
Identification and functional analysis of GNAI1 as a biomarker associated with immune-related genes in pediatric acute myeloid leukemia
Highlight box
Key findings
• GNAI1 was identified as a novel immune-related biomarker in pediatric acute myeloid leukemia (AML) with prognostic value. Silencing GNAI1 reduced AML cell proliferation while enhancing T cell activation and cytotoxicity, demonstrating its functional role in disease pathogenesis.
What is known and what is new?
• Immunotherapy is promising for AML treatment, and identifying immune-related biomarkers is crucial. GNAI1 participates in various intracellular signaling pathways across different cancers.
• This study identifies GNAI1 specifically as an AML biomarker linked to immune infiltration, revealing its involvement in oxidative phosphorylation and ubiquitin-mediated proteolysis pathways.
What is the implication, and what should change now?
• GNAI1 represents a potential therapeutic target in AML. Future studies should validate GNAI1 as a prognostic marker in larger cohorts and explore GNAI1-targeted therapies as part of personalized treatment approaches for pediatric AML patients.
Introduction
Background
Acute myeloid leukemia (AML) is a malignant neoplasm of the blood and the bone marrow, characterized by the uncontrolled proliferation of myeloid progenitor cells. A hallmark of AML is the accumulation of excessive immature leukocytes (blasts) in the bone marrow and circulating blood, impeding normal hematopoiesis (1). AML constitutes one of the predominant forms of acute leukemia in pediatric populations, accounting for nearly 20% of all childhood leukemia cases. The incidence of AML varies with age, peaking in children under 2 years old, remaining minimal in children aged 2 to 9 years, and increasing progressively during adolescence (2). Males are more susceptible to AML than females, with a prevalence ratio of 5:4.3. AML significantly impacts the well-being and quality of life of pediatric patients, posing substantial demands on healthcare systems. The primary treatment for AML is chemotherapy, often supplemented by targeted therapy or hematopoietic stem cell transplantation. However, AML treatment faces challenges including chemotherapy-related adverse effects, rising drug resistance, potential relapse, and complications from transplantation (3). Thus, exploring new biomarkers and understanding intricate pathways are crucial for revealing AML’s etiology and molecular mechanisms, providing essential targets for diagnosis and treatment innovations.
Rationale and knowledge gap
This research focuses on identifying and analyzing biomarkers linked to immune-related genes (IRGs) using weighted gene co-expression network analysis (WGCNA) and survival analysis techniques. The immune system plays a crucial role in recognizing and eliminating cells or entities that have undergone pathological alterations, involving a synergistic interplay of various cells and molecules. Recently, the role of immunity in cancer genesis and progression has garnered extensive scrutiny. The immune system can identify and eradicate mutated or neoplastic cells via its surveillance mechanism, impeding cancer initiation and dissemination (4). Conversely, cancer cells can evade immune system attacks through mechanisms such as expressing immune checkpoint molecules, secreting immune-suppressive factors, inducing immune tolerance, and promoting immunosuppressive environments. These strategies facilitate cancer progression and metastasis, highlighting the complex interplay between malignancies and host immune defenses (5). This multifaceted relationship offers novel insights and therapeutic targets in cancer diagnosis and therapy.
Immunity profoundly impacts the emergence and development of AML. Studies show that AML cells might express immune checkpoint molecules like programmed death-ligand 1 (PD-L1) and release immune-suppressive agents like transforming growth factor beta (TGF-β). They can promote immune tolerance and enhance the proliferation of immunosuppressive cells, including Treg cells and myeloid-derived suppressor cells (MDSCs), attenuating the immune system’s anti-neoplastic capabilities and aiding AML progression and recurrence (6). AML cells are vulnerable to immune system detection and elimination, especially through cytotoxic immune entities like natural killer (NK) cells, cytotoxic T lymphocytes (CTLs), and natural killer T (NKT) cells, which are instrumental in the immune-mediated elimination of AML cells (7,8). Thus, immunity serves dual roles in AML, both suppressing and promoting the disease’s onset and advancement. However, the specific molecular mechanisms underlying these dual roles are yet to be fully elucidated.
Objective
This study aimed to decode the role of immunity in pediatric AML and evaluate the effectiveness and safety of immunomodulatory treatments, providing innovative insights and approaches for diagnosing and treating childhood AML. Additionally, this investigation aimed to elucidate the functionalities and characteristics of immune infiltration associated with these biomarkers, offering novel perspectives and substantial groundwork for immunotherapy strategies for AML.
