
The prognostic and immune significance of fibronectin type III domain-containing 1 gene in pan-cancer and its relationship with proliferation and migration of stomach adenocarcinoma
Highlight box
Key findings
• Fibronectin type III domain containing 1 (FNDC1) is upregulated in Stomach adenocarcinoma (STAD) compared to adjacent normal tissues. Knocking down FNDC1 can decrease the proliferation, migration and invasion abilities of gastric cancer cells.
What is known and what is new?
• FNDC1 has received increasing attention in recent years due to its close association with the biological behavior of tumors.
• This study utilized bioinformatics approaches to investigate the genetic variations, mRNA expression levels, prognostic value, microsatellite instability, tumor mutation burden, immunological relevance, and pathway enrichment analysis of FNDC1 across different tumors. Building on the preliminary pan-cancer analysis, we focused on STAD and conducted in vitro validations using clinical samples and STAD cell lines to assess the expression level, prognostic value, and cellular functions of FNDC1 in STAD.
What is the implication, and what should change now?
• FNDC1 is highly expressed in various tumor tissues and may serve as a potential prognostic biomarker and immunotherapy target in cancer. It plays a crucial role in proliferation, migration and invasion of STAD cells.
Introduction
Stomach adenocarcinoma (STAD) is a prevalent malignant tumor disease globally, with poor prognosis posing a significant threat to human health. Related studies report that the incidence of gastric cancer varies globally, with high-risk regions including East Asia (Japan and Mongolia) and Eastern Europe, while the incidence is generally lower in Northern Europe, North America, and Africa (1,2). According to the latest statistics, there were approximately 1.033 million new cases of STAD worldwide and around 783,000 deaths in 2018. STAD ranks fifth in terms of incidence and second in terms of mortality among malignant tumors (3). In China, in 2020, gastric cancer ranked third in terms of incidence among malignant tumors, with 479,000 new cases. It ranked third in terms of mortality, with 374,000 deaths, and the mortality rate was 15.9 per 100,000. In recent years, there has been a continuous upward trend in both incidence and a younger age at diagnosis (4). In the early stages, STAD often presents no obvious symptoms, and symptomatic patients have mostly progressed to intermediate or advanced stages, showing manifestations such as weight loss, vomiting blood, and black stool, often accompanied by lymph node (LN) and distant metastasis. Reported high-risk factors have been identified to play important roles in promoting the occurrence and development of STAD, such as Helicobacter pylori infection, atrophic gastritis, high-salt diet, advanced age, low socioeconomic status, smoking, alcohol consumption, familial susceptibility, history of gastric surgery, pernicious anemia, obesity, and gastroesophageal reflux (5-8). Current commonly used treatments include endoscopic therapy, surgical resection, adjuvant chemotherapy, radiotherapy, molecular targeted drugs, and immune checkpoint inhibitors. However, the prognosis for advanced-stage STAD remains poor, with a subsequent decrease in quality of life and a 5-year overall survival (OS) rate of less than 50% (9). Therefore, STAD is a complex disease influenced by multiple factors, and the detailed mechanisms regulating its development and progression are not yet fully understood.
Fibronectin (FN) is a multi-domain glycoprotein with a molecular weight of approximately 450 kDa. It is a widely present and important component of the extracellular matrix, playing a crucial role in cell adhesion, migration, proliferation, and differentiation (10). The III type fibronectin domain is one of the important structural domains of FN. Fibronectin type III domain containing 1 (FNDC1) is a protein-coding and disease-related gene located on human chromosome 6q25.3, with a protein molecular weight of 205 kDa. FNDC1, also known as activator of G-protein signaling 8 (AGS8) or MEL4B, is a non-receptor-dependent activator of G-protein signaling, which can activate G-proteins and signal transduction through interaction with G-protein β and γ subunits (10-13). FNDC1 is reported to be expressed in various tissues, such as mesenchymal stem cells, synovium, fetal cartilage, thyroid, heart, kidney, adipose tissue, oral pharynx, and digestive tract, participating in protein polymerization (14). Studies have shown that high methylation of FNDC1 can promote apoptosis in human salivary adenoid cystic carcinoma cells (15). FNDC1, FN1, and androgen receptor (AR) have been found to be co-overexpressed in metastatic prostate cancer, and increased expression of microRNA-1207-3p significantly inhibits migration and proliferation. It directly targets FNDC1 to regulate FN1, thereby modulating proliferation, apoptosis, and migration in prostate cancer (12). It is believed that FNDC1 is closely related to the pathogenesis of acute otitis media (13). FNDC1 and MXRA5 have been reported as novel biomarkers for aortic valve stenosis (16).
Given the complex functional characteristics of FNDC1, this study utilized multiple databases to elucidate the potential functions of FNDC1 in pan-cancer and its impact on immune cell infiltration in pan-cancer. Furthermore, tissue microarray technology combined with STAD clinical samples was used to investigate the relationship between FNDC1, HER2, MMR protein expression, clinicopathological features, and prognosis. The results were further validated through in vitro cell experiments to confirm the effects of FNDC1 on STAD cell proliferation, invasion, and migration. The aim of this study is to provide assistance in identifying new therapeutic targets for STAD patients. We present this article in accordance with the REMARK and MDAR reporting checklists (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2279/rc).
Methods
Bioinformatics pan-cancer analysis
Mutation analysis
The multi-dimensional cancer gene set cBioPortal database (http://cbioportal.org) was used to analyze the mutation status of the FNDC1 gene and its mutation sites.
Gene expression analysis
Pan-cancer analysis of RNA-seq data and corresponding clinical information from 33 tumor types in The Cancer Genome Atlas (TCGA) database was conducted using the Sangerbox online analysis platform (http://vip.sangerbox.com) and The Human Protein Atlas (HPA) database (17). In the “Expression Analysis” section, a total of 10,228 samples from 33 types of cancer, including adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), acute myeloid leukemia (LAML), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), mesothelioma (MESO), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), sarcoma (SARC), skin cutaneous melanoma (SKCM), STAD, testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), thymoma (THYM), uterine corpus endometrial carcinoma (UCEC), uterine carcinosarcoma (UCS), and uveal melanoma (UVM), were analyzed for differential expression of FNDC1 mRNA in tumor tissues and corresponding adjacent tissues.
Prognosis analysis
RNA sequencing data and corresponding clinical information for 33 types of tumors were obtained from TCGA database (https://portal.gdc.com). Univariate Cox regression analysis was conducted, and a forest plot was generated using the “forestplot” R package to display the P values, hazard ratio (HR), and 95% confidence interval (CI). Statistical analysis was performed using R software version 4.0.3.
Tumor-related features
The expression of FNDC1 in pan-cancer was analyzed in relation to tumor mutation burden (TMB) and microsatellite instability (MSI) using the Sangerbox online analysis platform (http://vip.sangerbox.com). The TMB and MSI data were sourced from studies by Vesteinn Thorsson et al. (18) and Russell Bonneville et al. (19).
Immune infiltration and immune checkpoint analysis
To perform reliable immunological correlation assessment, we utilized the R software package “immunedeconv” and employed the xCell algorithm to evaluate the correlation between FNDC1 expression and immune infiltration scores in pan-cancer. Furthermore, we extracted the expression levels of eight transcripts associated with immune checkpoints, namely SIGLEC15, IDO1, CD274, HAVCR2, PDCD1, CTLA4, LAG3, and PDCD1LG2, to observe the expression patterns of immune checkpoint-related genes.
Pathway enrichment analysis
The STRING database (https://string-db.org/) was used to obtain a protein set with direct physical interactions with FNDC1, ranked in the top 10. Gene set enrichment analysis (GSEA) software was utilized to divide pan-cancer samples from TCGA into high and low expression groups based on FNDC1 expression levels, and to detect the impact of this gene on biological functions and signaling pathways within pan-cancer tissues. Enrichment analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) datasets.
