The different roles of USP20 may be attributed to the heterogeneity of different tumor types.
Colorectal cancer (CRC) is a leading cause of cancer-related mortality worldwide (14). To date, there are only a few reports about the effect of USP20 on CRC. It has been found that USP20 is upregulated in colon cancer cell lines, and significantly increases cell proliferation, migration, and invasion. In terms of tumor-promoting mechanisms, USP20 can regulate stabilities of β-catenin and SOX4 in colon cancer cells (6,9). In this study, we aim to discover the novel molecular mechanisms of USP20 in CRC by performing metabolomics study and experimental verification. We present this article in accordance with the MDAR and ARRIVE reporting checklists (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2194/rc).
Methods
Cohort and data collection
Data on the clinical characteristics and tumor and normal tissue transcriptome of patients with CRC were collected from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/analysis_page?app=Downloads). The transcriptome data format was processed standardized and matched with the clinical data. The transcriptome data of 647 tumor tissues and 51 normal tissues were obtained. Of these, 590 patients with CRC had survival data. In addition, 539 patients with CRC had complete clinical data including age, sex, and disease stage. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
Immune infiltration analysis
CAMOIP is a multi-omics pan-cancer analysis tool that facilitates the immune infiltration analysis of the TCGA cohort. It was used to compare differences in tumor immune infiltration between the high and low USP20 expression groups. The CIBERSORT method was utilized for the specific immune infiltration assessment used in this procedure.
Cell culture and USP20 knockout (USP20-KO)
Mouse colon cell line CT26 was purchased from ProCell (Wuhan, China). CT26 and 293T cells were cultured in Dulbecco’s Modified Eagle Medium (#1791922, Gibco, New York, USA) supplemented with 10% fetal bovine serum (#10099–141, Gibco), 100 U/mL penicillin and 100 µg/mL streptomycin (#P1400, Solarbio, Beijing, China). The incubator conditions were set to 5% CO2 at 37 ℃. The CRISPR-Cas9 knockout system was used to construct USP20-KO CT26 cells. Briefly, the backbone plasmid pLenti-Cas9-gRNA-puro was purchased from BioVector NTCC Inc. (Beijing, China). According to the designed sgRNA sequence, DNA was synthesized and inserted into pLenti-Cas9-gRNA-puro. The recombinant plasmid was subsequently transfected into 293T cells along with the pPAX2 and pMD2G plasmids (Invitrogen, Carlsbad, USA) using Lipofectamine 3000 reagent (#L3000015, Invitrogen), according to the manufacturer’s instructions. Successfully recombined lentiviruses were collected and used to infect CT26 cells. Finally, USP20-KO cells were screened out by 30 µg/mL puromycin (#P8230, Solarbio). Corresponding, negative control (NC) CT26 cells were established with the same procedure using plasmid pLenti-Cas9-gRNA-puro.
Animal study
Six- to eight-week-old male BALB/c mice were purchased from Huafukang Bioscience Co., Ltd. (Beijing, China). The mice were maintained in specific pathogen-free conditions in the animal facilities of Tianjin Nankai Hospital. Mouse CRC intraperitoneal metastasis models were established via the intraperitoneal injection of 5×105 NC or USP20-KO cells (10 mice per group). These mice were sacrificed 10 days after cell injection. Further, the ascites burden and peritoneal tumor burden were assessed. All animal experiments were performed under a project license (No. NKYY-DWLL-2024-138) granted by the Animal Ethics Committee of the Tianjin Nankai Hospital, in compliance with the local institutional guidelines for the care and use of animals. A protocol was prepared before the study without registration.