The pivotal finding of this research is the recognition of GNAI1 as an indicative biomarker linked with IRGs in AML. GNAI1, a member of the G protein α subunit lineage, is implicated in various intracellular signaling cascades, including cyclic adenosine monophosphate (cAMP), phosphoinositide 3-kinases/protein kinase B (PI3K/AKT), and rat sarcoma/extracellular signal-regulated kinases (RAS/ERK) pathways (9). The expression and functionality of GNAI1 vary across different cancer types, with some studies indicating its tumor-suppressive properties, while others highlight its role in tumor progression (10-12). In AML, GNAI1’s expression inversely associates with patient prognosis and survival, marking it as a potential adverse prognostic indicator. We present this article in accordance with the STROBE reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1595/rc).
Methods
Data source
The dataset GSE9476 (GPL96), acquired from the Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/gds), served as the training cohort and included 26 AML tissue specimens and 20 control samples. Additionally, a collection of 177 AML fragments per kilobase of exon per million reads mapped (FPKM) datasets was obtained from The Cancer Genome Atlas (TCGA)-AML archives, accessible via the University of California Santa Cruz (UCSC) Xena platform (http://xena.ucsc.edu/). These datasets were then converted to transcripts per million (TPM) data. This dataset was further integrated with TPM records from 70 normal specimens sourced from the Genotype-Tissue Expression (GTEx) repository (https://www.commonfund.nih.gov/). A differential expression analysis was conducted using a criterion of |log2 fold change (FC)| >1 and an adjusted P value <0.05 to identify differentially expressed genes 1 (DEGs 1). To enhance the accuracy and consistency of the analysis, normalization across arrays was performed using the normalizeBetweenArrays method. Additionally, a compendium of 1,793 IRGs was sourced from the Immunology Database and Analysis Portal (ImmPort) database (http://www.immport.org/).
Application of WGCNA
In this investigation, the WGCNA methodology was applied to the GSE9476 dataset using the WGCNA package (version 1.72-5) to identify AML-related gene modules (13). Initially, sample clustering analysis was performed to detect potential outliers. Subsequently, to ensure adherence to a scale-free topology, an optimal soft thresholding power was determined through gene clustering of all specimens. Following this, the resulting topological matrix underwent hierarchical clustering based on the dissimilarity measures among genes. Gene modules, with a minimum size of 100 genes per module, were delineated using the hybrid dynamic tree-cutting algorithm. Lastly, key gene modules were determined, emphasizing those with correlation coefficient absolute values exceeding 0.5 and P values below 0.05, indicating their association with AML.
Acquisition of key genes
Initially, differential expression analysis for AML versus normal samples in the GSE9476 dataset was conducted using the limma package (version 3.58.1), adopting a threshold of |log2FC| >1 and an adjusted P value <0.05 (13,14). Subsequently, the intersections of IRGs, module genes, and DEGs 2 from the GSE9476 dataset identified potential candidate genes. The comparative expression of these candidates between AML and normal cohorts was analyzed and illustrated. By establishing a definitive threshold for the expression levels of these candidates, individuals in the TCGA-AML dataset were categorized into groups of high or low expression. The prognostic implications of these candidate genes were determined via Kaplan-Meier (K-M) survival curves, focusing on genes that demonstrated significant survival differences, thereby designating them as key genes (15).
Independent prognostic analysis
To explore the prognostic implications of clinical characteristics, we conducted a univariate Cox regression analysis on variables such as age, gender, risk category, and pivotal genes within the TCGA-AML cohort (16). Additionally, to identify independent prognostic indicators, a multivariate Cox regression analysis was performed on variables that demonstrated significance (P<0.05) in the preliminary univariate analysis, utilizing a stepwise selection method (17). Genes identified among these independent prognostic indicators were recognized as biomarkers. Subsequently, we investigated the differential expression of these biomarkers across various clinical strata.
Enrichment analysis
To further elucidate the relevant signaling pathways and underlying biological mechanisms associated with the identified biomarkers, gene set enrichment analysis (GSEA) was employed (18). Initially, correlations between the biomarkers and other genes within the GSE9476 dataset were assessed to determine the correlation coefficients. Subsequently, using the c2.cp.kegg.v2023.1.Hs.symbols as the reference gene set, GSEA was performed based on the ranked gene correlation coefficients, applying a threshold of adjusted P value <0.05 and |normalized enrichment score (NES)| >1. Additionally, AML cohorts were classified into high or low expression groups based on the median expression values of the biomarkers. The gene set variation analysis (GSVA) package (version 1.50.0) was then utilized to identify differentially activated pathways between these expression groups for each biomarker (19).
