
Overexpression of LINC00880 promotes colorectal cancer growth
Highlight box
Key findings
• LINC00880 is upregulated in colorectal cancer (CRC) and promotes tumor growth.
What is known and what is new?
• LncRNA plays an extremely vital role in the occurrence and development of various human tumors.
• The role of LINC00880 in CRC progression was investigated for the first time.
What is the implication, and what should change now?
• LINC00880 is a potential biomarker and therapeutic target for CRC.
Introduction
Colorectal cancer (CRC) is a prevalent and deadly malignancy within the digestive system, currently ranking third in incidence and second in mortality globally (1). The complex pathogenesis of CRC involves genetic and lifestyle factors that contribute to the malignant transformation of polyps. Early detection through screening is crucial for asymptomatic early-stage CRC, but low screening compliance highlights the urgent need for innovative strategies (2). The treatment of CRC typically involves a combination of surgery, chemotherapy, radiation therapy, targeted therapy, and immunotherapy (3). Personalized medicine, based on tumor profiling, is an emerging approach to enhance treatment efficacy. A comprehensive understanding of CRC pathogenesis is essential for advancing early detection methods and refining treatments.
Long non-coding RNAs (lncRNAs) are RNA molecules that are longer than 200 nucleotides and do not encode proteins (4). They play a significant role in a variety of biological processes through regulating gene expression (5). In CRC, lncRNAs have emerged as significant modulators in tumorigenesis, invasion, and metastasis, with their dysregulation often indicating a poor prognosis (6-9). They regulate critical cellular processes, including the cell cycle, apoptosis, DNA repair, and epithelial-to-mesenchymal transition, pathways commonly deregulated in cancer (10-14). The distinct expression patterns of lncRNAs in CRC position them as potential biomarkers for both diagnosis and prognosis. Additionally, their roles in cancer progression also implicate them as therapeutic targets (15).
Despite the advances in lncRNA research, the specific role of LINC00880 in CRC, particularly its contribution to disease’s initiation and progression of CRC remains unknown. Prior study has reported that LINC00880 could play a crucial role in the development of lung adenocarcinoma by modulating the PI3K/AKT signaling pathway (16). Therefore, we conducted both in vivo and in vitro experiments to further explore the function of LINC00880 in CRC cells. We present this article in accordance with the MDAR and ARRIVE reporting checklists (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2025-54/rc).
Methods
Public genomic data repositories
Based on the lncRNA sequencing data from The Cancer Genome Atlas (TCGA) CRC cohort, this study systematically investigated the expression profile of LINC00880. Following the extraction of target lncRNA expression data using a standardized bioinformatics pipeline, the Wilcoxon signed-rank test was applied to assess the differential expression significance of LINC00880 across clinical subgroups, including tumor stage and molecular subtypes.
Cultured cell lines and clinical tissue samples
The normal human colonic epithelial cell line NCM460 and the CRC cell lines (RKO, HCT8, SW480, DLD1, and HCT116) were all obtained from the Cell Bank of the Chinese Academy of Medical Sciences (Shanghai, China). All cell lines were maintained in Dulbecco’s modified Eagle medium enriched with 10% heat-inactivated fetal bovine serum and 1% penicillin-streptomycin antibiotic cocktail, under standard culture conditions (37 ℃, 5% CO2, humidified atmosphere). This study utilized a CRC cohort consisting of 90 primary tumor tissues and their paired adjacent non-tumor tissues (NCTs). The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. Ethics approval for this investigation was formally granted by the Institutional Review Board of the Affiliated Hospital of Jiangnan University (No. WXSY-YXLL-AF/SC-11/02.0). Prior to sample collection and experimental procedures, written informed consent was acquired from all participating donors and their legal representatives. The research protocol was conducted in strict compliance with internationally recognized ethical standards for biomedical studies involving human subjects.
Small interfering RNAs (siRNAs) and transfection
siRNAs were commercially sourced from RiboBio (Guangzhou, China) and delivered into cells via GenMute™ siRNA Transfection Reagent (Signagen Laboratories, Beijing, China). Transfection efficiency was quantitatively assessed through quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis at 36 h post-transfection. The specific primer sequences employed for PCR amplification are detailed in Table 1.