Clinical sample assessment
Tissue samples were retrieved from the pathology department tissue core library information management system of Zhongshan Hospital, Fudan University, from STAD patients who underwent potentially curative gastrectomy and LN dissection in the Department of General Surgery from 2014 to 2016. According to the guidelines of the Japanese Gastric Cancer Association, all patients underwent total or subtotal gastrectomy and D2 LN dissection. Surgical samples were fixed in 10% buffered formalin and evaluated according to the standards of the American Pathological Association. Tumor staging was assessed according to the 8th edition of the tumor-node-metastasis (TNM) staging system for STAD by the International Union Against Cancer and the American Joint Committee on Cancer (UICC/AJCC). Inclusion criteria: (I) STAD; (II) D2 LN dissection; (III) R0 resection; (IV) tissue core points available for analysis. Exclusion criteria: (I) residual gastric tumor; (II) palliative resection; (III) metastatic malignant tumors; (IV) inability to obtain tissue core points and incomplete clinical and pathological information; (V) preoperative radiotherapy or chemotherapy; (VI) history of previous surgery for digestive system diseases; (VII) severe internal medical conditions. A total of 741 cases of gastric cancer diagnosed by histopathology were selected, with 741 core points, and 663 cases of normal gastric tissue adjacent to the cancer, with 663 core points. Basic clinical information including age, gender, location, tumor size, histological grade, and tumor stage was compiled. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study was approved by the ethics committee of Zhongshan Hospital (No. B2021-842R) and informed consent was taken from all the participants in our study.
The above tissue cores were aggregated and formed into tissue microarray blocks using the high-throughput tissue microarray method of our research group, and then sectioned. The tissue microarray was stained with hematoxylin and eosin (H&E), and at least two pathology experts performed microscopic quality control using an Olympus BX43F microscope to ensure the accuracy of each core point.
Immunohistochemical staining was performed for FNDC1, human epidermal growth factor receptor 2 (HER2), and DNA mismatch repair (MMR) proteins (MLH1, PMS2, MSH2, and MSH6). Immunohistochemical staining for FNDC1, MLH1, PMS2, MSH2, and MSH6 was performed using the DAKO immunohistochemical automatic staining machine (DAKO Omnis/GI100). Immunohistochemical staining for HER2 was performed using the fully automated immunohistochemistry system (Roche Benchmark XT), using the EnVision two-step method. The primary antibodies used were FNDC1 rabbit anti-human (PA5-56602, 1:250) purchased from Invitrogen, HER-2/NEU rabbit anti-human (4B5, working solution) purchased from Roche, PMS2 rabbit anti-human (EP51, 1:50), MLH1 rabbit anti-human (ES05, 1:50), MSH2 rabbit anti-human (RED2, 1:200), and MSH6 rabbit anti-human (EP49, 1:200) purchased from Zhongshan Golden Bridge Biotechnology Co., Ltd.
Immunohistochemical staining interpretation criteria
FNDC1 immunohistochemical staining interpretation
Two experienced pathologists independently evaluated the staining results using an Olympus BX43F microscope. FNDC1 positivity was located in the nucleus or cytoplasm of tumor cells, and 5 random 400× fields (hot spots) were selected for observation and counting. The staining intensity of positive cells was scored on a 4-point scale: no staining (negative) was scored as 0; light yellow (weakly positive) was scored as 1; tan-yellow (positive) was scored as 2; brown (strongly positive) was scored as 3. The percentage of positive cells under high magnification was classified into 4 levels: ≤25% was scored as 1; 26–50% was scored as 2; 51–75% was scored as 3; >75% was scored as 4. The final score was obtained by multiplying the percentage score of positive cells by the staining intensity score. A score higher than 0 was defined as positive, while a score lower than 0 was defined as negative.
MMR immunohistochemical staining interpretation criteria
The qualitative interpretation of MMR immunohistochemical reaction as positive, negative, or uncertain was based on the deposition of the staining product in the nuclei of cancer cells. Only when tumor cells were completely unstained in the nucleus, MLH1, MSH2, PMS2, or MSH6 expression was considered negative. Gastric adenocarcinoma lacking MLH1, MSH2, PMS2, or MSH6 expression was determined as mismatch repair deficient (dMMR), while gastric adenocarcinoma maintaining expression of all markers was interpreted as proficient mismatch repair (pMMR). Lymphocytes and normal epithelial cells served as positive internal controls.
HER2 immunohistochemical staining interpretation criteria
The interpretation was based on the Gastric Cancer HER2 Testing Guidelines (2016 Edition) (20): HER2 positivity was located on the tumor cell membrane, with no staining scored as 0; faint staining visible as 1+; weak to moderate staining as 2+; strong staining as 3+. Cases with immunohistochemical staining results of 2+ or 3+ did not require complete membrane staining. In gastric cancer, U-shaped staining (i.e., partial extracellular membrane staining) was considered positive. According to the HER2 guidelines for gastric cancer, cases with HER2 IHC 3+ or IHC 2+ with in situ hybridization (ISH) positive are classified as HER2-positive. Cases with IHC scores of 0 or 1+ are considered HER2-negative, and ISH testing is not required. For cases with an IHC score of 2+, further ISH testing is necessary to assess the HER2 gene amplification status (20,21).
Follow-up
Follow-up information was obtained by reviewing patient medical records and conducting telephone follow-ups with patients or their family members. Follow-up began after the pathological diagnosis was confirmed by surgical resection and was conducted monthly. The follow-up cut-off date was March 15, 2020. Follow-up was terminated for patients who were lost to follow-up or deceased. The study outcomes included OS and disease-free survival (DFS). DFS was defined as the time from the date of surgery to the first tumor metastasis/recurrence, or the last follow-up. OS was defined as the time from the date of surgery to the time of tumor-related death, or the last follow-up.
Observational indicators
(I) Comparison of FNDC1, HER2, and MMR protein expression in different groups; (II) analysis of the relationship between FNDC1, HER2, and MMR protein expression and the clinicopathological features of STAD; (III) analysis of prognostic survival based on patient follow-up, and evaluation of the predictive value of FNDC1, HER2, and MMR protein expression for the prognosis of STAD patients.
Statistical analysis
Statistical analysis was performed using SPSS 25.0 (IBM, China). Quantitative data were expressed as mean ± standard deviation (X±SD) and analyzed using independent sample t-tests. FNDC1, HER2 high expression or low expression, and MMR protein expression were expressed as absolute values or percentages [n (%)], and analyzed using the Chi-squared test. The correlation between the aforementioned immunohistochemical indicators and various clinicopathological parameters was analyzed using cross-tabulation and Fisher’s exact probability test, with Spearman’s nonparametric correlation coefficient used for testing. Kaplan-Meier survival curves and log-rank tests were used to assess high or low expression of FNDC1, HER2, and MMR protein in relation to disease outcomes (recurrence/metastasis, death). Multivariate analysis was performed using COX proportional hazards regression analysis. A P value <0.05 was considered statistically significant. The correlation coefficient r ranges between −1 and 1, with a larger absolute value indicating a stronger correlation.
Impact of FNDC1 on the biological functions of STAD Cells
Materials
This experiment included the normal gastric mucosal cell line GES-1 and the gastric cancer cell lines MNK-45, MGC-803, HGC-27, and BGC-823. All five cell lines were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, and maintained in the Center Laboratory of Zhongshan Hospital, Fudan University, at 37 ℃ in a humidified incubator with 5% CO2. The cells were cultured in RPMI1640/Ham’s F12 medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin. When the cell density reached 70–90%, passage culture was performed, and cell adhesion to the culture plates was observed using an inverted microscope.