Flow cytometry
The tumors were cut into approximately 1 mm3 pieces and digested with 0.05 mg/mL of type-IV collagenase (#C5138, Sigma, St. Louis, USA), hyaluronidase (#H3506, Sigma), and DNase I (#D5025, Sigma) at 37 ℃ for 60 min. Single cells were obtained by grinding and filtering through a 70-µm strainer. Subsequently, mononuclear cells were obtained by density gradient centrifugation using 40% and 80% Percoll (#17-0891-09, GE, Boston, USA). Single cell suspensions in ascites were obtained via centrifugation. Single-cell suspensions were treated with red blood cell lysate and stained with the following antibodies: phycoerythrin (PE)-mouse CD8 antibody (#100707, Biolegend, San Diego, USA) labelled CD8+ T cells; fluorescein isothiocyanate (FITC)-mouse CD11b antibody (#101205, Biolegend) and PE-mouse GR-1 antibody (#108407, Biolegend) labelled myeloid-derived suppressor cells (MDSCs). The stained cells were detected and analyzed using the NovoCyte Flow Cytometer (ACEA Biosciences, San Diego, USA).
Untargeted metabolomics
The NC and USP20-KO cell samples (n=3) were obtained via three independent experiments. If the cells were in good condition, the medium was removed, and the cells were washed with phosphate buffered saline (PBS). The cells were added with cold methanol and acetonitrile mixed solution (1:1) to remove proteins and obtain metabolites. After further centrifugation and drying, the samples were resolved in a solvent (acetonitrile/water, 1:1) and then analyzed using ultra-high-performance liquid chromatography coupled to quadrupole time-of-flight mass spectrometry (LC-MS/MS) in Shanghai Applied Protein Technology Co., Ltd. (Shanghai, China). The chromatographic and mass spectrometric parameters were set according to the organizational procedures. The mass spectrometer was operated in positive and negative ion modes. The original data were converted into the mzXML format. Next, the XCMS software was used for peak alignment, retention time correction, and peak area extraction. These data were further preprocessed and evaluated, and the R software was used for biological information and statistical analysis. Principal component analysis was performed to evaluate the overall distribution trend of samples between groups and the difference degree of samples between groups. The orthogonal partial least-squares discriminant analysis (OPLS-DA) model was established to explore the differential metabolic molecules. The model evaluation parameters (R2, Q2) were obtained via a seven-fold cross-validation. A Q2 of >0.5 indicates reliable stability; 0.3≤ Q2 ≤0.5, good stability; and Q2 of <0.3, low reliability. The permutation test was performed on the model to ensure its validity. Variable Importance for the Projection (VIP) obtained from the OPLS-DA model was used to measure the influence intensity and interpretation ability of each metabolite on the classification and discrimination of samples.
Enzyme-linked immunosorbent assay (ELISA)
NC or USP20-KO CT26 cells were isolated and seeded at 1×105 cells/mL respectively. After 3 days, the supernatants were harvested and the prostaglandin E2 (PGE2) levels were determined in triplicate by ELISA kit (#D751014, Sangon Biotech, Shanghai, China).
Western blotting
Whole‑cell proteins were extracted using radio-immunoprecipitation assay (RIPA) lysis buffer (#R0020, Solarbio), respectively. Protein concentration was quantified by bicinchoninic acid protein assay kit (#PC0020, Solarbio) and proteins (40 µg per lane) were separated in 12% polyacrylamide gel. After electrophoresis, the proteins were transferred into polyvinylidene fluoride (PVDF) membrane (#IPVH00010, EMD Millipore, Billerica, USA), followed by blocking with 5% bovine serum albumin (BSA) (#A8020, Solarbio) in tris-buffered saline (TBS) with 0.1% Tween‑20 (#T8220, Solarbio) at room temperature for 3 hours and then incubating with primary antibodies (1:1,000) at 4 ℃ overnight. Next, the membranes were incubated with horseradish peroxidase‑conjugated goat anti‑rabbit (1:2,000; #SE13, Solarbio) secondary antibodies for 1 hour at room temperature. The protein bands were visualized by BioVision ECL western blotting substrate kit (#K820‑500, BioVision, Palo Alto, USA). ImageJ software (version 1.5; National Institutes of Health, Bethesda, USA) was used to analyze the intensities of the band signals obtained. The primary antibodies of microsomal prostaglandin E synthase-1 (mPGES-1, #ab180589), microsomal prostaglandin E synthase-2 (mPGES-2, #ab300052), 15-hydroxyprostaglandin dehydrogenase (15-PGDH, #ab187161), and β-actin (#ab213262) were obtained from Abcam. The primary antibody of USP20 (#A301-189A) was obtained from Thermo Fisher Scientific (Waltham, USA).