Table 1
| Primers | Sequences-F | Sequences-R |
|---|---|---|
| Primers for real-time PCR | ||
| LINC00880 | GAGGCTCCGAACATCTGAGG | GGCCTGGCTTTGTCTGAGTC |
| β-actin | AGTGTGACGTGGACATCCGCAAAG | ATCCACATCTGCTGGAAGGTGGAC |
| CDC6 | GCGAGGCCTGAGCTGTG | GCTGAGAGGCAGGGCTTTTA |
| CDC25A | GTGGGAGAACAGCGAAGACA | AATCCAAACAAACGTGGCGG |
| CCNE1 | AGAGGAAGGCAAACGTGACC | TTTGCCCAGCTCAGTACAGG |
| MCM4 | CCCGAGCTTGTCCTTGTCG | GTGGAATCCTCGCCTCTACG |
| PCNA | GCCCTGGTTCTGGAGGTAAC | TAGCTGGTTTCGGCTTCAGG |
| MCM3 | GACGACGAGGCTTCTGAACA | GGACGACTTTGGGACGAACT |
| ORC6 | TGCTGAGGAAAGCAGAGGAG | ATCCAGGAAGCTGCAAGGTC |
| POLD3 | AGCTACTACTGGGGAACGC | GTATGTCACCCAAGGGACGC |
| PRIM2 | TGCCACCGTTTGTGTTTTCC | TTCAGAAGGTGGCTGCAAGT |
| U6 | CAGCACATATACTAAAATTGGAACG | ACGAATTTGCGTGTCATCC |
| Sequences of siRNA | ||
| si-NC | UUCUCCGAACGUGUCACGUTT | |
| si-LINC00880-1 | GCGCCUGAGUCAUUUGAAATT | |
| si-LINC00880-2 | GCAUUCGAUGUCAGGCCAUTT | |
| Related sequence information for plasmid construction | ||
| LINC00880-OE | GTGAACCGTCAGATCGAATTCGTGAAGAACAGTGTTTCTCTCCTACAG | TAATCCAGAGGTTGAGGATCCTCATTTCTTTCCTGTTTATTAACATTCA |
F, forward; NC, negative control; OE, overexpressing; PCR, polymerase chain reaction; R, reverse; si, small interfering; siRNA, small interfering RNA.
qRT-PCR
Total cellular RNA was isolated with TRIzol® reagent (Yeasen Biotechnology, Shanghai, China). First-strand complementary DNA (cDNA) synthesis was carried out using HiScript III RT SuperMix for qPCR (+gDNA wiper) (Vazyme Biotech, Nanjing, China). Quantitative PCR amplification of target and reference genes was performed with Hieff® qPCR SYBR Green Master Mix (No Rox) (Yeasen Biotechnology), and β-actin expression served as the endogenous control for normalization of relative gene expression data.
Plasmid construction, lentivirus packaging, and infection
The full-length LINC00880 sequence was PCR-amplified and subcloned into the pLenti-EF1a-EGFP-F2A-Puro-CMV-MCS lentiviral vector via the ClonExpress™ II One-Step Cloning System (Vazyme Biotech). Plasmid transfection was mediated by LipoFiter™ Liposomal Transfection Reagent (HanBio Technology, Shanghai, China) following the manufacturer’s protocol. Lentiviral particles (PackGene Biotech, Guangzhou, China) were subsequently generated to transduce HCT116 cells for stable cell line establishment. Transduction efficiency was validated 72 h post-infection, followed by puromycin selection (2 µg/mL; Beyotime Biotechnology, Shanghai, China) to isolate stably transduced clones.
Cell Counting Kit-8 (CCK-8) and colony formation assays
A total of 2,000 cells were seeded into 96-well plates. After the cells adhered, CCK-8 was added to measure cell viability according to the manufacturer’s instructions. For colony formation assays, 1,000 cells were seeded into each well of a six-well plate and incubated for 14 days. The colonies were then fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The number of colonies was counted using an inverted microscope, and images were captured for documentation.
5-ethynyl-2’-deoxyuridine (EdU) assay
Cell proliferation was assessed using the BeyoClick™ EdU Assay Kit (Beyotime Biotechnology). CRC cell lines (DLD1, RKO, and HCT116) were pretreated and incubated with 10 µM EdU under standard culture conditions (37 ℃) for 2 h. Following labeling, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and processed for fluorescent detection using the Click-iT® Plus EdU Alexa Fluor™ 488 Imaging Kit (Beyotime Biotechnology, Shanghai, China) according to the manufacturer’s protocol. Proliferation rates were quantified by calculating the ratio of EdU-positive cells to total nuclei across three randomly selected fields using an inverted fluorescence microscope.