Immune infiltration analysis
Initially, single-sample GSEA (ssGSEA) was employed to calculate differential activity scores of immune cells between groups with high and low biomarker expression (20). Subsequently, a correlation analysis was performed between the biomarkers and differentially expressed immune cells. The results were visualized using a lollipop chart to clearly represent the associations.
Statement of ethics
This study involved the extraction of CD8+ T cells from the peripheral blood of 6 healthy pediatric volunteers (n=6). All volunteers were informed of the purpose, procedure and possible risks of the study before sampling, and signed an informed consent form. Personal information of the participants was kept strictly confidential. The sampling process was carried out by professional medical personnel under sterile conditions, using disposable sterile equipment, and controlling the amount of blood collected each time within a reasonable range to ensure the safety of volunteers. Volunteers could withdraw from the study at any time without prejudice to their medical rights. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study protocol was reviewed and approved by the Institutional Review Board of the Affiliated Children’s Hospital of Kunming Medical University/Kunming Children’s Hospital (approval No. 2024-05-035-K01) to ensure the safety and rights of volunteers.
Isolation and induction culture of CD8+ T cells
Begin by diluting fresh anticoagulated whole blood with phosphate-buffered saline (PBS). Add a gradient separation solution and carefully layer the diluted blood atop the separation solution. Centrifuge at room temperature using a horizontal rotor at 500–1,000 g for 20–30 minutes. Post-centrifugation, carefully harvest the leukocyte-rich buffy coat layer, located between the plasma and separation solution, into a sterile 15 mL centrifuge tube. Wash the leukocyte layer with 10 mL of PBS or a designated cell wash solution and centrifuge at 250 g for 10 minutes. Discard the supernatant and resuspend the cell pellet in 5 mL of PBS or cell wash solution, followed by another centrifugation at 250 g for 10 minutes. Isolate CD8+ T cells using the EasySep™ Direct Human CD8+ T Cell Isolation Kit (Stemcell, Vancouver, Canada, 1966). Prepare a solution of Ultra-LEAF™ Purified anti-human CD28 antibody (BioLegend, San Diego, USA, 302934) and Ultra-LEAF™ Purified anti-human CD3 antibody (BioLegend, 317326) in PBS to a concentration of 5 µg/mL. Add this antibody mixture to the CD8+ T cell culture plate and incubate at 37 ℃ for 2 hours. After incubation, decant the supernatant. Finally, resuspend the isolated CD8+ T cells in Roswell Park Memorial Institute (RPMI) 1640 complete culture medium and transfer them onto a culture plate for subsequent expansion.
Characterization of CD8+ T cells via flow cytometry
Relocate the cell suspension and culture medium into Eppendorf (EP) tubes and centrifuge at 300 g for 5 minutes to sediment the cells. Afterward, remove the culture medium and collect the cell pellet. Add precooled PBS to the cells, ensuring they are resuspended by gentle agitation or careful pipetting, then centrifuge again at 300 g for 5 minutes to wash the cells thoroughly; repeat this step twice. Add 100 µL of 1× Binding Buffer to the cell pellet and resuspend thoroughly. Add 5 µL of anti-CD8 antibody to each sample tube, mix well, and incubate at room temperature, protected from light, for 15 minutes. Then, add 400 µL of 1× Binding Buffer to each tube and mix thoroughly. Prior to analysis, filter the cell suspension through a nylon mesh into flow cytometry tubes to obtain a single-cell suspension. Use flow cytometry (BD, FACSCalibur, Franklin Lakes, USA) to acquire data from 10,000 cells per stained sample and analyze the data using FlowJo software Version 10 (FlowJo V10) for data acquisition.
Instantaneous transfection of si-GNAI1 and reverse transcription polymerase chain reaction (RT-PCR) detection of transfection efficiency
Begin by diluting 50 nM small interfering RNA (siRNA) into 100 µL of serum-free medium, ensuring thorough mixing to achieve a homogenous siRNA solution. Next, add 100 µL of polyethylenimine (PEI) with a molecular weight of 40,000 (PEI 40000) to the siRNA solution, ensuring gentle yet thorough integration. Allow the mixture to rest at ambient temperature for 10–15 minutes to facilitate the formation of a positively charged nucleic acid transfection reagent complex. When the cells reach 70–80% confluency, introduce 2 mL of pre-warmed complete culture medium to each well. Add 100 µL of the nucleic acid-PEI complex to the cells, ensuring uniform distribution by gently agitating the culture plate. Continue to culture the cells in a 37 ℃, 5% CO2 incubator. Post-transfection, the expression of the target gene can be detected as early as 5 hours.