For the inhibition of the proteasome, USP20-KO CT26 cells were treated by 5 µM MG-132 (#M7449, Sigma) for 24 hours, and then the Western blotting was performed as described above.
Statistical analysis
For the clinical data, the differences in the USP20 expression between tumor and normal tissues were compared using the unpaired and paired t-test, respectively. Patients with CRC were divided into the high or low USP20 expression groups according to the median USP20 expression level in tumors. The overall survival (OS) of these two groups was compared using the Kaplan-Meier survival curve. Statistical analysis was conducted using the log-rank test. The Chi-squared test was utilized to analyze the correlation between the USP20 expression and the clinical characteristics. For cellular and animal experiments, at least three biological replicates were performed. Data between two groups were analyzed using the student’s t-test. These analyses were performed using GraphPad Prism 8. A P value of <0.05 was considered statistically significant.
For the data of metabolomics, the student’s t-test was used to determine significance differences between two groups. An OPLS-DA VIP of >1 and a P value of <0.05 were the screening criteria for significantly different metabolites.
Results
USP20 expression in tumor tissue is correlated to poor prognosis of CRC
Based on the TCGA-CRC data, the USP20 expression was explored. We found that the USP20 expression in CRC tumor tissues (n=647) was significantly upregulated compared with that in normal tissues (n=51) (Figure 1A). Moreover, the USP20 expression in tumor tissues (n=50) was also upregulated compared with that in the corresponding CRC normal tissues (n=50) (Figure 1B). Based on the Kaplan-Meier survival curve, there was a remarkable difference in terms of OS between the high and low USP20 expression groups (Figure 1C). The high USP20 expression indicated a worse OS. In addition, the correlation between USP20 expression and clinical characteristics in patients with CRC was analyzed. Results showed that the USP20 expression was correlated with T stage, N stage, and M stage. Further, the high USP20 expression group had a higher proportion of patients with advanced-stage disease than the low USP20 expression group (Table 1).
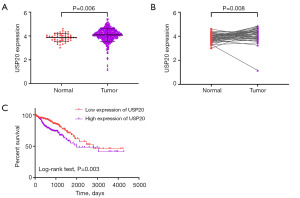
Table 1
| Clinical characteristics | n (%) | USP20 expression | P | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | 0.61 | |||
| <50 | 64 (11.9) | 30 | 34 | |
| ≥50 | 475 (88.1) | 239 | 236 | |
| Gender | 0.32 | |||
| Female | 254 (47.1) | 121 | 133 | |
| Male | 285 (52.9) | 148 | 137 | |
| Stage | 0.01 | |||
| I/II | 304 (56.4) | 166 | 138 | |
| III/IV | 235 (43.6) | 103 | 132 | |
| T | 0.04 | |||
| T1/2 | 109 (20.2) | 64 | 45 | |
| T3/4 | 430 (79.8) | 205 | 225 | |
| N | 0.03 | |||
| N0 | 314 (58.3) | 170 | 144 | |
| N1 | 128 (23.7) | 61 | 67 | |
| N2 | 97 (18.0) | 38 | 59 | |
| M | 0.04 | |||
| M0 | 453 (84.0) | 235 | 218 | |
| M1 | 86 (16.0) | 34 | 52 | |
USP20, ubiquitin-specific peptidase 20.
Knockout of USP20 in CRC cell inhibits intraperitoneal metastasis in mice
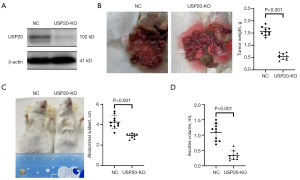
To validate the effect of USP20 on tumor progression, the USP20-KO cell was established on CT26 cell line (Figure 2A). And then, the NC and USP20-KO CT26 cells were injected respectively for intraperitoneal metastasis mice model of CRC. The models were subjected to tumor burden assessment on the day 10 after cell injection. It was found that the intraperitoneal tumor weight (Figure 2B), abdominal diameter (Figure 2C) and volume of ascites (Figure 2D) of the USP20-KO group decreased significantly compared with the NC group. The intraperitoneal tumor weight, abdominal diameter and volume of ascites of the USP20-KO group was 0.55±0.13 g, 2.93±0.23 cm and 0.34±0.14 mL respectively. The counterpart of NC group was 1.57±0.17 g, 4.25±0.63 cm and 1.09±0.28 mL.