Fluorescence in situ hybridization (FISH)
LINC00880 and 18S ribosomal RNA FISH probes were commercially obtained from RiboBio. The FISH assay was performed in strict accordance with the manufacturer’s protocol. Briefly, cellular specimens were fixed with 4% paraformaldehyde for 15 min at room temperature, followed by permeabilization using a solution containing 0.5% Triton X-100 in phosphate-buffered saline (PBS) at 4 ℃ for 5 min. Following three washes with PBS, cellular specimens were subjected to prehybridization in buffer (37 ℃, 30 min). Subsequently, a 20 µM probe cocktail in hybridization buffer [10% dextran sulfate, 2× saline sodium citrate (SSC)] was applied to the samples for overnight incubation at 37 ℃. Fluorescent imaging was performed using an Olympus DP80-Cellsens system (Olympus Corporation, Tokyo, Japan) after nuclear counterstaining with 1 µg/mL 4’-6-diamidino-2-phenylindole (DAPI; 10 min) and post-stain washes in PBS-Tween 20 (PBS-T; 0.1% Tween 20).
Subcellular fractionation
Subcellular RNA fractionation was performed using the Ambion™ PARIS™ Kit (Thermo Fisher Scientific) in strict accordance with the manufacturer’s protocol. Cytoplasmic and nuclear RNA extracts were isolated and subjected to qRT-PCR analysis to determine the expression disparity of LINC00880 between these two subcellular compartments.
Detection of the cell cycle
The cells were collected and washed twice with pre-cooled PBS. They were then fixed with pre-cooled 70% ethanol and stored at 4 ℃ overnight. After staining with propidium iodide (Beyotime Biotechnology), the cell cycle distributions of these cells were analyzed using a FACSCanto II flow cytometer (Agilent, Santa Clara, CA, USA).
Western blot
Protein extraction was performed using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime Biotechnology) supplemented with 1× protease inhibitor cocktail (MedChemExpress, Shanghai, China). Protein samples were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently transferred onto polyvinylidene fluoride (PVDF) membranes (Merck Millipore, Burlington, MA, USA) using wet transfer method. The membranes were processed sequentially as follows: (I) blocking with 5% non-fat dry milk in TBST for 1 h at room temperature; (II) incubation with primary antibodies against MCM3 (1:2,000; Starter, Hangzhou, China) and β-actin (1:10,000; ABclonal, Wuhan, China) overnight at 4 ℃; (III) three 5-min washes with TBST under gentle agitation; (IV) incubation with horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (IgG) secondary antibody (1:10,000; Jackson ImmunoResearch, West Grove, PA, USA) for 1 h at room temperature with shaking. Finally, protein signals were detected using enhanced chemiluminescence (ECL) prime chemiluminescent substrate (Vazyme Biotech; Cat# E412-02) and imaged with a ChemiDoc MP Imaging System (Bio-Rad Laboratories, Hercules, CA, USA).
Tumor formation in nude mouse models
LINC00880-knockdown or control cell suspensions were subcutaneously administered into the right axillary region of 5-week-old male BALB/c nude mice (thymus-deficient; n=8 per cohort). Tumor progression was quantitatively monitored by caliper measurements every 72 h post-inoculation until study termination. At the experimental endpoint (4 weeks post-implantation), animals were humanely euthanized via CO2 asphyxiation followed by cervical dislocation. Excised xenografts were systematically documented through macroscopic imaging and precision-weighted. All animal experiments were conducted under the Project License (No. JN.20241030b0161220 and Approval No. 577) granted by the Medical Experimental Animal Ethics Committee of Jiangnan University, in compliance with institutional guidelines for the care and use of laboratory animals. A protocol was prepared before the study without registration.
Immunohistochemistry (IHC) staining
Mouse tumor sections were deparaffinized and rehydrated, and then antigen retrieval using sodium citrate solution (Beyotime Biotechnology). Next, the sections were blocked and incubated overnight at 4 ℃ with the following primary antibodies: anti-Ki-67 (1:100), anti-vimentin (1:100), and anti-MCM3 (1:100). Finally, the sections were stained using diaminobenzidine (DAB) reagent kit (Gene Tech, Shanghai, China) and hematoxylin. Images were captured with an upright IHC microscope (Nikon, Tokyo, Japan).