Execution of CCK-8 assay
Initiate the experiment by selecting logarithmic growth phase Kasumi-1 cells and co-culturing them with CD8+ T cells at ratios of 3:1 (9,000 Kasumi-1 cells to 3,000 CD8+ T cells) and 5:1 (15,000 Kasumi-1 cells to 3,000 CD8+ T cells). Position the Kasumi-1 cells in the apical chamber and the CD8+ T cells in the basal chamber of a co-culture device. Use the previously described method for the transfection of Kasumi-1 cells. After a 48-hour co-culture period, collect 100 µL of the co-cultured tumor and CD8+ T cells and transfer them into 96-well plates, establishing six replicates for each condition. Add 10 µL of Cell Counting Kit-8 (CCK-8) solution to each well and incubate the plate in a 37 ℃, 5% CO2 incubator for 2–4 hours. The optical density (OD) was then measured at 450 nm at 0, 24, 48, and 72 hours to evaluate cellular viability.
Utilization of flow cytometry for quantifying CD8+ and CD8+PD-1+ T cell populations
Begin by transferring the cellular suspension and medium into EP tubes and centrifuge at 300 g for 5 minutes to sediment the cells. Then, discard the supernatant, gather the cellular sediment, and resuspend it in precooled PBS with gentle agitation or pipetting. Centrifuge once more at 300 g for 5 minutes, repeating the wash cycle twice. Subsequently, reconstitute the cellular sediment in 100 µL of 1× Binding Buffer to prepare the cell suspension. Introduce 5 µL each of CD8 (Biogene, Portland, USA) and PD-1 (Elabscience, Houston, USA) antibodies to the respective si-NC and si-GNAI1 tubes in both 3:1 and 5:1 ratios, ensuring gentle homogenization. Allow incubation at ambient temperature, shielded from light, for 15 minutes. Following incubation, add 400 µL of 1× Binding Buffer to each tube and ensure a homogeneous mixture. Strain the cell suspension through nylon mesh into flow cytometry tubes to prepare for analysis, aiming for a uniform single-cell suspension. Analyze 10,000 cells per sample using a flow cytometer (BD, FACSCalibur) to acquire the stained sample data, and interpret the results with the aid of FlowJo software (FlowJo V10).
Statistical analysis
Bioinformatic evaluations were performed utilizing the R 4.3.1 programming environment. Variations among distinct groups were deemed statistically significant at a threshold where P<0.05. For multiple comparisons, adjusted P values were calculated. Statistical significance levels are indicated in figures as follows: * denotes P<0.05, ** denotes P<0.01, and *** denotes P<0.001. All specific P values are reported with appropriate precision—with three decimal places for values between 0.001 and 0.01, and two decimal places for values ≥0.01 (with three decimal places for values near 0.05).
Results
A total of 3,611 modular genes were screened through WGCNA
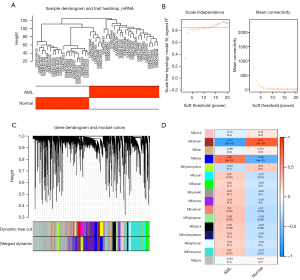
To identify modular genes associated with AML, WGCNA was conducted. Initially, cluster analysis indicated that no samples needed to be eliminated (Figure 1A). Subsequently, the optimal soft-thresholding power (β) was selected to be 13 when the scale-free topology fit index (R2) reached 0.85, at which point the network approximated a scale-free distribution (Figure 1B). Sixteen co-expression gene modules were then identified (Figure 1C). Among these, the MEbrown module [correlation (cor) =−0.77] and the MEblue module (cor =0.65) were considered key gene modules (Figure 1D). A total of 3,611 genes from these key modules were identified for further analysis.