USP20 affects immune infiltration in CRC ascites and tumor tissue
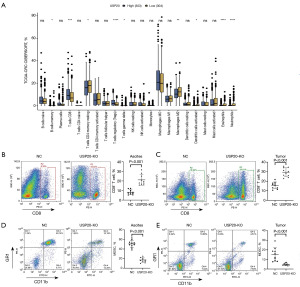
The immune cell infiltration played an important role in the metastasis of CRC. Therefore, we compared the immune cell infiltration between the high and low USP20 expression groups based on CIBERSORT. As shown in Figure 3A, the proportions of CD8+ T cells and regulatory T cells were down-regulated, but the proportion of M2 macrophages, eosinophils, and neutrophils were increased in the low USP20 expression group compared with those in the high USP20 expression group. To validate the relationship of USP20 and immune cell infiltration, the ascites and tumor tissue of the intraperitoneal metastasis mice model mentioned above were collected and the immune cells in them were analyzed by flow cytometry. The proportions of CD8+ T cells in the ascites and tumor of the USP20-KO group significantly increased compared with those in the NC group (Figure 3B,3C). However, this result was not in accordance with the data based on CIBERSORT. Additionally, we found that the proportions of MDSCs in the ascites and tumor were decreased remarkably when USP20 was knocked out (Figure 3D,3E).
USP20 affects the metabolome of CRC cells
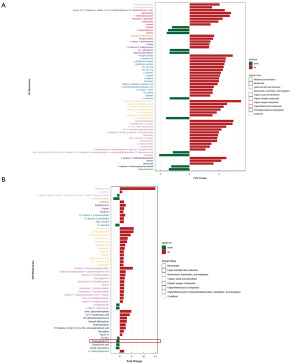
To investigate the novel mechanism of USP20 on immune infiltration, LC-MS/MS-based metabolomics analysis was performed between NC and USP20-KO CT26 cells. Based on the screening criteria (an OPLS-DA VIP of >1 and a P value of <0.05), 59 differential metabolites were screened in positive ion mode and 50 differential metabolites in negative ion mode. The histograms showed the significant differences, and the differential metabolites were categorically attributed according to superclass (Figure 4A,4B). In negative ion mode, we found that PGE2, which was recognized as an immunosuppressive molecule, was reduced in the USP20-KO cells.
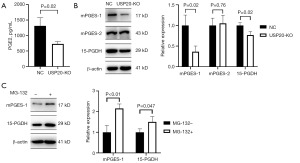
USP20 regulates the synthesis and degradation of PGE2
In order to verify the metabolomics analysis of PGE2, the culture supernatants of NC and USP20-KO CT26 cells were detected by PGE2 ELISA kit. The concentration of PGE2 in USP20-KO cell culture supernatant was really lower than in NC cell culture supernatant (Figure 5A). Subsequently, the expressions of PGE2-metabolizing enzymes, including and 15-PGDH, mPGES-1 and mPGES-2, were compared between the NC and USP20-KO cells. It was found that the expressions of mPGES-1 and 15-PGDH were remarkably down-regulated in the USP20-KO cells (Figure 5B). To discover whether the down-regulation of the PGE2-metabolizing enzymes by USP20 was related to the protein degradation mediated by ubiquitin-proteasome pathway, the USP20-KO cells were treated by a proteasome inhibitor, MG-132, and then the expressions of mPGES-1 and 15-PGDH were compared. It was shown that the protein levels of mPGES-1 and 15-PGDH in USP20-KO cells were recovered after the cells were treated by MG-132 (Figure 5C).