Statistical analyses
We conducted statistical analyses using GraphPad Prism 9.0. The associations between LINC00880 expression levels and clinical features were analyzed using the Chi-squared test or Fisher’s exact test. Intergroup comparisons were assessed using two-tailed Student’s t-test. For multigroup comparisons against a shared control, Dunnett’s test was applied with multiplicity adjustment. Prognostic significance of LINC00880 in CRC patients was analyzed through Kaplan-Meier curves, with survival differences determined by log-rank testing. Statistical significance was defined as two-tailed P<0.05 throughout the study. All experiments were conducted in three independent replicates, and all data are presented as mean ± standard deviation (SD).
Results
LINC00880 is overexpressed in CRC and is associated with poor prognosis
Comprehensive analysis of RNA sequencing (RNA-seq) profiles across 33 TCGA tumor cohorts (https://portal.gdc.cancer.gov) revealed consistent LINC00880 upregulation in multiple malignancies, particularly CRC, gastric carcinoma, and rectal tumors, when compared with matched normal counterparts (Figure 1A). From the TCGA-colon adenocarcinoma (COAD) and TCGA-rectum adenocarcinoma (READ) datasets, we directly analyzed the expression levels of LINC00880 in 50 paired samples. The results revealed a significant upregulation of LINC00880 in CRC tissues compared with their corresponding normal tissues (Figure 1B).
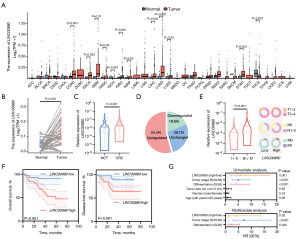
To investigate the potential correlation between LINC00880 expression levels and the prognosis of CRC patients, we initially validated the expression of LINC00880 in 90 paired CRC tissues and NCTs by qRT-PCR. Our findings revealed that LINC00880 expression was increased by more than 1.5-fold in 54.4% (49/90) of the CRC tissues compared with their corresponding NCTs (Figure 1C). Subsequently, we stratified these CRC patients into high- and low-expression groups on the basis of the median expression value of LINC00880 and examined the correlation between LINC00880 and various clinicopathological parameters (Figure 1D). The results indicated that the expression levels of LINC00880 were positively correlated with tumor stage and that high expression of LINC00880 was associated with low survival rates, suggesting a poor prognosis (Figure 1E,1F). Both univariate and multivariate analyses, indicated that LINC00880 might serve as a potential independent prognostic indicator for CRC (Figure 1G). Collectively, these findings underscore the significant role of LINC00880 in CRC pathogenesis and its potential relevance in predicting poor prognosis.
LINC00880 is located primarily in the nucleus of CRC cells
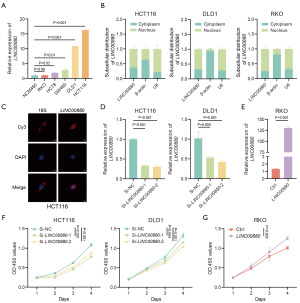
To understand the role of LINC00880 in CRC, we initially examined its expression across various CRC cell lines and discovered that LINC00880 was more highly expressed in these lines than in the normal colonic epithelial cell line NMC460 (Figure 2A). Subsequently, using qRT-PCR and RNA-FISH, we explored the subcellular distribution of LINC00880 in CRC cells. According to the results, LINC00880 was chiefly situated in the nucleus of CRC cells. (Figure 2B,2C).