FLT3, IL3RA, PTX3, KLRC3 and TGFBR3 were reserved as key genes
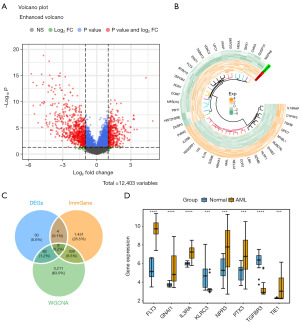
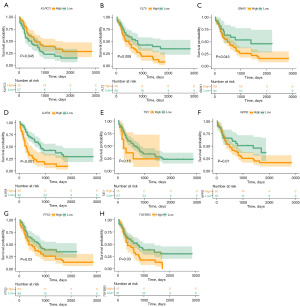
Firstly, a total of 7,133 DEGs1 were identified through differential analysis of combined AML data from the UCSC Xena and GTEx databases. Among these, 3,991 genes were upregulated, and 3,142 genes were downregulated in AML. Next, 1,189 DEGs2 (427 upregulated genes and 762 downregulated genes) were identified in the GSE9476 dataset (Figure 2A,2B). These DEGs2 were intersected with 1,793 IRGs, 3,611 modular genes, and 7,133 DEGs1 to identify 8 candidate genes (Figure 2C). Among these candidate genes, FLT3, GNAI1, IL3RA, NPR3, PTX3, and TIE1 were significantly upregulated in AML, whereas TGFBR3 and KLRC3 were downregulated in the AML group (Figure 2D). Among the 8 candidate genes, the expression levels of 6 genes showed significant differences in survival according to K-M survival curves (Figure 3). Notably, low expression of KLRC3 was associated with lower survival, while high expression of FLT3, IL3RA, PTX3, and TGFBR3 was associated with worse prognosis. Therefore, these 6 genes were considered key genes.
Utilizing age and risk category as distinct prognostic indicators in AML
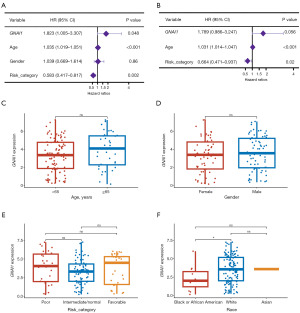
The pivotal genes and clinical attributes from the TCGA-AML database were subjected to univariate Cox regression analysis to determine their prognostic independence. As depicted in Figure 4A, the P values for GNAI1, age, and risk category were all below the 0.05 threshold, necessitating their inclusion in subsequent multivariate Cox regression analysis. This comprehensive analysis revealed that both age and risk category serve as distinct prognostic indicators in AML. Notably, GNAI1 displayed marginal significance, with a P value of 0.056, and was thus recognized as a potential biomarker (Figure 4B). Additionally, an exploration into the expression variances of GNAI1 across diverse clinical demographics was undertaken (Figure 4C-4F), revealing a pronounced elevation in expression uniquely within the White racial group (Figure 4F).
The signaling pathways significantly associated with GNAI1 included oxidative phosphorylation, ubiquitin mediated proteolysis
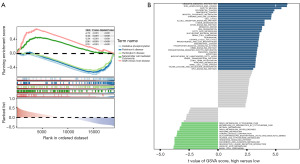
To further explore the signaling pathways associated with the biomarkers, functional enrichment analyses were performed. First, GSEA revealed that the top five signaling pathways enriched by GNAI1 included oxidative phosphorylation and NK cell-mediated cytotoxicity, among others (Figure 5A). Next, GSVA was used to identify differentially activated signaling pathways between high and low GNAI1 expression groups. These pathways included ubiquitin-mediated proteolysis, Wnt signaling pathways, and others (Figure 5B).
Ten differential immune cells were identified that were significantly associated with GNAI1
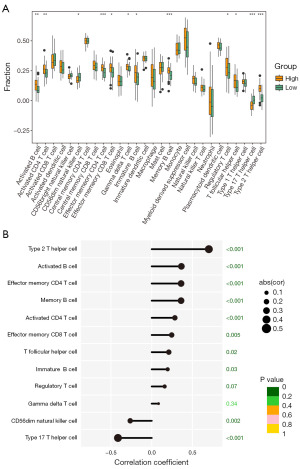
To elucidate the impact of immune cells on AML, immune cell infiltration was analyzed. In this cohort, aside from NK cells and T helper 17 (Th17) cells, which exhibited notably lower expression in the high GNAI1 expression group, ten additional cell types (including activated B cells, activated CD4 T cells, effector memory CD4 T cells, effector memory CD8 T cells, gamma delta T cells, immature B cells, memory B cells, regulatory T cells, T follicular helper cells, and T helper 2 (Th2) cells demonstrated elevated expression within the high-risk category (see Figure 6A). Furthermore, a detailed correlation analysis revealed a significant association of these ten distinct immune cell types with GNAI1. Notably, CD56dim NK cells and Th17 cells showed a substantial negative correlation with GNAI1 expression, while the other ten cell types, such as activated B cells and memory B cells, exhibited a positive correlation (Figure 6B).