Discussion
Patients with early-stage CRC did not commonly present with evident symptoms. Due to the low rate of regular colonoscopy screening, patients are commonly diagnosed with late-stage CRC (15). Therefore, novel prognostic markers should be investigated for evaluating the patient’s prognostic status and screening new treatment targets. Via an in-depth exploration of data in the TCGA database, we found that USP20 was highly expressed in colon cancer tissues than in normal tissues. But in a previous paper, it was reported that the expression of USP20 was lower in cancer tissues than in normal tissues from CRC patients (4). Interestingly, that paper agreed with us on the viewpoint that high expression of USP20 was closely related to lymph node and distant metastases in patients with CRC. Furthermore, in our animal experiments, it was demonstrated that knockout of USP20 significantly reduced the risk of intraperitoneal metastasis in tumor-bearing mice.
Based on the CIBERSORT analysis and the fluorescence-activated cell sorting (FACS) assay on the mouse model, we concluded that the expression of USP20 significantly affected the immune infiltration. However, there was a disagreement about the relationship of the USP20 expression and the CD8+ T cell infiltration between the results of the two research methods. Additionally, through the animal experiment, we found that knockout of USP20 in CRC cell could reduce the infiltrations of MDSCs in tumor and ascites. Therefore, it is more rational that the increase of CD8+ T cells cooperated with the decrease of the MDSCs to cause the inhibition of intraperitoneal metastasis.
For exploring the novel mechanism of USP20 in regulating immune cell, the LC-MS/MS-based metabolomics analysis was performed between NC and USP20-KO cells. The result indicated that PGE2 should be the key regulator for immune cells. According to the previous studies, PGE2 plays an important role in CRC development by binding to the downstream receptors EP1, EP2, EP3, and EP4, thereby regulating different immune cell functions (16,17). Moreover, tumor-derived PGE2 could promote MDSC cell differentiation (18). Subsequently, we confirmed the result of the metabolomics analysis by detecting the PGE2 in the culture supernatant of the NC and USP20-KO CT26 cells.
In order to investigate the mechanism of reduction of PGE2 level, the expressions of three PGE2-metabolizing enzymes were measured. Wherein mPGES-1 and mPGES-2 were terminal enzymes that synthesis PGE2 (19), and 15-PGDH was the degrading enzyme of PGE2 that converts PGE2 to inactivate 15-keto-PGE2 (20). We found that expressions of mPGES-1 and 15-PGDH were both down-regulated in the USP20-KO cells. Although the down-regulation of 15-PGDH should increase the level of PGE2, maybe the mPGES-1 was the key regulatory enzyme of PGE2 production. To this day, there is no evidence that these enzymes could be regulated by the protein degradation mediated by ubiquitin-proteasome pathway. Therefore, the proteasome inhibitor, MG-132, was used to initially verify that the proteasome plays an important role in regulating the two PGE2-metabolizing enzymes by USP20. In future works, we would elaborately explore the degradation mechanism of these enzymes
Conclusions
The USP20 expression is significantly upregulated in CRC, and can promote tumor metastasis by affecting immune cell infiltration. As an important tumor metabolite which can influence immune cell infiltration, PGE2 is regulated by USP20 through the protein degradation of mPGES-1 and 15-PGDH mediated by proteasome.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2194/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2194/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2194/prf