LINC00880 promotes CRC growth in vitro and in vivo
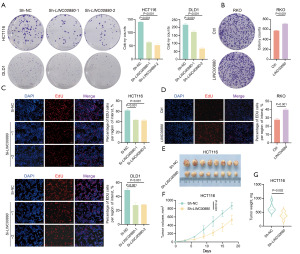
We selected HCT116 and DLD1 cell lines for LINC00880 knockdown and the RKO cell line for overexpression to explore LINC00880’s function in CRC (Figure 2D,2E). First, CCK-8 assays were conducted to determine the effect of LINC00880 on cell growth. Our results revealed a significant inhibition of proliferation in both HCT116 and DLD1 cells following LINC00880 knockdown, while LINC00880 overexpression led to significant promotion of proliferation in RKO cells (Figure 2F,2G). Afterward, we developed stable CRC cell lines with LINC00880 suppression and examined their potential to form colonies. The results revealed that LINC00880 knockdown weakened clonogenic potential, while overexpressing enhanced colony formation ability (Figure 3A,3B). These results were further supported by the results of the EdU assays (Figure 3C,3D). To further explore the tumorigenic potential of LINC00880 in vivo, xenograft tumor models were set up. Significantly, reducing LINC00880 expression led to a notable slowdown in tumor growth and a considerable decrease in tumor size compared to the control group. (Figure 3E,3F). Additionally, compared to the control group, the LINC00880 knockdown group exhibited a substantial decrease in tumor weight (Figure 3G). These findings indicate that LINC00880 is vital for enhancing cell proliferation and tumor development in CRC.
LINC00880 promotes cell cycle progression in CRC
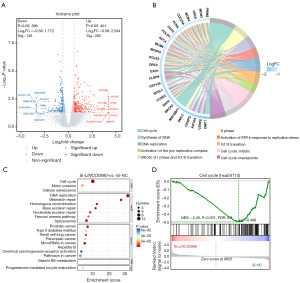
To investigate how LINC00880 might function in CRC, we carried out RNA-seq and differentially expressed gene (DEG) analyses on HCT116 cells where LINC00880 was knocked down. The RNA-seq analysis revealed significant changes in gene expression following the down-regulation of LINC00880 in HCT116 cells: 146 genes were significantly down-regulated, while 282 genes were significantly up-regulated (Figure 4A). Reactome and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses showed that these DEGs positively regulated by LINC00880 were enriched in pathways associated with the cell cycle and DNA replication (Figure 4B,4C). To further investigate these findings, gene set enrichment analysis (GSEA) was conducted, confirming that LINC00880 knockdown significantly inhibited the cell cycle pathway compared to the control group (Figure 4D). Subsequently, we assessed whether LINC00880 knockdown or overexpression influences the cell cycle in CRC cells. The results indicated that LINC00880 suppression induced a decrease in the number of cells in the S phase, whereas LINC00880 overexpression promotes the transition from the G1 to S phase of the cell cycle (Figure 5A,5B). Collectively, these results suggest that LINC00880 promotes CRC cell proliferation through cell-cycle modulation.
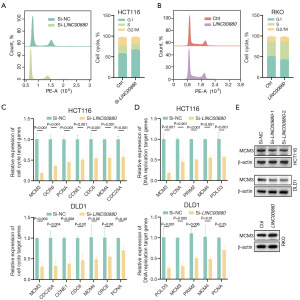
MCM3 as a potential target of LINC00880
To validate the RNA-seq results, we performed qRT-PCR on the downstream targets enriched in the cell cycle and DNA replication pathways identified by KEGG. Our findings indicated that LINC00880 knockdown resulted in the repression of the majority of genes associated with the cell cycle and DNA replication pathways. Notably, the expression cell cycle-related genes, such as MCM4, OCR6, PCNA, CCNE1, CDC6, MCM3, and CDC25A, tended to decrease in HCT116 and DLD1 cells following LINC00880 knockdown (Figure 5C). Moreover, the expression of DNA replication-related genes, such as MCM4, PCNA, PRIM2, MCM3, and POLD3, were downregulated in these cells (Figure 5D). Based on the qRT-PCR results, MCM3 was selected as the downstream effector of LINC00880 for Western blot validation. Experimental validation showed that silencing LINC00880 led to decreased MCM3 expression, while LINC00880 overexpression resulted in increased MCM3 expression (Figure 5E). IHC analysis of tumors resected from BALB/c nude mice demonstrated that LINC00880 knockdown significantly downregulated both of Ki-67 expression levels and MCM3 protein expression compared to the control group (Figure S1). Collectively, these findings suggest that LINC00880 may promote tumor growth by facilitating cell cycle progression and DNA replication through targeting MCM3.