Effects of GNAI1 silencing on CD8+ T cell activation
Initially, following isolation and induction procedures, we verified the presence of CD8+ T cells, constituting approximately 46% of the total cell population in pediatric samples, as shown in Figure 7A. Kasumi-1 cells were then transiently transfected with si-GNAI1. RT-PCR analysis revealed that si-GNAI1-001, si-GNAI1-002, and si-GNAI1-003 significantly reduced GNAI1 expression, with si-GNAI1-001 and si-GNAI1-002 showing the most effective gene silencing. Therefore, si-GNAI1-001 or si-GNAI1-002 were selected for further gene silencing experiments, as illustrated in Figure 7B.
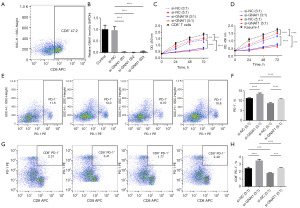
In co-culture experiments of Kasumi-1 cells with CD8+ T cells at various ratios, we observed a marked decrease in the viability of CD8+ T cells in both si-NC (3:1) and si-NC (5:1) groups compared to the CD8+ T cell control group. This decline in viability was even more pronounced in the si-GNAI1 (3:1) and si-GNAI1 (5:1) groups compared to the si-NC (3:1) group, indicating a significant impact of GNAI1 silencing on cell survival. A similar trend was noted for the viability of Kasumi-1 cells across the respective groups, as depicted in Figure 7C,7D.
Furthermore, we assessed the proportion of PD-1+ T cells and PD-1+CD8+ T cells in co-cultures of Kasumi-1 cells with CD8+ T cells at different ratios. The results revealed that, in comparison to the si-NC (3:1) group, the si-GNAI1 (3:1) group exhibited a significant increase in the ratios of PD-1+ T cells and PD-1+CD8+ T cells, whereas the si-NC (5:1) group showed a notable decrease. In contrast, the si-GNAI1 (5:1) group demonstrated a significant rise in both PD-1+ T cell and PD-1+CD8+ T cell ratios compared to the si-NC (5:1) group, as shown in Figure 7E-7H.
Discussion
Key findings
In this study, we identified GNAI1 as a promising immune-related biomarker in AML, exhibiting a significant correlation with patient prognosis and immune cell infiltration. From the GSE9476 dataset, we identified the brown and pink modules containing genes correlated with AML prognosis and differentiation, which extensively intersected with IRGs. From the TCGA-AML dataset, we identified eight candidate genes (GNAI1, GNAI3, GNG2, GNG7, GNGT1, GNGT2, RGS10, RGS18) involved in the G protein signaling pathway that modulates cellular processes such as proliferation, migration, apoptosis, and differentiation. Elevated expression of six key genes (GNAI1, GNAI3, GNG2, GNG7, GNGT1, RGS10) was associated with reduced overall survival in AML patients. Notably, GNAI1 expression demonstrated marginal significance (P=0.056) concerning AML prognosis.
Functional analyses revealed GNAI1’s involvement in pathways such as oxidative phosphorylation, ubiquitin-mediated protein degradation, metabolism, apoptosis, and chemoresistance in AML cells. GNAI1 expression significantly correlated with the infiltration of ten immune cell types, showing an inverse relationship with NK cells and Th17 cells, and a positive correlation with Th2 cells and activated B cells. These findings suggest that GNAI1 may contribute to immune evasion and suppression mechanisms in AML.
Strengths and limitations
The present study aimed to delineate biomarkers intertwined with IRGs in AML by conducting bioinformatics analyses on two AML datasets (GSE9476 and TCGA-AML) and 1,793 IRGs. We employed WGCNA to identify AML-related module genes, performed differential and survival analyses to ascertain candidate and key genes, and conducted functional enrichment and immune infiltration analyses to elucidate the potential mechanisms of the identified biomarkers.
However, this study has limitations, including a small sample size, lack of confirmatory experimental data, and exclusion of other contributory factors. Regarding familial predisposition to GNAI1 expression, there is limited data available in the literature. Further research in larger familial cohorts is necessary to elucidate the potential hereditary aspects of GNAI1 expression and its impact on AML risk.
Comparison with similar researches
AML is a prevalent form of leukemia in children, constituting 15% to 20% of all pediatric leukemia instances (21). Despite significant advancements in treating pediatric AML, approximately 30% of patients relapse post-treatment, with subsequent prognoses being notably dire (22). The immune system substantially influences the progression of AML, impacting the proliferation, invasion, dissemination, and chemotherapy response of AML cells. Recent advancements in high-throughput sequencing technologies have unveiled an array of IRGs linked to the initiation and prognosis of AML, such as PD-L1, CD47, and CD33, which serve as potential targets for immunotherapy or diagnostic markers. However, the expression patterns and functional implications of IRGs in pediatric AML remain largely unexplored (23).