Real-time quantitative polymerase chain reaction (RT-qPCR) was used to detect the mRNA levels of the FNDC1 gene in each cell line, and Western blot was used to assess the protein expression of FNDC1 in each cell line
RT-qPCR was used to detect the expression of FNDC1 mRNA. Total RNA from each group of cells was extracted using TRIzol reagent (Ambion, USA) and its purity was verified. Total cDNA was synthesized using the TaKaRa Reverse Transcription Kit. The primer sequences used were as follows: forward primer: 5'-CAACATTGCCTATGGGAAGTCA-3', reverse primer: 5'-CTCGATCCATTCACCTCCAG-3'. β-Actin was used as the internal reference gene with the following primer sequences: forward primer: 5'-AGGCACCAGGGCGTGAT-3', reverse primer: 5'-GCCCACATAGGAATCCTTCTGAC-3'. The PCR program consisted of an initial denaturation at 95 ℃ for 5 minutes, followed by denaturation at 95 ℃ for 10 seconds, annealing at 60 ℃ for 45 seconds, and extension at 95 ℃ for 15 seconds, repeated for a total of 45 cycles. The experiment was repeated three times. The results were presented as 2−ΔΔCt.
For FNDC1 siRNA transfection of MGC-803 cells, the cells were washed three times with PBS and collected by centrifugation. RIPA lysis buffer (MeilunBio, China) and PMSF (phenylmethylsulfonyl fluoride) were used to extract total cellular proteins. According to the instructions, Western blot was performed using FNDC1 (ES16354, ELK Biotechnology, 1:1,500) and β-Actin (Abmart, 1:10,000) antibodies.
Cell line construction, RT-qPCR, and Western blot to verify transfection efficiency
According to the instructions of Lipofectamine 8000 transfection reagent, synthetic siRNA-FNDC1 plasmids and control plasmids with unrelated sequences were transfected into MGC-803 cells. This established the siRNA-FNDC1 group, control vector transfected group, and a blank control group. After 24 hours of siRNA transfection, RT-qPCR was performed to measure the mRNA levels of the target gene. After 72 hours of siRNA transfection, Western blot was conducted to assess the protein expression in the cells. The methods were the same as previously described.
Cell Counting Kit-8 (CCK8) assay
MGC-803 and HGC-27 cells transfected with siRNA-FNDC1 were in the logarithmic growth phase. Cells were digested with trypsin and prepared as a cell suspension, which was then seeded in a 96-well plate (4,000 cells/well) with 6 replicate wells per group. The plate was incubated at 37 ℃ in a 5% CO2 incubator. When the cells adhered evenly to the bottom of the wells (around 70% confluence), transfection was performed according to the experimental groups. After 24 hours of incubation, 10 µL of CCK-8 reagent (DOJINDO LABORATORIES, Japan) was added to each well at 0, 24, 48, and 72 h. The plate was incubated for an additional 2 hours in the incubator, and the absorbance was measured at 450 nm using an enzyme-linked immunosorbent assay (ELISA) reader. The obtained absorbance values at each time point were compared to the 0 h values to generate a cell proliferation curve.
Cell scratch assay
MGC-803 and HGC-27 cells were seeded in a 6-well plate and transfected with FNDC1 siRNA. A vertical scratch was made on the back of the plate using a 200-µL pipette tip. The wells were washed three times with 1× PBS and then cultured in DMEM medium containing 2% serum. Images of the scratches were taken and recorded under a 40× microscope to ensure consistent positioning. Subsequently, the cells were incubated at 37 ℃ in a 5% CO2 incubator for 6, 12, and 24 h, and photographs were taken and recorded. The scratch area of each group was analyzed using Image J software to calculate the relative migration rate at 24 hours.
Transwell invasion assay
The day before the experiment, thaw Matrigel gel from −20 ℃ freezer to 4 ℃ refrigerator overnight. Mix Matrigel gel with pre-chilled serum-free culture medium in a 1:8 ratio. In each well of the Transwell chamber, seed 200 µL of serum-free culture cell suspension containing 2.5×105 cells/mL. Add 500 µL of culture medium containing 10% FBS to the lower chamber of the 24-well plate. Incubate at 37 ℃, 5% CO2 in a regular incubator for 48 hours. Fix the cells in the upper chamber with 4% paraformaldehyde at room temperature for 15 minutes. Stain with 1 mL of 0.1% crystal violet for 10 minutes. Capture images using a microscope. Analyze and count the cells using Image J software.
Results
Genetic variation analysis of FNDC1 gene in pan-cancer
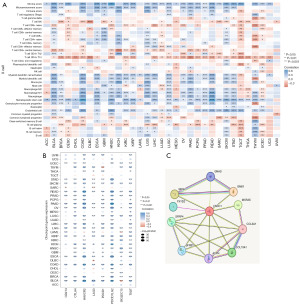
In a pan-cancer analysis, we found that among 10,967 samples, 438 samples (4%) had mutations in the FNDC1 gene. The main genetic alterations of FNDC1 include mutations (in green) and deep deletions (in blue), and the mutation frequency of the FNDC1 gene varies with tumor type. The top two tumors with a mutation frequency of 10% are malignant melanoma of the skin (15.77%) and endometrial cancer (12.48%). In the TCGA database, the most common type of FNDC1 gene alteration is mutation, with the highest frequencies observed in skin melanoma (14.64%) and endometrial cancer (12.1%) (Figure 1A). Further exploration revealed that the main mutation of the FNDC1 gene in pan-cancer is missense mutation, with the A624T/V/E mutation occurring most frequently, observed in colorectal adenocarcinoma, low-grade glioma, and STAD.
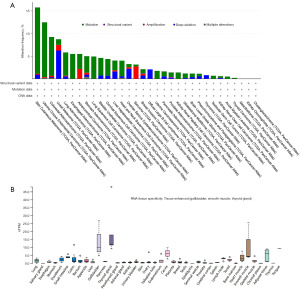
Analysis of FNDC1 messenger RNA (mRNA) expression in normal tissues and pan-cancer
The mRNA expression levels of FNDC1 in various normal tissues were analyzed using the HPA database, showing that FNDC1 is highly expressed in the gallbladder, thyroid, and smooth muscle tissues in the normal human body (Figure 1B). Analysis of FNDC1 expression levels in 33 different types of tumors using the TCGA database revealed that FNDC1 expression is tumor-specific, with the highest expression observed in thyroid cancer (Figure 2A). Differential expression of FNDC1 mRNA in 21 types of tumors was analyzed using the GENE DENOVO database, demonstrating that in 10 types of tumor tissues, FNDC1 mRNA expression levels were higher than in normal tissues, while in 3 types of tumor tissues, they were lower than in normal tissues, and these differences were statistically significant (P<0.05) (Figure 2B). Furthermore, analysis using the Sangerbox online analysis platform showed that in comparison to normal tissues, FNDC1 mRNA expression was upregulated in seven types of tumor tissues and downregulated in two types of tumor tissues among the 33 tumor types analyzed (P<0.05) (Figure 2C,2D). Integrating the results from the analysis of these two databases, it is evident that FNDC1 mRNA expression levels were higher than those in normal tissues in seven types of tumor tissues, including BRCA, CHOL, COAD, ESCA, HNSC, KIRC, and STAD, indicating that FNDC1 is generally highly expressed in various tumor tissues.
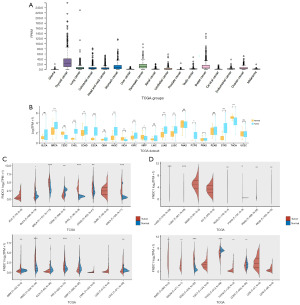
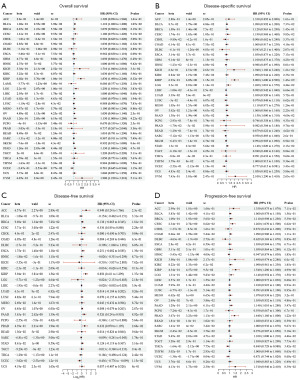
Prognostic analysis of FNDC1 in pan-cancer
The Kaplan-Meier survival analysis investigated the clinical prognostic relevance of FNDC1 in different types of tumors. For instance, the high expression of FNDC1 was associated with poor OS in BLCA, KICH, KIRP, LGG, PAAD, STAD, and UVM, but correlated with favorable prognosis in UCEC (Figure 3A). The high expression of FNDC1 was correlated with poor disease-specific survival (DSS) in KICH, KIRP, LGG, PAAD, STAD, and UVM, but associated with favorable prognosis in UCEC (Figure 3B). The high expression of FNDC1 was associated with poor DFS in ACC, CESC, KIRP, PAAD, and PRAD, but correlated with favorable prognosis in UCEC (Figure 3C). The high expression of FNDC1 was associated with poor progression-free survival (PFS) in ACC, BRCA, CESC, KICH, KIRP, PAAD, PRAD, and STAD, but correlated with favorable prognosis in THCA and UCEC (Figure 3D). These results indicate that FNDC1 expression has a strong prognostic predictive ability in different tumors, with high FNDC1 expression often associated with poor prognosis.