Discussion
In the digestive system, CRC is one of the most widespread malignant tumors. Despite recent advancements in treatment, the prognosis for CRC patients remains unfavorable. The discovery of novel therapy targets is crucial for the development of innovative CRC therapies (17). LINC00880 has been implicated in the pathogenesis of lung adenocarcinoma (16). However, its role in CRC remains unknown. Our research showed that LINC00880 is notably increased in CRC tissues and correlates with lower survival rates in CRC patients. Functionally, we found that LINC00880 promotes tumor growth both in vitro and in vivo, suggesting its potential as an oncogene and a therapeutic target in CRC. Consistent with our observations, Feng and colleagues reported that LINC00880 contributes to the malignancy of lung adenocarcinoma (16). These findings further support the notion that LINC00880 may play a broader role in cancer development across different types of tumors.
To further explore the mechanistic role of LINC00880, we employed KEGG pathway enrichment analysis and GSEA to investigate the impact of LINC00880 on cellular processes. These analyses revealed that LINC00880 plays a significant role in regulating numerous cancer-related pathways, including the cell cycle, DNA replication, and DNA repair. These pathways are known to be critical for tumor development, highlighting the potential importance of LINC00880 in cancer progression (18-20). The significance of the cell cycle in tumorigenesis is highlighted by its role in regulating cell proliferation and division. Abnormalities in cell cycle control can lead to the uncontrolled proliferation of tumor cells (21,22). LncRNAs can either promote or inhibit tumor cell proliferation by modulating key regulatory proteins and signaling pathways associated with the cell cycle, thereby influencing tumor development (23-25). The relationship between DNA replication and tumorigenesis is intricate and complex. Tumor formation typically arises from a combination of multiple factors, such as errors in DNA replication, diminished expression of tumor suppressor genes, disruptions in the balance between cell apoptosis and proliferation, and defects in DNA repair (26-29). Therefore, maintaining the accuracy of DNA replication, preserving the stability of the genome, and enhancing DNA repair capabilities are highly important for preventing the occurrence of tumors (30,31).
Our study reveals that silencing LINC00880 in CRC cells leads to reduced expression of genes associated with the cell cycle and DNA replication. In particular, MCM3, a key gene in both pathways, appears to be a key downstream target mediating the regulatory function of LINC00880. MCM3 belongs to the MCM family, initially discovered via a yeast genetic screen. Nan et al. reported that the m6A demethylase FTO stabilizes LINK-A to exert oncogenic effects via MCM3-mediated cell-cycle progression and HIF-1α activation (32). Multiple studies have highlighted the overexpression of MCM3 in various cancers, such as CRC and breast cancer, and is closely associated with tumor cell proliferation, metastasis, and poor prognosis (33-35). The high expression of MCM3 suggests its important role in the development and progression of cancer, potentially serving as a biomarker for cancer diagnosis and prognosis (33). The results indicate that LINC00880 might be involved in controlling the expression of genes related to the cell cycle, with a specific focus on its complex interaction with MCM3. Given that reduced MCM3 expression is associated with increased sensitivity to helicase or replication inhibitors, we investigated whether LINC00880 expression affects sensitivity to these drugs. Our data revealed a positive correlation between LINC00880 expression levels and half-maximal inhibitory concentration (IC50) values for both drugs (data not shown), suggesting that elevated LINC00880 expression diminishes drug sensitivity. Further investigation is needed to elucidate how LINC00880 regulates these genes and its broader implications in cancer biology.
Collectively, we have demonstrated that LINC00880, a lncRNA that plays a critical oncogenic role in CRC, drives tumor progression by transcriptionally activating MCM3 and the cell cycle pathway. These findings not only highlight the significant potential of LINC00880 as a prognostic marker and therapeutic target but also reveal its crucial role in regulating the CRC cell cycle and promoting disease progression. Future research will focus on validating these mechanisms in clinical samples and further exploring the specific mechanisms by which LINC00880 regulates downstream targets of the cell cycle pathway. This aims to establish LINC00880 as a novel diagnostic and prognostic biomarker as well as a therapeutic target for CRC, thereby providing new perspectives for CRC treatment strategies.