Explanations of findings
GNAI1, a member of the G protein α subunit lineage, is implicated in various intracellular signaling cascades that influence AML development. Interestingly, platelets, which exhibit dysfunction in myeloproliferative disorders, also express GNAI proteins that mediate thrombosis and inflammation (24). Moreover, BCR-ABL tyrosine kinase inhibitors used in the treatment of chronic myeloid leukemia also target GNAI2 (25), highlighting the significance of GNAI signaling in myeloid malignancies.
Some studies have investigated the role of genetic polymorphisms in G protein-related genes in various diseases. These genetic variations may affect the expression levels of GNAI1 and could potentially contribute to the susceptibility to AML (26). These genetic variants may influence the expression levels of GNAI1 and potentially contribute to AML susceptibility.
Implications and actions needed
Biomarkers play a crucial role in disease diagnosis, prognosis, and monitoring treatment response (27). Consequently, identifying molecular markers for AML to tailor personalized treatment strategies is pivotal to augment survival rates and enhance the quality of life for children afflicted with AML (28). Our findings suggest that GNAI1 could be a valuable target for developing immunotherapeutic strategies in pediatric AML.
Nevertheless, further validation in larger cohorts and a deeper exploration of GNAI1’s molecular mechanisms in AML are necessary to provide more valuable information for clinical treatment. Additional research should focus on understanding how GNAI1 modulates the immune microenvironment in AML and whether targeting GNAI1 could enhance anti-tumor immune responses.
Conclusions
In conclusion, our study identified GNAI1 as a novel immune-related biomarker in pediatric AML. GNAI1 expression was significantly associated with patient prognosis and immune cell infiltration. Functional analyses revealed GNAI1’s involvement in key signaling pathways and immune regulation in AML. Knocking down GNAI1 using siRNA reduced AML cell viability and enhanced T cell activation, as evidenced by increased proportions of PD-1+ T cells and PD-1+CD8+ T cells in co-culture experiments. These findings provide new insights into the role of GNAI1 in AML pathogenesis and highlight its potential as a therapeutic target. However, further validation in larger cohorts and deeper mechanistic exploration are warranted. Future studies should investigate the clinical utility of GNAI1 as a prognostic marker and evaluate the therapeutic efficacy of targeting GNAI1, which may ultimately inform personalized treatment strategies and improve outcomes for children with AML.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1595/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1595/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1595/prf
Funding: The support for this study was generously provided by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1595/coif). All authors report that this study was supported by the Sichuan Science and Technology Program (Nos. 2022YFS0622 and 2022YFS0622-B4) and the Yunnan Zhu Xiaofan Expert Workstation (No. 202105AF150070). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study protocol was reviewed and approved by the Institutional Review Board of the Affiliated Children’s Hospital of Kunming Medical University/Kunming Children’s Hospital (approval No. 2024-05-035-K01) to ensure the safety and rights of volunteers. Informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bhansali RS, Pratz KW, Lai C. Recent advances in targeted therapies in acute myeloid leukemia. J Hematol Oncol 2023;16:29. [Crossref] [PubMed]
- Tseng S, Lee ME, Lin PC. A Review of Childhood Acute Myeloid Leukemia: Diagnosis and Novel Treatment. Pharmaceuticals (Basel) 2023;16:1614. [Crossref] [PubMed]
- Zarnegar-Lumley S, Caldwell KJ, Rubnitz JE. Relapsed acute myeloid leukemia in children and adolescents: current treatment options and future strategies. Leukemia 2022;36:1951-60. [Crossref] [PubMed]
- Wei J, Li W, Zhang P, et al. Current trends in sensitizing immune checkpoint inhibitors for cancer treatment. Mol Cancer 2024;23:279. [Crossref] [PubMed]