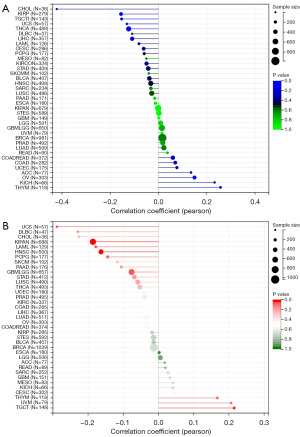
Analysis of FNDC1 and tumor-related features in pan-cancer
We calculated the Pearson correlation between FNDC1 and TMB in each tumor type. We observed a significant positive correlation between FNDC1 expression and TMB in two types of tumors, including THYM (N=118) (R=0.26, P=0.005) and OV (N=303) (R=0.15, P=0.009). There was a significant negative correlation in four types of tumors, including KIRP (N=279) (R=−0.16, P=0.009), LIHC (N=357) (R=−0.11, P=0.04), THCA (N=489) (R=−0.12, P=0.006), CHOL (N=36) (R=−0.42, P=0.01) (Figure 4A). FNDC1 expression was significantly positively correlated with MSI in TGCT (N=148) (R=0.21, P=0.009) and significantly negatively correlated in four types of tumors, including LAML (N=129) (R=−0.18, P=0.04), pan-kidney cohort (KIPAN) (N=688) (R=−0.19, P<0.001), HNSC (N=500) (R=−0.16, P<0.001), and UCS (N=57) (R=−0.29, P=0.03) (Figure 4B).
Correlation analysis of FNDC1 expression with immune cell infiltration and immune checkpoints
FNDC1 exhibits strong positive correlations with immune cells and stromal cells in various tumors, including BLCA, COAD, ESCA, HNSC, and KICH. In STAD, FNDC1 shows significant correlations with cell infiltrations such as endothelial cells, eosinophils, granulocyte-monocyte progenitors, hematopoietic stem cells, macrophage M1, macrophage M2, monocytes, activated myeloid dendritic cells, and myeloid dendritic cells (Figure 5A). Considering the importance of immune checkpoint regulators in tumor immunity, we analyzed the expression of eight immune checkpoint-associated genes in different tumors. In STAD, FNDC1 exhibits significant positive correlations with HAVCR2 and PDCD1LG2 (Figure 5B).
FNDC1-associated proteins and signaling pathways
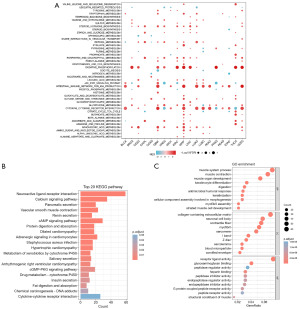
Proteins that are similar to the FNDC1 expression pattern and are in the top 100 list include GNAS, GNB1, MXRA5, COL3A1, COL10A1, ASPN, SFRP2, SFRP4, FXYD2, and GNG2. The reported associated signaling pathways include the Wnt pathway and mTOR pathway (Figure 5C). Using GSEA analysis in pan-cancer, we found that the expression level of FNDC1 in pan-cancer is associated with several immune response and metabolic pathways, such as cytokine-cytokine receptor interaction, intestinal-immune-network-for-IgA-production, oxidative-phosphorylation, arachidonic-acid-metabolism, and retinol-metabolism (Figure 6A). Further KEGG pathway enrichment analysis revealed that the FNDC1-related genes in STAD are involved in pathways such as Neuroactive ligand-receptor interaction, Calcium signaling pathway, cAMP signaling pathway, Vascular smooth muscle contraction, and Pancreatic secretion (Figure 6B). GO functional enrichment analysis results showed that the FNDC1-related genes in STAD are associated with pathways such as muscle system process, collagen-containing extracellular matrix, and receptor ligand activity (Figure 6C).
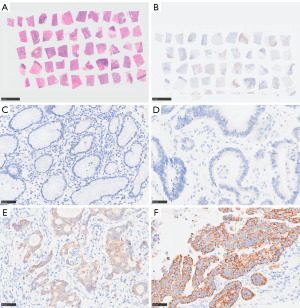
Expression of FNDC1, HER2, and MMR proteins in STAD samples
In this study, ISH testing was performed on 67 STAD samples, revealing HER2 amplification in 26 cases and no amplification in 41 cases. Statistical analysis indicated a strong correlation between HER2 amplification and positive HER2 protein expression, with a significant consistency observed (P<0.001). Therefore, we subsequently used the results of HER2 immunohistochemistry for statistical analysis. See Table 1.
Table 1
| HER-2 IHC | HER-2 ISH | Total | P value | χ2 | |
|---|---|---|---|---|---|
| Non-amplification | Amplification | ||||
| Negative | 35 | 1 | 36 | <0.001 | 42.530 |
| Positive | 6 | 25 | 31 | ||
| Total | 41 | 26 | 67 | ||
HER2, human epidermal growth factor receptor 2; IHC, immunohistochemistry; ISH, in situ hybridization; STAD, stomach adenocarcinoma.
The positive expression rates of FNDC1 and dMMR in STAD group were higher than those in the control group, and the difference was statistically significant (P<0.001). However, there was no significant statistical difference in the positive expression rate of HER2 between the gastric adenocarcinoma group and the control group. See Table 2, Figure 7A-7F.
Table 2
| Proteins | STAD group (n=741) | Control group (n=663) | χ2 | P value |
|---|---|---|---|---|
| FNDC1 | 336 | <0.001 | ||
| Positive | 453 (61.13) | 89 (13.42) | ||
| Negative | 288 (38.87) | 574 (86.58) | ||
| HER2 | 0.09 | 0.76 | ||
| Negative | 667 (90.01) | 600 (90.50) | ||
| Positive | 74 (9.99) | 63 (9.50) | ||
| MMR | 28.8 | <0.001 | ||
| dMMR | 40 (5.40) | 3 (0.45) | ||
| pMMR | 701 (94.60) | 660 (99.55) |
Data are presented as n (%). FNDC1, fibronectin type III domain-containing 1; HER2, human epidermal growth factor receptor 2; MMR, mismatch repair; dMMR, mismatch repair deficient; pMMR, proficient mismatch repair.
Relationship between the expression levels of FNDC1, HER2, and MMR proteins and the clinical pathological features of STAD
A total of 741 cases of gastric cancer were analyzed for various pathological parameters: age, gender, tumor location, tumor size, differentiation degree, Lauren classification, lymphovascular invasion, neural invasion, signet ring cell carcinoma component, tumor deposit, postoperative recurrence, T staging, N staging, M staging, and TNM staging. We used statistical methods to analyze the relationship between the expression of FNDC1, HER2, and MMR proteins and the aforementioned clinical pathological parameters in gastric adenocarcinoma. The statistical analysis results showed that the difference in the expression level of the FNDC1 protein was significantly associated with age, Lauren classification, lymphovascular invasion, neural invasion, the presence of signet ring cell carcinoma component, T staging, and TNM staging (P<0.05). This indicates that the expression level of FNDC1 is higher in older gastric cancer patients, those with more diffuse or mixed Lauren classification, lymphovascular invasion, neural invasion, signet ring cell carcinoma component, higher T staging, and higher postoperative pathological TNM staging. However, the difference in the expression level of the FNDC1 protein was not statistically significant in relation to gender, tumor location, differentiation degree, tumor deposit, postoperative recurrence, N staging, and M staging. The positive expression of HER2 was statistically significantly different from negative/low expression in terms of Lauren classification (P<0.001), signet ring cell carcinoma component (P<0.001), T staging (P=0.03), TNM staging (P=0.04), and postoperative recurrence (P=0.03), but had no significant correlation with other pathological parameters. The difference in the expression level of MMR protein was statistically significant in comparison with gender (P=0.04), tumor location (P=0.01), tumor size (P=0.004), neural invasion (P<0.001), signet ring cell carcinoma component (P=0.02), and postoperative recurrence (P=0.009), but had no significant correlation with other pathological parameters. See Tables 3-5.