Conclusions
This study demonstrates that the lncRNA LINC00880 exhibits marked overexpression in CRC, where it transcriptionally activates the MCM3 gene and modulates cell cycle-related signaling pathways, thereby promoting tumor cell proliferation and driving disease progression. These findings not only provide novel insights into the molecular mechanisms underlying CRC pathogenesis but also elucidate the potential value of this lncRNA as an innovative prognostic biomarker and therapeutic target from an epigenetic regulatory perspective.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2025-54/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2025-54/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-2025-54/prf
Funding: This study was partially supported by grants from
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2025-54/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. Ethics approval for this investigation was formally granted by the Institutional Review Board of the Affiliated Hospital of Jiangnan University (No. WXSY-YXLL-AF/SC-11/02.0). Prior to sample collection and experimental procedures, written informed consent was acquired from all participating donors and their legal representatives. The research protocol was conducted in strict compliance with internationally recognized ethical standards for biomedical studies involving human subjects. All animal experiments were conducted under Project License (No. JN.20241030b0161220 and Approval No. 577) granted by the Medical Experimental Animal Ethics Committee of Jiangnan University, in compliance with institutional guidelines for the care and use of laboratory animals. A protocol was prepared before the study without registration.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Giaquinto AN, Sung H, Newman LA, et al. Breast cancer statistics 2024. CA Cancer J Clin 2024;74:477-95. [Crossref] [PubMed]
- Mauri G, Patelli G, Crisafulli G, et al. Tumor "age" in early-onset colorectal cancer. Cell 2025;188:589-93. [Crossref] [PubMed]
- Spaander MCW, Zauber AG, Syngal S, et al. Young-onset colorectal cancer. Nat Rev Dis Primers 2023;9:21. [Crossref] [PubMed]
- Herman AB, Tsitsipatis D, Gorospe M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol Cell 2022;82:2252-66. [Crossref] [PubMed]
- Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol 2021;220:e202009045. [Crossref] [PubMed]
- Bian Z, Yang F, Xu P, et al. LINC01852 inhibits the tumorigenesis and chemoresistance in colorectal cancer by suppressing SRSF5-mediated alternative splicing of PKM. Mol Cancer 2024;23:23. [Crossref] [PubMed]
- Xu P, Gao G, Yang C, et al. Overexpression of LINC00853 enhances tumorigenesis and metastasis of gastric cancer. Pathol Res Pract 2024;253:155065. [Crossref] [PubMed]
- Chia L, Abu N, Masre SF. The roles of lncRNA XIST in cancer chemoresistance. ExRNA 2023;5:1-2. [Crossref]
- Zhang Z, Zeng C, Zhang W. Considerations before the application of 5-hydroxymethylation levels of long non-coding RNAs for non-invasive cancer diagnosis. Extracell Vesicles Circ Nucl Acids 2022;3:10-3. [Crossref] [PubMed]
- Singh DK, Cong Z, Song YJ, et al. MANCR lncRNA Modulates Cell-Cycle Progression and Metastasis by Cis-Regulation of Nuclear Rho-GEF. Mol Cell Biol 2024;44:372-90. [Crossref] [PubMed]
- Gupta S, Silveira DA, Lorenzoni PR, et al. LncRNA PTENP1/miR-21/PTEN Axis Modulates EMT and Drug Resistance in Cancer: Dynamic Boolean Modeling for Cell Fates in DNA Damage Response. Int J Mol Sci 2024;25:8264. [Crossref] [PubMed]
- Bian Z, Zhou M, Cui K, et al. SNHG17 promotes colorectal tumorigenesis and metastasis via regulating Trim23-PES1 axis and miR-339-5p-FOSL2-SNHG17 positive feedback loop. J Exp Clin Cancer Res 2021;40:360. [Crossref] [PubMed]
- Wang X, Cheng H, Zhao J, et al. Long noncoding RNA DLGAP1-AS2 promotes tumorigenesis and metastasis by regulating the Trim21/ELOA/LHPP axis in colorectal cancer. Mol Cancer 2022;21:210. [Crossref] [PubMed]
- Zhang J, Cui K, Huang L, et al. SLCO4A1-AS1 promotes colorectal tumourigenesis by regulating Cdk2/c-Myc signalling. J Biomed Sci 2022;29:4. [Crossref] [PubMed]