- Alsaafeen BH, Ali BR, Elkord E. Resistance mechanisms to immune checkpoint inhibitors: updated insights. Mol Cancer 2025;24:20. [Crossref] [PubMed]
- Penter L, Wu CJ. Therapy response in AML: a tale of two T cells. Blood 2024;144:1134-6. [Crossref] [PubMed]
- Myers JA, Miller JS. Exploring the NK cell platform for cancer immunotherapy. Nat Rev Clin Oncol 2021;18:85-100. [Crossref] [PubMed]
- Raskov H, Orhan A, Salanti A, et al. Natural Killer Cells in Cancer and Cancer Immunotherapy. Cancer Lett 2021;520:233-42. [Crossref] [PubMed]
- Jian J, Yuan C, Hao H. Identifying key genes and functionally enriched pathways in acute myeloid leukemia by weighted gene co-expression network analysis. J Appl Genet 2025;66:347-62. [Crossref] [PubMed]
- Lv Y, Wang Y, Song Y, et al. LncRNA PINK1-AS promotes Gαi1-driven gastric cancer tumorigenesis by sponging microRNA-200a. Oncogene 2021;40:3826-44. [Crossref] [PubMed]
- Raza A, Yen MC, Anuraga G, et al. Comparative Analysis of the GNAI Family Genes in Glioblastoma through Transcriptomics and Single-Cell Technologies. Cancers (Basel) 2023;15:5112. [Crossref] [PubMed]
- Wu Y, Zhang J, Li G, et al. Exosomal miR-320d promotes angiogenesis and colorectal cancer metastasis via targeting GNAI1 to affect the JAK2/STAT3 signaling pathway. Cell Death Dis 2024;15:913. [Crossref] [PubMed]
- Wang X, Bajpai AK, Gu Q, et al. Weighted gene co-expression network analysis identifies key hub genes and pathways in acute myeloid leukemia. Front Genet 2023;14:1009462. [Crossref] [PubMed]
- Zhao Y, Zhang X, Zhao Y, et al. Identification of potential therapeutic target genes, key miRNAs and mechanisms in acute myeloid leukemia based on bioinformatics analysis. Med Oncol 2015;32:152. [Crossref] [PubMed]
- Lai B, Lai Y, Zhang Y, et al. Survival prediction in acute myeloid leukemia using gene expression profiling. BMC Med Inform Decis Mak 2022;22:57. [Crossref] [PubMed]
- Gao Y, Jia Y, Yu Z, et al. Analysis of the differential expression and prognostic relationship of DEGs in AML based on TCGA database. Eur J Med Res 2023;28:103. [Crossref] [PubMed]
- Zhang J, Chen Q, Zhang Y, et al. Construction of a random survival forest model based on a machine learning algorithm to predict early recurrence after hepatectomy for adult hepatocellular carcinoma. BMC Cancer 2024;24:1575. [Crossref] [PubMed]
- Wang Y, Armendariz DA, Wang L, et al. Enhancer regulatory networks globally connect non-coding breast cancer loci to cancer genes. Genome Biol 2025;26:10. [Crossref] [PubMed]
- Li H, Xu Y, Wang A, et al. Integrative bioinformatics and machine learning approach unveils potential biomarkers linking coronary atherosclerosis and fatty acid metabolism-associated gene. J Cardiothorac Surg 2025;20:70. [Crossref] [PubMed]
- He Y, Lai J, Wang Q, et al. ssMutPA: single-sample mutation-based pathway analysis approach for cancer precision medicine. Gigascience 2024;13:giae105. [Crossref] [PubMed]
- Creutzig U, van den Heuvel-Eibrink MM, Gibson B, et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood 2012;120:3187-205. [Crossref] [PubMed]
- Curran E, Corrales L, Kline J. Targeting the innate immune system as immunotherapy for acute myeloid leukemia. Front Oncol 2015;5:83. [Crossref] [PubMed]
- Restelli C, Ruella M, Paruzzo L, et al. Recent Advances in Immune-Based Therapies for Acute Myeloid Leukemia. Blood Cancer Discov 2024;5:234-48. [Crossref] [PubMed]
- Repsold L, Joubert AM. Platelet Function, Role in Thrombosis, Inflammation, and Consequences in Chronic Myeloproliferative Disorders. Cells 2021;10:3034. [Crossref] [PubMed]
- Nagel S, Haake J, Pommerenke C, et al. Establishment of the Myeloid TBX-Code Reveals Aberrant Expression of T-Box Gene TBX1 in Chronic Myeloid Leukemia. Int J Mol Sci 2023;25:32. [Crossref] [PubMed]
- Thompson MD, Hendy GN, Percy ME, et al. G protein-coupled receptor mutations and human genetic disease. Methods Mol Biol 2014;1175:153-87. [Crossref] [PubMed]
- Zakari S, Niels NK, Olagunju GV, et al. Emerging biomarkers for non-invasive diagnosis and treatment of cancer: a systematic review. Front Oncol 2024;14:1405267. [Crossref] [PubMed]
- Guo X, Zhou X. Risk stratification of acute myeloid leukemia: Assessment using a novel prediction model based on ferroptosis-immune related genes. Math Biosci Eng 2022;19:11821-39. [Crossref] [PubMed]