Table 3
| Clinical pathological characteristics | Case | FNDC1 expression | χ2 | P value | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Age (years) | 9.622 | 0.002 | |||
| ≤60 | 303 | 165 | 138 | ||
| >60 | 438 | 288 | 150 | ||
| Sex | 2.349 | 0.13 | |||
| Male | 518 | 326 | 192 | ||
| Female | 223 | 127 | 96 | ||
| Tumor location | 1.764 | 0.18 | |||
| Lower part of the stomach | 307 | 179 | 128 | ||
| Other location | 434 | 274 | 160 | ||
| Tumor size (cm) | 0.588 | 0.44 | |||
| ≤5 | 581 | 351 | 230 | ||
| >5 | 160 | 102 | 58 | ||
| Differentiation degree | 0.112 | 0.74 | |||
| Well/moderately | 86 | 54 | 32 | ||
| Poorly | 655 | 399 | 256 | ||
| Lauren classification | 14.623 | <0.001 | |||
| Intestinal type | 233 | 166 | 67 | ||
| Diffuse/mixed type | 508 | 287 | 221 | ||
| Lymphovascular invasion | 16.788 | <0.001 | |||
| Yes | 332 | 230 | 102 | ||
| No | 409 | 223 | 186 | ||
| Neural invasion | 0.083 | 0.77 | |||
| Yes | 396 | 244 | 152 | ||
| No | 345 | 209 | 136 | ||
| Signet ring cell carcinoma | 39.454 | <0.001 | |||
| Yes | 251 | 114 | 137 | ||
| No | 490 | 339 | 151 | ||
| Tumor deposit | 2.272 | 0.13 | |||
| Yes | 152 | 101 | 51 | ||
| No | 589 | 352 | 237 | ||
| Postoperative recurrence | 0.152 | 0.70 | |||
| Yes | 274 | 170 | 104 | ||
| No | 467 | 283 | 184 | ||
| T staging | 5.446 | 0.02 | |||
| T1 | 153 | 81 | 72 | ||
| T2–4 | 588 | 372 | 216 | ||
| N staging | 3.56 | 0.059 | |||
| N0 | 260 | 147 | 113 | ||
| N1–3 | 481 | 306 | 175 | ||
| M staging | 0.003 | 0.96 | |||
| M0 | 736 | 450 | 286 | ||
| M1 | 5 | 3 | 2 | ||
| TNM staging | 4.28 | 0.04 | |||
| I–II staging | 343 | 196 | 147 | ||
| III–IV staging | 398 | 257 | 141 | ||
FNDC1, fibronectin type III domain-containing 1; STAD, stomach adenocarcinoma; TNM, tumor-node-metastasis.
Table 4
| Clinical pathological characteristics | Case | HER2 expression | χ2 | P value | |
|---|---|---|---|---|---|
| Negative | Positive | ||||
| Age (years) | 3.273 | 0.07 | |||
| ≤60 | 303 | 280 | 23 | ||
| >60 | 438 | 387 | 51 | ||
| Sex | 1.301 | 0.26 | |||
| Male | 518 | 462 | 56 | ||
| Female | 223 | 205 | 18 | ||
| Tumor location | 0.027 | 0.87 | |||
| Lower part of the stomach | 307 | 277 | 30 | ||
| Other location | 434 | 390 | 44 | ||
| Tumor size (cm) | 0.093 | 0.76 | |||
| ≤5 | 581 | 524 | 57 | ||
| >5 | 160 | 143 | 17 | ||
| Differentiation degree | 2.848 | 0.09 | |||
| Well/moderately | 86 | 73 | 13 | ||
| Poorly | 655 | 594 | 61 | ||
| Lauren classification | 24.435 | <0.001 | |||
| Intestinal type | 233 | 191 | 42 | ||
| Diffuse/mixed type | 508 | 476 | 32 | ||
| Lymphovascular invasion | 0.282 | 0.60 | |||
| Yes | 332 | 301 | 31 | ||
| No | 409 | 366 | 43 | ||
| Neural invasion | 0.127 | 0.72 | |||
| Yes | 396 | 355 | 41 | ||
| No | 345 | 312 | 33 | ||
| Signet ring cell carcinoma | 13.261 | <0.001 | |||
| Yes | 251 | 240 | 11 | ||
| No | 490 | 427 | 63 | ||
| Tumor deposit | 0.062 | 0.80 | |||
| Yes | 152 | 136 | 16 | ||
| No | 589 | 531 | 58 | ||
| Postoperative recurrence | 4.806 | 0.03 | |||
| Yes | 274 | 238 | 36 | ||
| No | 467 | 429 | 38 | ||
| T staging | 4.855 | 0.03 | |||
| T1 | 153 | 145 | 8 | ||
| T2–4 | 588 | 522 | 66 | ||
| N staging | 0.061 | 0.80 | |||
| N0 | 260 | 235 | 25 | ||
| N1–3 | 481 | 432 | 49 | ||
| M staging | 0.558 | 0.46 | |||
| M0 | 736 | 662 | 74 | ||
| M1 | 5 | 5 | 0 | ||
| TNM staging | 4.071 | 0.04 | |||
| I staging | 181 | 170 | 11 | ||
| II–IV staging | 560 | 497 | 63 | ||
HER2, human epidermal growth factor receptor 2; STAD, stomach adenocarcinoma; TNM, tumor-node-metastasis.
Table 5
| Clinical pathological characteristics | Cases | MMR protein expression | χ2 | P value | |
|---|---|---|---|---|---|
| pMMR | dMMR | ||||
| Age (years) | 3.137 | 0.08 | |||
| ≤60 | 303 | 292 | 11 | ||
| >60 | 438 | 409 | 29 | ||
| Sex | 4.465 | 0.04 | |||
| Male | 518 | 496 | 22 | ||
| Female | 223 | 205 | 18 | ||
| Tumor location | 6.009 | 0.01 | |||
| Lower part of the stomach | 307 | 283 | 24 | ||
| Other location | 434 | 418 | 16 | ||
| Tumor size (cm) | 8.462 | 0.004 | |||
| ≤5 | 581 | 557 | 24 | ||
| >5 | 160 | 144 | 16 | ||
| Differentiation degree | 0.166 | 0.77 | |||
| Well/moderately | 86 | 82 | 4 | ||
| Poorly | 655 | 619 | 36 | ||
| Lauren classification | 2.398 | 0.12 | |||
| Intestinal type | 233 | 216 | 17 | ||
| Diffuse/mixed type | 508 | 485 | 23 | ||
| Lymphovascular invasion | 0.462 | 0.50 | |||
| Yes | 332 | 312 | 20 | ||
| No | 409 | 389 | 20 | ||
| Neural invasion | 13.746 | <0.001 | |||
| Yes | 396 | 386 | 10 | ||
| No | 345 | 315 | 30 | ||
| Signet ring cell carcinoma | 5.06 | 0.02 | |||
| Yes | 251 | 244 | 7 | ||
| No | 490 | 457 | 33 | ||
| Tumor deposit | 0.007 | 0.93 | |||
| Yes | 152 | 144 | 8 | ||
| No | 589 | 557 | 32 | ||
| Postoperative recurrence | 6.883 | 0.009 | |||
| Yes | 274 | 267 | 7 | ||
| No | 467 | 434 | 33 | ||
| T staging | 0.823 | 0.36 | |||
| T1 | 153 | 147 | 6 | ||
| T2–4 | 588 | 554 | 34 | ||
| N staging | 0.448 | 0.50 | |||
| N0 | 260 | 244 | 16 | ||
| N1–3 | 481 | 457 | 24 | ||
| M staging | 0.287 | 0.59 | |||
| M0 | 736 | 696 | 40 | ||
| M1 | 5 | 5 | 0 | ||
| TNM staging | 1.291 | 0.26 | |||
| I–II staging | 343 | 321 | 22 | ||
| III–IV staging | 398 | 380 | 18 | ||
dMMR, mismatch repair deficient; MMR, mismatch repair; pMMR, proficient mismatch repair; STAD, stomach adenocarcinoma; TNM, tumor-node-metastasis.