- Liu Y, Liu B, Jin G, et al. An Integrated Three-Long Non-coding RNA Signature Predicts Prognosis in Colorectal Cancer Patients. Front Oncol 2019;9:1269. [Crossref] [PubMed]
- Feng Y, Zhang T, Zhang Z, et al. The super-enhancer-driven lncRNA LINC00880 acts as a scaffold between CDK1 and PRDX1 to sustain the malignance of lung adenocarcinoma. Cell Death Dis 2023;14:551. [Crossref] [PubMed]
- Strickler JH, Yoshino T, Graham RP, et al. Diagnosis and Treatment of ERBB2-Positive Metastatic Colorectal Cancer: A Review. JAMA Oncol 2022;8:760-9. [Crossref] [PubMed]
- Li P, Liu S, Wang T, et al. Multisite DNA methylation alterations of peripheral blood mononuclear cells serve as novel biomarkers for the diagnosis of AIS/stage I lung adenocarcinoma: a multicenter cohort study. Int J Surg 2025;111:40-54. [Crossref] [PubMed]
- Shen JZ, Qiu Z, Wu Q, et al. FBXO44 promotes DNA replication-coupled repetitive element silencing in cancer cells. Cell 2021;184:352-369.e23. [Crossref] [PubMed]
- Yao Y, Liu C, Wang B, et al. HOXB9 blocks cell cycle progression to inhibit pancreatic cancer cell proliferation through the DNMT1/RBL2/c-Myc axis. Cancer Lett 2022;533:215595. [Crossref] [PubMed]
- Zatulovskiy E, Zhang S, Berenson DF, et al. Cell growth dilutes the cell cycle inhibitor Rb to trigger cell division. Science 2020;369:466-71. [Crossref] [PubMed]
- Yang H, Zhen X, Yang Y, et al. ERCC6L facilitates the onset of mammary neoplasia and promotes the high malignance of breast cancer by accelerating the cell cycle. J Exp Clin Cancer Res 2023;42:227. [Crossref] [PubMed]
- Cai Z, Shi Q, Li Y, et al. LncRNA EILA promotes CDK4/6 inhibitor resistance in breast cancer by stabilizing cyclin E1 protein. Sci Adv 2023;9:eadi3821. [Crossref] [PubMed]
- Wang D, Chen J, Li B, et al. A noncoding regulatory RNA Gm31932 induces cell cycle arrest and differentiation in melanoma via the miR-344d-3-5p/Prc1 (and Nuf2) axis. Cell Death Dis 2022;13:314. [Crossref] [PubMed]
- Niinuma T, Kitajima H, Sato T, et al. LINC02154 promotes cell cycle and mitochondrial function in oral squamous cell carcinoma. Cancer Sci 2025;116:393-405. [Crossref] [PubMed]
- Cleary JM, Aguirre AJ, Shapiro GI, et al. Biomarker-Guided Development of DNA Repair Inhibitors. Mol Cell 2020;78:1070-85. [Crossref] [PubMed]
- Jardim DL, Goodman A, de Melo Gagliato D, et al. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021;39:154-73. [Crossref] [PubMed]
- Drew Y, Zenke FT, Curtin NJ. DNA damage response inhibitors in cancer therapy: lessons from the past, current status and future implications. Nat Rev Drug Discov 2025;24:19-39. [Crossref] [PubMed]
- Dong S, Li A, Pan R, et al. Carboplatin-resistance-related DNA damage repair prognostic gene signature and its association with immune infiltration in breast cancer. Front Immunol 2025;16:1522149. [Crossref] [PubMed]
- Tubbs A, Nussenzweig A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017;168:644-56. [Crossref] [PubMed]
- Basu AK. DNA Damage, Mutagenesis and Cancer. Int J Mol Sci 2018;19:970. [Crossref] [PubMed]
- Nan Y, Liu S, Luo Q, et al. m(6)A demethylase FTO stabilizes LINK-A to exert oncogenic roles via MCM3-mediated cell-cycle progression and HIF-1α activation. Cell Rep 2023;42:113273. [Crossref] [PubMed]
- Zhu Z, Li M, Weng J, et al. LncRNA GAS6-AS1 contributes to 5-fluorouracil resistance in colorectal cancer by facilitating the binding of PCBP1 with MCM3. Cancer Lett 2024;589:216828. [Crossref] [PubMed]
- Løkkegaard S, Elias D, Alves CL, et al. MCM3 upregulation confers endocrine resistance in breast cancer and is a predictive marker of diminished tamoxifen benefit. NPJ Breast Cancer 2021;7:2. [Crossref] [PubMed]
- Chen Y, Li LY, Li JD, et al. Expression, potential biological behaviour and clinical significance of MCM3 in pancreatic adenocarcinoma: a comprehensive study integrating high throughput sequencing, CRISPR screening and in-house immunohistochemistry. Ann Med 2024;56:2405879. [Crossref] [PubMed]