Funding: This study was funded by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2194/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. All animal experiments were performed under a project license (No. NKYY-DWLL-2024-138) granted by the Animal Ethics Committee of the Tianjin Nankai Hospital, in compliance with the local institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Park JJ, Yun JH, Baek KH. Polyclonal and monoclonal antibodies specific for ubiquitin-specific protease 20. Monoclon Antib Immunodiagn Immunother 2013;32:193-9. [Crossref] [PubMed]
- Qin B, Zhou L, Wang F, et al. Ubiquitin-specific protease 20 in human disease: Emerging role and therapeutic implications. Biochem Pharmacol 2022;206:115352. [Crossref] [PubMed]
- Li Q, Ye C, Tian T, et al. The emerging role of ubiquitin-specific protease 20 in tumorigenesis and cancer therapeutics. Cell Death Dis 2022;13:434. [Crossref] [PubMed]
- Jin R, Luo Z. USP20 is a predictor of poor prognosis in colorectal cancer and associated with lymph node metastasis, immune infiltration and chemotherapy resistance. Front Oncol 2023;13:1023292. [Crossref] [PubMed]
- Huang ML, Shen GT, Li NL. Emerging potential of ubiquitin-specific proteases and ubiquitin-specific proteases inhibitors in breast cancer treatment. World J Clin Cases 2022;10:11690-701. [Crossref] [PubMed]
- Guan Y, Jiang SR, Liu JG, et al. USP20 regulates the stability of EMT transcription factor SOX4 and influences colorectal cancer metastasis. Pathol Res Pract 2022;233:153879. [Crossref] [PubMed]
- Wang C, Yang C, Ji J, et al. Deubiquitinating enzyme USP20 is a positive regulator of Claspin and suppresses the malignant characteristics of gastric cancer cells. Int J Oncol 2017;50:1136-46. [Crossref] [PubMed]
- Chen W, Wu S, Chen Y, et al. USP20 mediates malignant phenotypic changes in bladder cancer through direct interactions with YAP1. Neoplasia 2025;60:101102. [Crossref] [PubMed]
- Wu C, Luo K, Zhao F, et al. USP20 positively regulates tumorigenesis and chemoresistance through β-catenin stabilization. Cell Death Differ 2018;25:1855-69. Erratum in: Cell Death Differ 2023;30:857-8. [Crossref] [PubMed]
- Ha J, Kim M, Seo D, et al. The Deubiquitinating Enzyme USP20 Regulates the TNFα-Induced NF-κB Signaling Pathway through Stabilization of p62. Int J Mol Sci 2020;21:3116. [Crossref] [PubMed]
- Feng J, Liu P, Li X, et al. The deubiquitinating enzyme USP20 regulates the stability of the MCL1 protein. Biochem Biophys Res Commun 2022;593:122-8. [Crossref] [PubMed]
- Mathien S, Déléris P, Soulez M, et al. Deubiquitinating Enzyme USP20 Regulates Extracellular Signal-Regulated Kinase 3 Stability and Biological Activity. Mol Cell Biol 2017;37:e00432-16. [Crossref] [PubMed]
- Kim JH, Seo D, Kim SJ, et al. The deubiquitinating enzyme USP20 stabilizes ULK1 and promotes autophagy initiation. EMBO Rep 2018;19:e44378. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- He J, Wu F, Han Z, et al. Biomarkers (mRNAs and Non-Coding RNAs) for the Diagnosis and Prognosis of Colorectal Cancer - From the Body Fluid to Tissue Level. Front Oncol 2021;11:632834. [Crossref] [PubMed]
- Mizuno R, Kawada K, Sakai Y. Prostaglandin E2/EP Signaling in the Tumor Microenvironment of Colorectal Cancer. Int J Mol Sci 2019;20:6254. [Crossref] [PubMed]
- Wang D, Fu L, Sun H, et al. Prostaglandin E2 Promotes Colorectal Cancer Stem Cell Expansion and Metastasis in Mice. Gastroenterology 2015;149:1884-1895.e4. [Crossref] [PubMed]
- Porta C, Consonni FM, Morlacchi S, et al. Tumor-Derived Prostaglandin E2 Promotes p50 NF-κB-Dependent Differentiation of Monocytic MDSCs. Cancer Res 2020;80:2874-88. [Crossref] [PubMed]
- Xu D, Cai J, Wan ZK, et al. Pathophysiological role of prostaglandin E synthases in liver diseases. Prostaglandins Other Lipid Mediat 2021;154:106552. [Crossref] [PubMed]
- Palla AR, Ravichandran M, Wang YX, et al. Inhibition of prostaglandin-degrading enzyme 15-PGDH rejuvenates aged muscle mass and strength. Science 2021;371:eabc8059. [Crossref] [PubMed]