Correlation analysis of FNDC1, HER2, and MMR protein expression in STAD tissue
In STAD tissue, a correlation analysis between FNDC1 and HER2 showed a positive correlation between their expression levels (r=0.109, P=0.003). However, a correlation analysis between FNDC1 and MMR in STAD tissue showed that the expression levels of the two were not significantly correlated (r=0.068, P=0.06). No clear statistical significance was found in the correlation analysis of HER2 and MMR expression levels in STAD tissue. See Tables 6,7.
Table 6
| FNDC1 | Positive (n=453) | Negative (n=288) | r | P value |
|---|---|---|---|---|
| HER2 | 0.109 | 0.003 | ||
| Negative | 396 | 271 | ||
| Positive | 57 | 17 | ||
| MMR | 0.068 | 0.06 | ||
| dMMR | 30 | 10 | ||
| pMMR | 423 | 278 |
FNDC1, fibronectin type III domain-containing 1; HER2, human epidermal growth factor receptor 2; MMR, mismatch repair; dMMR, mismatch repair deficient; pMMR, proficient mismatch repair; STAD, stomach adenocarcinoma.
Table 7
| MMR protein expression | HER2 | Total | r | P value | |
|---|---|---|---|---|---|
| Negative | Positive | ||||
| pMMR | 629 | 72 | 701 | −0.04 | 0.42 |
| dMMR | 38 | 2 | 40 | ||
HER2, human epidermal growth factor receptor 2; MMR, mismatch repair; dMMR, mismatch repair deficient; pMMR, proficient mismatch repair; STAD, stomach adenocarcinoma.
Analysis of risk factors affecting the prognosis and mortality of STAD patients
In order to further explore the relationship between the expression level of FNDC1 and the prognosis of gastric adenocarcinoma patients, we performed statistical analysis on the immunohistochemical results of FNDC1 in 741 cases of gastric cancer tissue. However, the statistical results showed that there was no significant statistical difference in the comparison of DFS and OS between the high expression group of FNDC1 and the low expression group. Univariate analysis results showed that age (P<0.001), tumor size (P<0.001), differentiation degree (P<0.001), Lauren classification (P=0.001), lymphovascular invasion (P<0.001), neural invasion (P<0.001), tumor deposit (P<0.001), postoperative recurrence (P<0.001), T staging (P<0.001), N staging (P<0.001), M staging (P<0.001), TNM staging (P<0.001), HER2 expression status (P=0.01), and MMR expression status (P=0.02) were associated with poor prognosis of gastric adenocarcinoma patients (P<0.05). Further multivariate Cox regression analysis showed that age (P=0.046), tumor size (P=0.050), Lauren classification (P=0.02), lymphovascular invasion (P=0.01), neural invasion (P=0.03), and postoperative recurrence (P<0.001) were independent risk factors affecting the prognosis. See Tables 8,9.
Table 8
| Variables | Single factor | |
|---|---|---|
| Mean (95% CI) | P value | |
| Age (years) | <0.001 | |
| ≤60 | 72.257 (69.207–75.307) | |
| >60 | 62.010 (58.932–65.087) | |
| Sex | 0.18 | |
| Male | 1.379 (62.964–68.372) | |
| Female | 67.638 (63.632–71.644) | |
| Tumor location | 0.07 | |
| Lower part of the stomach | 67.760 (64.392–71.127) | |
| Other location | 65.197 (62.265–68.128) | |
| Tumor size (cm) | <0.001 | |
| ≤5 | 69.883 (67.534–73.231) | |
| >5 | 54.090 (48.819–59.362) | |
| Differentiation degree | <0.001 | |
| Well/moderately | 65.197 (62.387–68.006) | |
| Poorly | 64.580 (62.171–66.988) | |
| Lauren classification | 0.001 | |
| Intestinal type | 57.763 (55.334–60.192) | |
| Diffuse/mixed type | 64.068 (61.313–66.822) | |
| Lymphovascular invasion | <0.001 | |
| Yes | 57.646 (54.047–61.245) | |
| No | 73.088 (70.550–75.626) | |
| Neural invasion | <0.001 | |
| Yes | 56.152 (52.902–59.402) | |
| No | 77.832 (75.436–80.228) | |
| Signet ring cell carcinoma | 0.52 | |
| Yes | 64.902 (60.925–68.879) | |
| No | 66.990 (64.362–69.619) | |
| Tumor deposit | <0.001 | |
| Yes | 44.521 (39.688–49.355) | |
| No | 72.036 (69.772–74.300) | |
| Postoperative recurrence | <0.001 | |
| Yes | 31.368 (28.819–33.916) | |
| No | 87.719 (87.312–88.125) | |
| T staging | <0.001 | |
| T1 | 82.833 (80.086–85.580) | |
| T2–4 | 62.083 (59.483–64.684) | |
| N staging | <0.001 | |
| N0 | 80.423 (77.988–82.858) | |
| N1–3 | 58.664 (55.737–61.591) | |
| M staging | <0.001 | |
| M0 | 66.823 (64.595–69.050) | |
| M1 | 26.000 (9.288–42.712) | |
| TNM staging | <0.001 | |
| I–II staging | 80.412 (78.211–82.613) | |
| III–IV staging | 54.260 (51.044–57.476) | |
| FNDC1 | 0.91 | |
| Positive | 66.653 (63.850–69.456) | |
| Negative | 65.826 (62.251–69.401) | |
| HER2 expression | 0.01 | |
| Positive | 50.813 (44.898–56.728) | |
| Negative | 67.427 (65.112–69.741) | |
| MMR protein expression | 0.02 | |
| pMMR | 65.779 (63.460–68.097) | |
| dMMR | 79.340 (73.810–84.870) | |
CI, confidence interval; FNDC1, fibronectin type III domain-containing 1; HER2, human epidermal growth factor receptor 2; MMR, mismatch repair; dMMR, mismatch repair deficient; pMMR, proficient mismatch repair; STAD, stomach adenocarcinoma; TNM, tumor-node-metastasis.
Table 9
| Covariates | SE | Wald | Exp(B) | 95 % CI of Exp(B) | P value | |
|---|---|---|---|---|---|---|
| Lower limit | Upper limit | |||||
| Age | 0.144 | 3.972 | 0.751 | 0.566 | 0.995 | 0.046 |
| Tumor size | 0.15 | 3.85 | 0.746 | 0.556 | 1 | 0.050 |
| Lauren classification | 0.178 | 5.434 | 1.515 | 1.068 | 2.147 | 0.02 |
| Lymphovascular invasion | 0.147 | 6.214 | 0.692 | 0.519 | 0.924 | 0.01 |
| Neural invasion | 0.186 | 4.564 | 0.673 | 0.468 | 0.968 | 0.03 |
| Postoperative recurrence | 0.591 | 96.725 | 0.003 | 0.001 | 0.009 | <0.001 |
CI, confidence interval; FNDC1, fibronectin type III domain-containing 1; STAD, stomach adenocarcinoma.
Expression levels of FNDC1 in human STAD cell lines
Western blot analysis of FNDC1 protein levels
We selected human gastric cancer cell lines (MNK-45, HGC-27, MGC-803, BGC-823) and GES-1 for analysis. The results showed that the protein expression levels of FNDC1 in gastric cancer cell lines were significantly higher than in GES-1 cells, with the highest expression levels observed in MGC-803 and HGC-27 cells, while BGC-823 cells had relatively lower expression levels (Figure 8A).
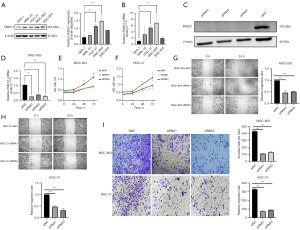
RT-qPCR analysis of FNDC1 mRNA levels
We selected human gastric cancer cell lines (MNK-45, HGC-27, MGC-803, BGC-823) and GES-1 for analysis. The results showed that the mRNA transcription levels of FNDC1 in gastric cancer cell lines were significantly higher than in GES-1 cells, with the highest transcription levels observed in MGC-803 and HGC-27 cells, while BGC-823 cells had relatively lower transcription levels (Figure 8B).
Effects of FNDC1 on proliferation, migration and invasion of gastric cancer cells
Based on the above results, to further investigate the biological functions of FNDC1 in gastric cancer, we transfected MGC-803 and HGC-27 cells with three siRNA knockdown vectors and verified the transfection efficiency through RT-PCR and Western blot. The transfected groups showed lower mRNA and protein expression levels, with siRNA1 and siRNA3 showing higher knockdown efficiency (Figure 8C,8D). Therefore, we selected siRNA1 and siRNA3 for further experiments. We used the CCK8 assay kit to detect the cell proliferation curves. The results showed that compared to the MGC-803-siRNC group, the cell proliferation rates in the MGC-803-siRNA1-FNDC1 group and the MGC-803-siRNA3-FNDC1 group were significantly reduced, and a similar trend was observed in the HGC-27 cells (Figure 8E,8F). Cell scratch assay results showed that after FNDC1 knockdown, the healing and migration abilities of MGC-803 and HGC-27 cells were significantly weakened (Figure 8G,8H). The results of the Transwell experiments showed that, compared to the MGC-803-siRNC group, the invasion ability of cells in the MGC-803-siRNA1-FNDC1 group and the MGC-803-siRNA3-FNDC1 group was significantly reduced, and a similar trend was observed in the HGC-27 cells (Figure 8I).
Discussion
STAD has a high incidence globally, and early detection is difficult, leading to poor prognosis and posing a serious threat to human life and health. While various comprehensive treatment options such as surgical resection, immunotherapy, targeted therapy, chemotherapy, and radiotherapy can prolong patient survival, many patients still experience recurrence or metastasis, leading to death. Therefore, early detection, early diagnosis, and the search for appropriate biomarkers play a crucial role in the active treatment and prognosis of patients. In recent years, proteins such as FNDC1, HER2, and MMR have become hotspots of research focus in the field of oncology.
FNDC1 is a gene that has received increasing attention in recent years due to its close association with the biological behavior of tumors. It is well known that the destruction of the extracellular matrix and the generation of new blood vessels in tumor tissue are important conditions for the aggressive metastasis of tumors. FNDC1 is one of the main structural domains of FN, and FN, as an extracellular matrix protein, plays a key role in the destruction of the extracellular matrix. Hayashi et al. (22) found that reducing the expression of FNDC1 can inhibit angiogenesis, as well as cell growth, migration, and proliferation. Increasingly, research results from both domestic and international sources indicate that FNDC1 is overexpressed in various types of tumor tissues. Chen et al. (23) found that overexpression of FNDC1 promotes resistance of colorectal cancer cells to radiotherapy and chemotherapy, leading to increased survival of residual tumor cells. Hong et al. (24) demonstrated that high expression of FNDC1 in lung adenocarcinoma patients is closely associated with the occurrence, progression, and prognosis of the tumor. Yunwen et al. (25) suggested that downregulation of FNDC1 expression can inhibit the proliferation, migration, and invasion of breast cancer cells. Additionally, upregulation of expression has also been observed in esophageal cancer and inflammatory bowel disease (26,27). Undoubtedly, some researchers have conducted small-sample studies on the expression and prognosis of FNDC1 in gastric cancer. The results indicate that high expression of FNDC1 is associated with poor prognosis in gastric cancer (28-30). Therefore, this study primarily investigates the prognostic and immunological functions of FNDC1 in human tumors, moving from pan-cancer analysis to validation in STAD. The overall framework starts with a pan-cancer analysis using public databases, followed by clinical sample and cell line functional validation focused on STAD.
This study also used tissue microarray technology to perform immunohistochemical staining and statistical analysis on 741 cases of gastric cancer tissue and 663 cases of normal gastric tissue. The results showed a higher positive expression rate of FNDC1 in gastric cancer tissue, indicating a higher FNDC1 protein content. Furthermore, as the T stage increased and TNM stage progressed, the expression rate of FNDC1 also increased, suggesting that FNDC1 is a cancer-promoting gene, consistent with previous literature reports. In this study, the immunohistochemical staining results of FNDC1 were interpreted based on the positive range and intensity, using all the criteria reported in previous literature and the receiver operating characteristic (ROC) curve to define the meaningful cutoff value for high or low expression of FNDC1 associated with OS and DFS. After multiple attempts, this study temporarily did not find a statistically significant difference in OS and DFS associated with high expression of FNDC1 in gastric cancer. However, the results of univariate survival analysis showed that older age, larger tumor diameter, lower differentiation degree, more diffuse or mixed type in Lauren classification, presence of vascular cancer embolus, nerve bundle invasion, and cancer nodes were associated with poor prognosis in gastric cancer patients. Further multivariate Cox regression analysis revealed that the differentiation degree, T stage, N stage, M stage, and TNM stage were omitted, and only age, tumor size, Lauren classification, vascular cancer embolus, nerve bundle invasion, and postoperative recurrence were related to OS. The omission of differentiation degree, T stage, N stage, M stage, and TNM stage was primarily due to the significant correlation found in the correlation analysis between these parameters and many clinical pathological parameters, leading to automatic omission in the statistical analysis. Although both univariate and multivariate analyses in this study did not show a significant statistical significance between high expression of FNDC1 and poor prognosis in gastric adenocarcinoma patients, its significant correlation with clinical pathological parameters such as age, Lauren classification, vascular cancer emboli, neural bundle invasion, presence of signet ring cell carcinoma component, T stage, TNM stage, etc., suggests a certain correlation between FNDC1 and the progression, invasion, and migration of gastric cancer. This preliminary finding is further supported by the in vitro experiments conducted in this study.
HER2, a transmembrane protein located on human chromosome 17 (17q21), is one of the human proto-oncogenes. Previous studies have shown that HER2 can be overexpressed or amplified in a variety of malignant tumors, such as breast cancer, prostate cancer, lung cancer, and bladder cancer, and is closely related to the occurrence and development of tumors. A large-scale multicenter study evaluating HER2 in 1,148 patients who underwent gastric cancer resection surgery found that HER2 overexpression is an important factor for poor prognosis in gastric cancer patients (31). As the earliest discovered therapeutic target in gastric cancer, there are currently classic anti-HER2 drugs, such as trastuzumab. However, the positive expression rate of HER2 in gastric cancer is not consistent (32,33). In this study, the positive expression rate of HER2 in gastric cancer was 9.99%, slightly lower than reported in the literature. Statistical results showed that HER2-positive patients had a later T stage and TNM stage, and a higher postoperative recurrence rate compared to HER2-negative/low-expression patients. Correlation analysis between FNDC1 and HER2 in gastric cancer tissue showed a positive correlation between their expression levels, indicating that higher expression of FNDC1 was associated with a higher HER2 positivity rate, while lower expression of FNDC1 was associated with a lower HER2 positivity rate. Survival analysis in this study indicated that HER2-positive patients had shorter OS and DFS compared to HER2-negative/low-expression patients, suggesting a greater likelihood of poor prognosis with overexpression of FNDC1 and HER2 in gastric cancer patients.
However, there are limitations in this study: firstly, as a retrospective study, the sample size related to FNDC1 expression in gastric cancer is relatively small, and gene testing was not conducted to verify the results of immunohistochemistry, requiring further experimental studies for validation. Secondly, there were only about 40 articles related to FNDC1, and reports on human tumors are rare, and its mechanism in the occurrence and development of gastric cancer still needs further exploration and verification at the cellular and experimental animal levels.
Conclusions
In summary, this study utilized bioinformatics approaches to investigate the genetic variations, mRNA expression levels, prognostic value, MSI, TMB, immunological relevance, and pathway enrichment analysis of FNDC1 across different tumors. Building on the preliminary pan-cancer analysis, we focused on STAD and conducted in vitro validations using clinical samples and STAD cell lines to assess the expression level, prognostic value, and cellular functions of FNDC1 in STAD. The aforementioned results suggest that the expression level of FNDC1 has the potential to serve as an important biomarker for predicting prognosis and evaluating the effectiveness of immunotherapy in human cancers.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the REMARK and MDAR reporting checklists. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2279/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2279/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2279/prf
Funding: The study was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2279/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study was approved by the ethics committee of Zhongshan Hospital (No. B2021-842R) and informed consent was taken from all the participant.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229-63. [Crossref] [PubMed]
- Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol 2019;14:26-38. [Crossref] [PubMed]
- Chinese Society of Clinical Oncology, Chinese Medical Association, Chinese Medical Association Publishing House. Clinical diagnosis and treatment guidelines for gastric cancer of Chinese Medical Association (2021 edition). Chinese Medical Journal 2022;102:1169-89.
- Yaghoobi M, McNabb-Baltar J, Bijarchi R, et al. What is the quantitative risk of gastric cancer in the first-degree relatives of patients? A meta-analysis. World J Gastroenterol 2017;23:2435-42. [Crossref] [PubMed]
- Murphy G, Dawsey SM, Engels EA, et al. Cancer Risk After Pernicious Anemia in the US Elderly Population. Clin Gastroenterol Hepatol 2015;13:2282-9.e1-4.
- Morgagni P, Gardini A, Marrelli D, et al. Gastric stump carcinoma after distal subtotal gastrectomy for early gastric cancer: experience of 541 patients with long-term follow-up. Am J Surg 2015;209:1063-8. [Crossref] [PubMed]
- Wroblewski LE, Peek RM Jr. Helicobacter pylori, Cancer, and the Gastric Microbiota. Adv Exp Med Biol 2016;908:393-408. [Crossref] [PubMed]
- Chon SH, Berlth F, Plum PS, et al. Gastric cancer treatment in the world: Germany. Transl Gastroenterol Hepatol 2017;2:53. [Crossref] [PubMed]
- Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci 2002;115:3861-3. [Crossref] [PubMed]
- Sato M, Cismowski MJ, Toyota E, et al. Identification of a receptor-independent activator of G protein signaling (AGS8) in ischemic heart and its interaction with Gbetagamma. Proc Natl Acad Sci U S A 2006;103:797-802. [Crossref] [PubMed]
- Das DK, Naidoo M, Ilboudo A, et al. miR-1207-3p regulates the androgen receptor in prostate cancer via FNDC1/fibronectin. Exp Cell Res 2016;348:190-200. [Crossref] [PubMed]
- van Ingen G, Li J, Goedegebure A, et al. Genome-wide association study for acute otitis media in children identifies FNDC1 as disease contributing gene. Nat Commun 2016;7:12792. [Crossref] [PubMed]
- Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648-60. [Crossref] [PubMed]
- Bell A, Bell D, Weber RS, et al. CpG island methylation profiling in human salivary gland adenoid cystic carcinoma. Cancer 2011;117:2898-909. [Crossref] [PubMed]
- Bouchareb R, Guauque-Olarte S, Snider J, et al. Proteomic Architecture of Valvular Extracellular Matrix: FNDC1 and MXRA5 Are New Biomarkers of Aortic Stenosis. JACC Basic Transl Sci 2021;6:25-39. [Crossref] [PubMed]
- Frost FG, Cherukuri PF, Milanovich S, et al. Pan-cancer RNA-seq data stratifies tumours by some hallmarks of cancer. J Cell Mol Med 2020;24:418-30. [Crossref] [PubMed]
- Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2019;51:411-2. [Crossref] [PubMed]
- Bonneville R, Krook MA, Kautto EA, et al. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis Oncol 2017;2017:PO.17.00073.
- Guideline Recommendations for HER2 Detection in Gastric Cancer Group. Guidelines for HER2 detection in gastric cancer(2016). Zhonghua Bing Li Xue Za Zhi 2016;45:528-32. [PubMed]
- Chinese Anti-Cancer Association Gastric Cancer Professional Committee, Chinese Anti-Cancer Association Oncology Endoscopy Professional Committee, Liang Han, et al. Consensus of Chinese Experts on HER-2 Testing for Gastric Cancer Gastric Biopsy Specimens (2023 Edition). Chinese Journal of Clinical Oncology 2023;50:973-82.
- Hayashi H, Al Mamun A, Sakima M, et al. Activator of G-protein signaling 8 is involved in VEGF-mediated signal processing during angiogenesis. J Cell Sci 2016;129:1210-22. [Crossref] [PubMed]
- Chen L, Liu J, Wang L, et al. Up-regulated FNDC1 accelerates stemness and chemoradiation resistance in colorectal cancer cells. Biochem Biophys Res Commun 2022;602:84-90. [Crossref] [PubMed]
- Hong H, Zhu H, Li C, et al. FNDC1 is highly expressed in lung adenocarcinoma and closely related with poor prognosis. Nan Fang Yi Ke Da Xue Xue Bao 2022;42:1182-90. [PubMed]
- Yunwen C, Shanshan G, Zhifei B, et al. The silencing of FNDC1 inhibits the tumorigenesis of breast cancer cells via modulation of the PI3K/Akt signaling pathway. Mol Med Rep 2021;23:479. [Crossref] [PubMed]
- Xu H, Meng XR, Zhou Y, et al. Expression of FNDC1 protein in esophageal cancer tissue and the impact of interfering with FNDC1 expression on the biological behavior of esophageal cancer cells . Journal of Zhengzhou University 2020;55:445-50. (Medical Sciences).
- Wuensch T, Wizenty J, Quint J, et al. Expression Analysis of Fibronectin Type III Domain-Containing (FNDC) Genes in Inflammatory Bowel Disease and Colorectal Cancer. Gastroenterol Res Pract 2019;2019:3784172. [Crossref] [PubMed]
- Zhong M, Zhang Y, Yuan F, et al. High FNDC1 expression correlates with poor prognosis in gastric cancer. Exp Ther Med 2018;16:3847-54. [Crossref] [PubMed]
- Ren J, Niu G, Wang X, et al. Overexpression of FNDC1 in Gastric Cancer and its Prognostic Significance. J Cancer 2018;9:4586-95. [Crossref] [PubMed]
- Fu YY. Expression and clinical significance of the FNDC1 gene in gastric adenocarcinoma tissue. Anhui: Anhui Medical University, 2019.
- Kurokawa Y, Matsuura N, Kimura Y, et al. Multicenter large-scale study of prognostic impact of HER2 expression in patients with resectable gastric cancer. Gastric Cancer 2015;18:691-7. [Crossref] [PubMed]
- Huang D, Lu N, Fan Q, et al. HER2 status in gastric and gastroesophageal junction cancer assessed by local and central laboratories: Chinese results of the HER-EAGLE study. PLoS One 2013;8:e80290. [Crossref] [PubMed]
- Sheng WQ, Huang D, Ying JM, et al. HER2 status in gastric cancers: a retrospective analysis from four Chinese representative clinical centers and assessment of its prognostic significance. Ann Oncol 2013;24:2360-4. [Crossref] [PubMed]


